Detection and Molecular Characterization of Rotavirus Infections in Children and Adults with Gastroenteritis from Vojvodina, Serbia
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. RNA Extraction, Detection, and Genetic Characterization of Rotavirus
2.3. Group A Rotavirus G Typing
2.4. Group A Rotavirus P Typing
2.5. Agarose Gel Electrophoresis
2.6. DNA Sequencing and Phylogenetic Analysis
2.7. Protein Structure Analysis of VP4 and VP7
2.8. Statistical Analysis
3. Results
3.1. Study Population
3.2. Rotavirus Prevalence
