2,5-Diketo-D-Gluconate Hyperproducing Gluconobacter sphaericus SJF2-1 with Reporting Multiple Genes Encoding the Membrane-Associated Flavoprotein-Cytochrome c Complexed Dehydrogenases
Abstract
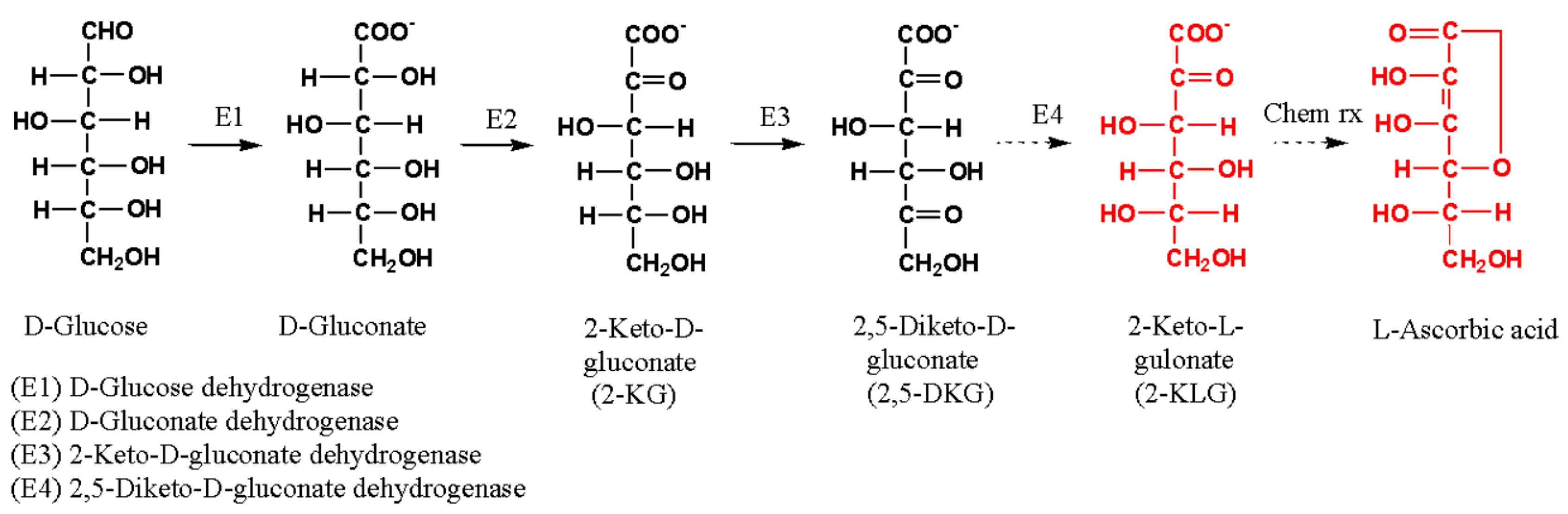
1. Introduction
2. Materials and Methods
2.1. Strain Isolation and Bacteria Culture
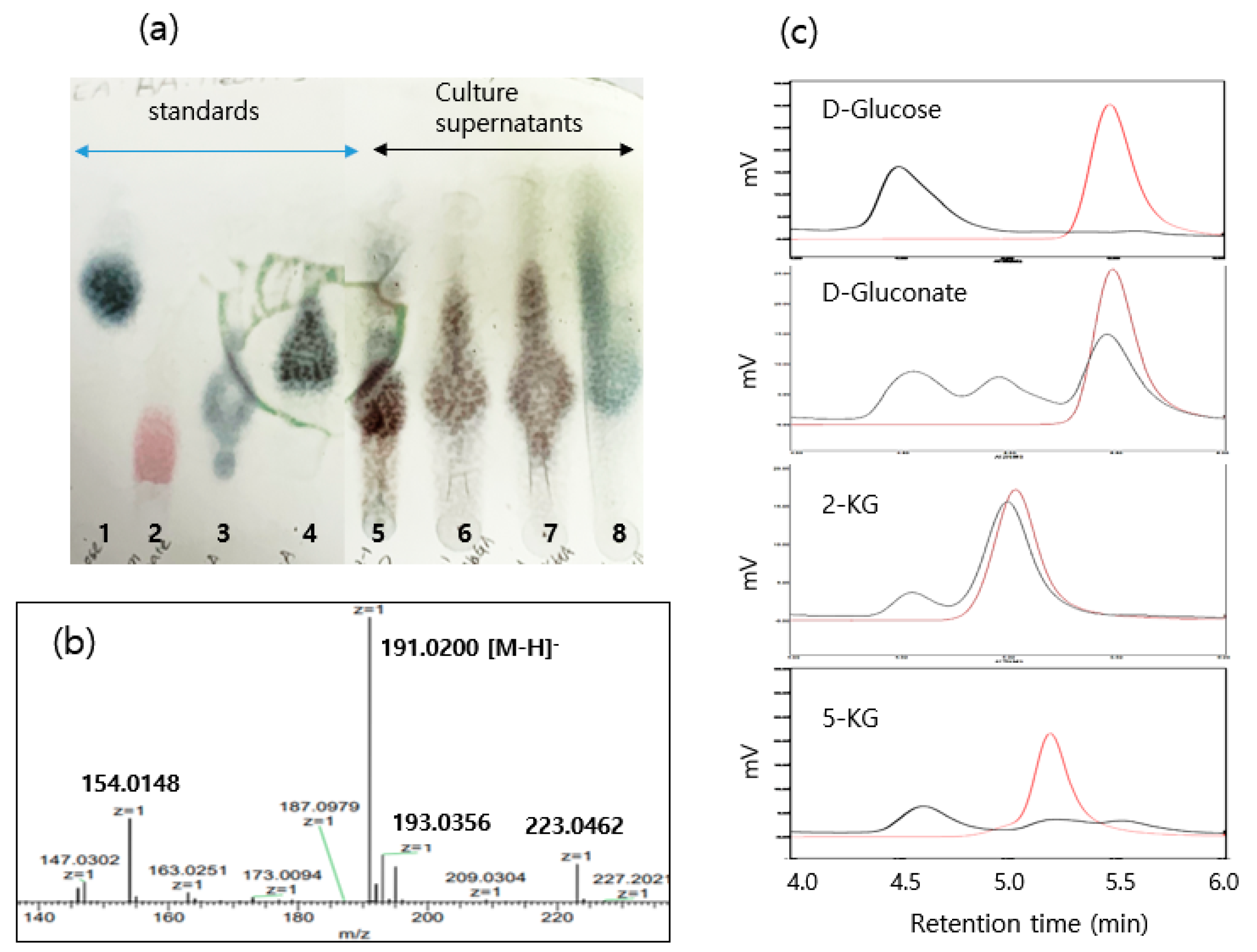
2.2. Analyses of Metabolites
2.3. Genome Sequencing and Analyses
3. Results and Discussion
3.1. Identification of Strain SJF2-1
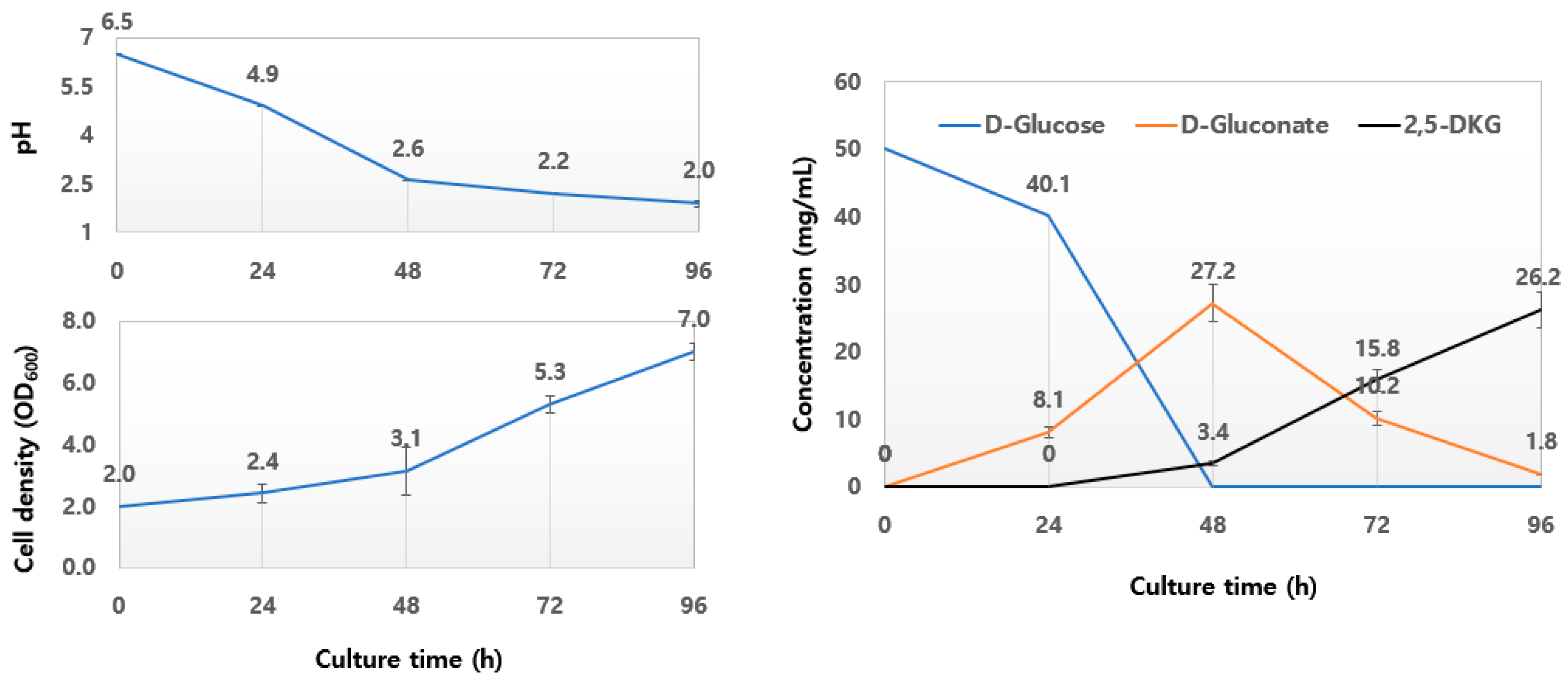
3.2. Biotransformation of Glucose to 2,5-DKG and Metabolite Analyses
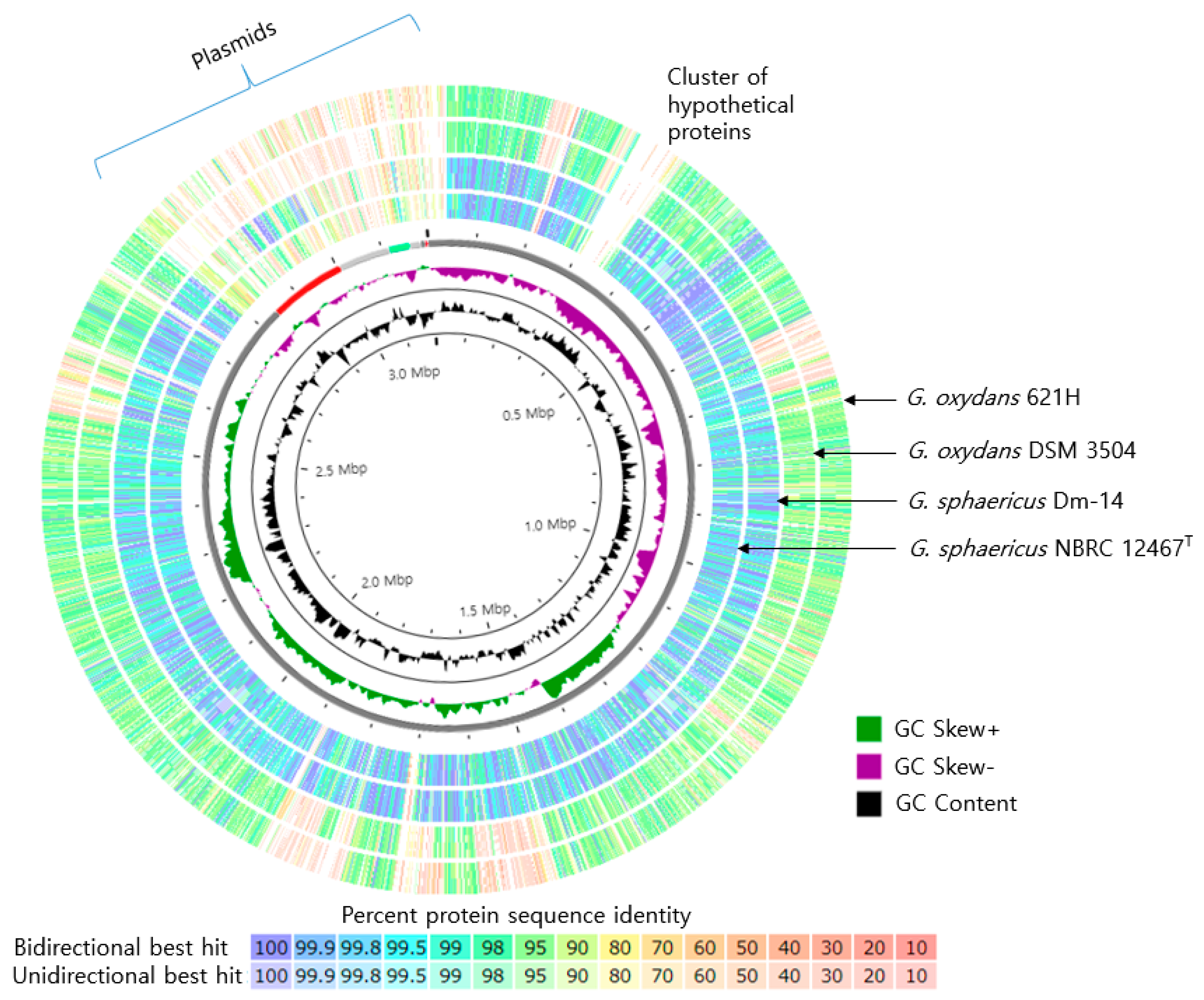
3.3. Genome Features of Strain SJF2-1 and Comparative Genome Analysis
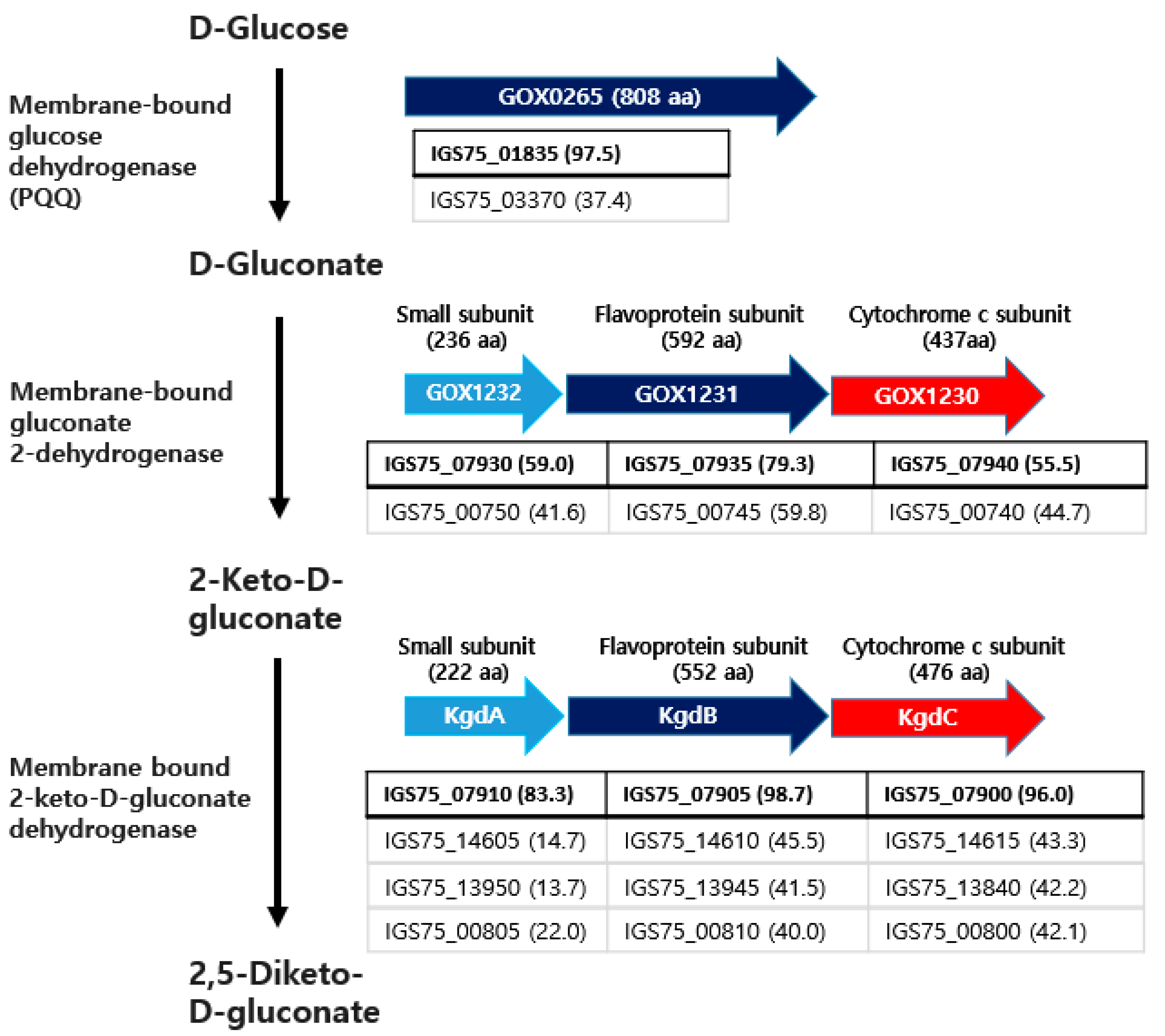
3.4. Genome Mining for Genes Related to Biotransformation of Glucose to 2,5-DKG
3.4.1. Genes Related to PQQ (Pyrroloquinoline Quinone)-Dependent Glucose Dehydrogenase
3.4.2. Genes Related to Membrane-Bound Gluconate 2-Dehydrogenase
3.4.3. Genes Related to the Membrane-Bound 2-KG Dehydrogenase
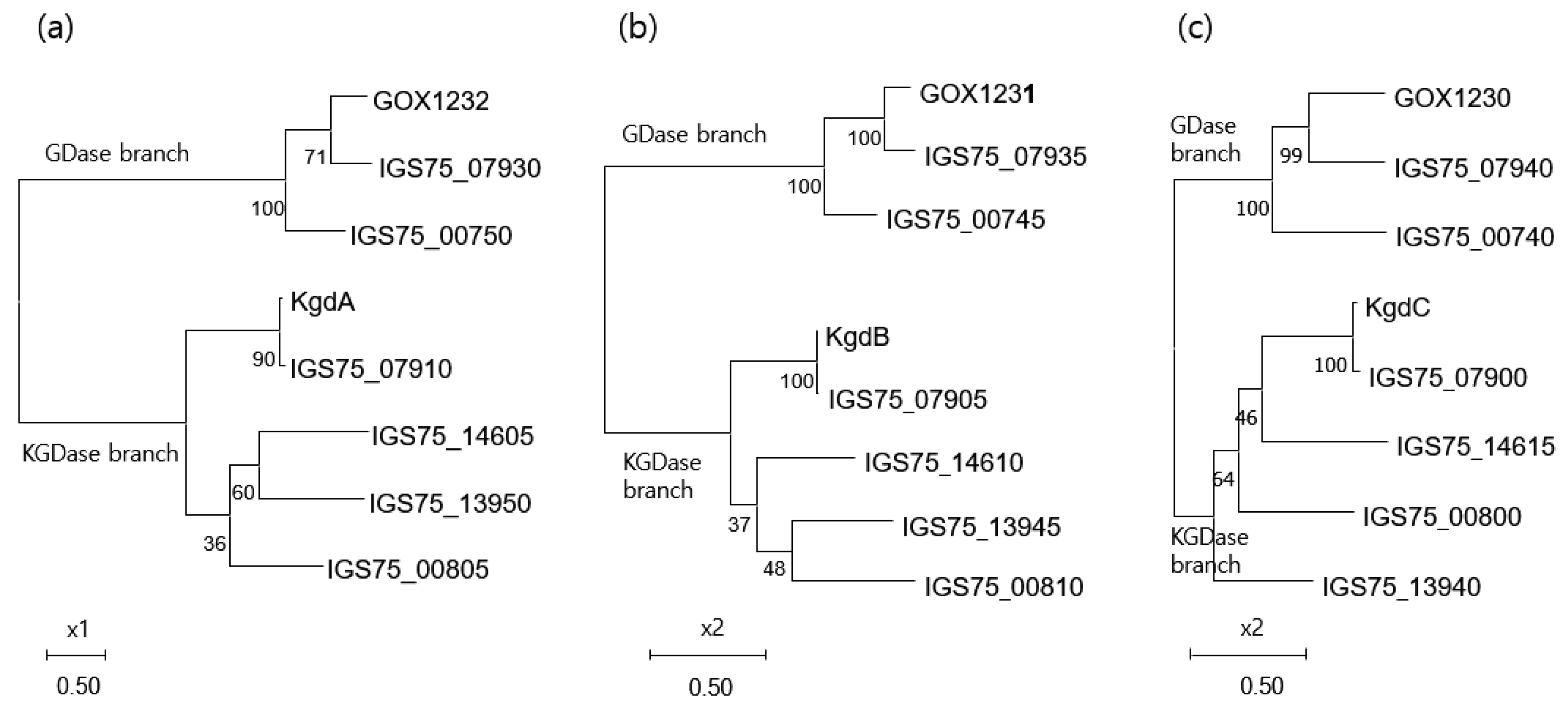
3.4.4. Phylogeny of the Membrane-Bound Flavoprotein-Cytochrome c Complexed Dehydrogenases
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Sauberlich, H.E. Pharmacology of vitamin C. Annu. Rev. Nutr. 1994, 14, 371–391. [Google Scholar] [CrossRef] [PubMed]
- Vitamin C Market Size to Worth Around USD 15.7 Billion by 2030. Precedence Research July 05, 2022. Available online: https://www.globenewswire.com/en/news-release/2022/07/05/2473878/0/en/Vitamin-C-Market-Size-to-Worth-Around-USD-15-7-Billion-by-2030.html (accessed on 20 December 2020).
- Wang, P.; Zeng, W.; Xu, S.; Du, G.; Zhou, J.; Chen, J. Current challenges facing one-step production of L-ascorbic acid. Biotechnol. Adv. 2018, 36, 1882–1899. [Google Scholar] [CrossRef] [PubMed]
- Pappenberger, G.; Hohmann, H.-P. Industrial production of L-ascorbic acid (vitamin C) and D-isoascorbic acid. In Biotechnology of Food and Feed Additives; Zorn, H., Czermak, P., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 143–188. [Google Scholar]
- Sonoyama, T.; Tani, H.; Matsuda, K.; Kageyama, B.; Tanimoto, M.; Kobayashi, K.; Yagi, S.; Kyotani, H.; Mitsushima, K. Production of 2-keto-L-gulonic acid from D-glucose by two-stage fermentation. Appl. Environ. Microbiol. 1982, 43, 1064–1069. [Google Scholar] [CrossRef] [PubMed]
- Aiguo, J.; Peiji, G. Synthesis of 2-keto-L-gulonic acid from gluconic acid by co-immobilized Gluconobacter oxydans and Corynebacterium sp. Biotechnol. Lett. 1998, 20, 939–942. [Google Scholar] [CrossRef]
- Anderson, S.; Marks, C.B.; Lazarus, R.; Miller, J.; Stafford, K.; Seymour, J.; Light, D.; Rastetter, W.; Estell, D. Production of 2-keto-L-gulonate, an intermediate in L-ascorbate synthesis, by a genetically modified Erwinia herbicola. Science 1985, 230, 144–149. [Google Scholar] [CrossRef]
- Grindley, J.; Payton, M.; Van de Pol, H.; Hardy, K. Conversion of glucose to 2-keto-L-gulonate, an intermediate in L-ascorbate synthesis, by a recombinant strain of Erwinia citreus. Appl. Environ. Microbiol. 1988, 54, 1770–1775. [Google Scholar] [CrossRef]
- Li, G.; Shan, X.Y.; Zeng, W.; Yu, S.; Zhang, G.; Zhou, J. The efficient production of 2, 5-diketo-D-gluconic acid by reducing browning levels during Gluconobacter oxydans ATCC 9937 fermentation. Front. Bioeng. Biotechnol. 2022, 10, 918277. [Google Scholar] [CrossRef] [PubMed]
- Qazi, G.; Parshad, R.; Verma, V.; Chopra, C.; Buse, R.; Träger, M.; Onken, U. Diketo-gluconate fermentation by Gluconobacter oxydans. Enzyme Microb. Technol. 1991, 13, 504–507. [Google Scholar] [CrossRef]
- He, Y.; Xie, Z.; Zhang, H.; Liebl, W.; Toyama, H.; Chen, F. Oxidative fermentation of acetic acid bacteria and its products. Front. Microbiol. 2022, 13, 879246. [Google Scholar] [CrossRef]
- Kataoka, N.; Matsutani, M.; Yakushi, T.; Matsushita, K. Efficient production of 2, 5-diketo-D-gluconate via heterologous expression of 2-ketogluconate dehydrogenase in Gluconobacter japonicus. Appl. Environ. Microbiol. 2015, 81, 3552–3560. [Google Scholar] [CrossRef]
- Ameyama, M. Gluconobacter oxydans subsp. sphaericus, new subspecies isolated from grapes. Int. J. Syst. Evol. Microbiol. 1975, 25, 365–370. [Google Scholar]
- Malimas, T.; Yukphan, P.; Takahashi, M.; Muramatsu, Y.; Kaneyasu, M.; Potacharoen, W.; Tanasupawat, S.; Nakagawa, Y.; Tanticharoen, M.; Yamada, Y. Gluconobacter sphaericus (Ameyama 1975) comb. nov. a brown pigment-producing acetic acid bacterium in the Alphaproteobacteria. J. Gen. Appl. Microbiol. 2008, 54, 211–220. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gupta, A.; Singh, V.K.; Qazi, G.; Kumar, A. Gluconobacter oxydans: Its biotechnological applications. J. Mol. Microbiol. Biotechnol. 2001, 3, 445–456. [Google Scholar]
- da Silva, G.A.R.; Oliveira, S.S.d.S.; Lima, S.F.; do Nascimento, R.P.; Baptista, A.R.d.S.; Fiaux, S.B. The industrial versatility of Gluconobacter oxydans: Current applications and future perspectives. World J. Microbiol. Biotechnol. 2022, 38, 1–20. [Google Scholar] [CrossRef]
- Bailey, R.; Bourne, E. Colour reactions given by sugars and diphenylamine-aniline spray reagents on paper chromatograms. J. Chromatogr. 1960, 4, 206–213. [Google Scholar] [CrossRef]
- Zielinski, A.A.F.; Braga, C.M.; Demiate, I.M.; Beltrame, F.L.; Nogueira, A.; Wosiacki, G. Development and optimization of a HPLC-RI method for the determination of major sugars in apple juice and evaluation of the effect of the ripening stage. Food Sci. Technol. 2014, 34, 38–43. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 20 December 2020).
- Lee, K.; Badaya, S.K.; Singh, R.; Lim, J.Y. Complete genome sequence of Gordonia sp. strain JH63, isolated from human skin. Microbiol. Resour. Announc. 2020, 9, e00059–20. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Richter, M.; Rossello-Mora, R.; Oliver Glockner, F.; Peplies, J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Carbasse, J.S.; Peinado-Olarte, R.L.; Göker, M. TYGS and LPSN: A database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res. 2021, 50, D801–D807. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M. The RAST server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Grant, J.R.; Stothard, P. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008, 36, W181–W184. [Google Scholar] [CrossRef]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotech. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; Von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Kumar, S.; Tamura, K.; Nei, M. MEGA: Molecular evolutionary genetics analysis software for microcomputers. Bioinformatics 1994, 10, 189–191. [Google Scholar] [CrossRef]
- Pujol, C.J.; Kado, C.I. Genetic and biochemical characterization of the pathway in Pantoea citrea leading to pink disease of pineapple. J. Bacteriol. 2000, 182, 2230–2237. [Google Scholar] [CrossRef]
- Chun, J.; Oren, A.; Ventosa, A.; Christensen, H.; Arahal, D.R.; da Costa, M.S.; Rooney, A.P.; Yi, H.; Xu, X.W.; De Meyer, S.; et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 2018, 68, 461–466. [Google Scholar] [CrossRef]
- Prust, C.; Hoffmeister, M.; Liesegang, H.; Wiezer, A.; Fricke, W.F.; Ehrenreich, A.; Gottschalk, G.; Deppenmeier, U. Complete genome sequence of the acetic acid bacterium Gluconobacter oxydans. Nat. Biotech. 2005, 23, 195–200. [Google Scholar] [CrossRef]
- Pronk, J.; Levering, P.; Olijve, W.; Van Dijken, J. Role of NADP-dependent and quinoprotein glucose dehydrogenases in gluconic acid production by Gluconobacter oxydans. Enzyme Microb. Technol. 1989, 11, 160–164. [Google Scholar] [CrossRef]
- Anthony, C.; Ghosh, M. The structure and function of the PQQ-containing quinoprotein dehydrogenases. Prog. Biophys. Mol. Biol. 1998, 69, 1–21. [Google Scholar] [CrossRef]
- Shinagawa, E.; Ano, Y.; Yakushi, T.; Adachi, O.; Matsushita, K. Solubilization, purification, and properties of membrane-bound D-glucono-δ-lactone hydrolase from Gluconobacter oxydans. Biosci. Biotechnol. Biochem. 2009, 73, 241–244. [Google Scholar] [CrossRef][Green Version]
- Meyer, M.; Schweiger, P.; Deppenmeier, U. Effects of membrane-bound glucose dehydrogenase overproduction on the respiratory chain of Gluconobacter oxydans. Appl. Microbiol. Biotech. 2013, 97, 3457–3466. [Google Scholar] [CrossRef]
- Shinagawa, E.; Matsushita, K.; Adachi, O.; Ameyama, M. D-Gluconate dehydrogenase, 2-keto-D-gluconate yielding, from Gluconobacter dioxyacetonicus: Purification and characterization. Agric. Biol. Chem. 1984, 48, 1517–1522. [Google Scholar] [CrossRef]
- Elfari, M.; Ha, S.-W.; Bremus, C.; Merfort, M.; Khodaverdi, V.; Herrmann, U.; Sahm, H.; Görisch, H. A Gluconobacter oxydans mutant converting glucose almost quantitatively to 5-keto-D-gluconic acid. Appl. Microbiol. Biotech. 2005, 66, 668–674. [Google Scholar] [CrossRef]
- Barker, P.D.; Ferguson, S.J. Still a puzzle: Why is haem covalently attached in c-type cytochromes? Structure 1999, 7, R281–R290. [Google Scholar] [CrossRef][Green Version]
- Dym, O.; Eisenberg, D. Sequence-structure analysis of FAD-containing proteins. Protein Sci. 2001, 10, 1712–1728. [Google Scholar] [CrossRef]
- Kataoka, N.; Saichana, N.; Matsutani, M.; Toyama, H.; Matsushita, K.; Yakushi, T. Characterization of 3 phylogenetically distinct membrane-bound D-gluconate dehydrogenases of Gluconobacter spp. and their biotechnological application for efficient 2-keto-D-gluconate production. Biosci. Biotechnol. Biochem. 2022, 86, 681–690. [Google Scholar] [CrossRef]
- Qin, Z.; Yu, S.; Chen, J.; Zhou, J. Dehydrogenases of acetic acid bacteria. Biotechnol. Adv. 2022, 54, 107863. [Google Scholar] [CrossRef]

| Genome Name | GenBank Accession | Genome Size (bp) | GC (%) | No. of Coding Sequence | No. of rRNA | No. of tRNA | No. of Gene | No. of Pseudogene |
|---|---|---|---|---|---|---|---|---|
| Chromosome | CP068419.1 | 2,844,550 | 58.5 | 2518 | 12 | 56 | 2693 | 103 |
| pSJF2-a | CP068420.1 | 162,291 | 55.7 | 177 | - * | - | 193 | 16 |
| pSJF2-b | CP068421.1 | 109,107 | 55.2 | 102 | - | - | 115 | 13 |
| pSJF2-c | CP068422.1 | 44,465 | 53.0 | 31 | - | - | 39 | 8 |
| pSJF2-d | CP068423.1 | 22,858 | 49.3 | 19 | - | - | 29 | 10 |
| pSJF2-e | CP068424.1 | 9589 | 52.0 | 13 | - | - | 13 | - |
| pSJF2-f | CP068425.1 | 5226 | 56.3 | 7 | - | - | 7 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Son, H.; Han, S.-U.; Lee, K. 2,5-Diketo-D-Gluconate Hyperproducing Gluconobacter sphaericus SJF2-1 with Reporting Multiple Genes Encoding the Membrane-Associated Flavoprotein-Cytochrome c Complexed Dehydrogenases. Microorganisms 2022, 10, 2130. https://doi.org/10.3390/microorganisms10112130
Son H, Han S-U, Lee K. 2,5-Diketo-D-Gluconate Hyperproducing Gluconobacter sphaericus SJF2-1 with Reporting Multiple Genes Encoding the Membrane-Associated Flavoprotein-Cytochrome c Complexed Dehydrogenases. Microorganisms. 2022; 10(11):2130. https://doi.org/10.3390/microorganisms10112130
Chicago/Turabian StyleSon, Haelim, Sang-Uk Han, and Kyoung Lee. 2022. "2,5-Diketo-D-Gluconate Hyperproducing Gluconobacter sphaericus SJF2-1 with Reporting Multiple Genes Encoding the Membrane-Associated Flavoprotein-Cytochrome c Complexed Dehydrogenases" Microorganisms 10, no. 11: 2130. https://doi.org/10.3390/microorganisms10112130
APA StyleSon, H., Han, S.-U., & Lee, K. (2022). 2,5-Diketo-D-Gluconate Hyperproducing Gluconobacter sphaericus SJF2-1 with Reporting Multiple Genes Encoding the Membrane-Associated Flavoprotein-Cytochrome c Complexed Dehydrogenases. Microorganisms, 10(11), 2130. https://doi.org/10.3390/microorganisms10112130






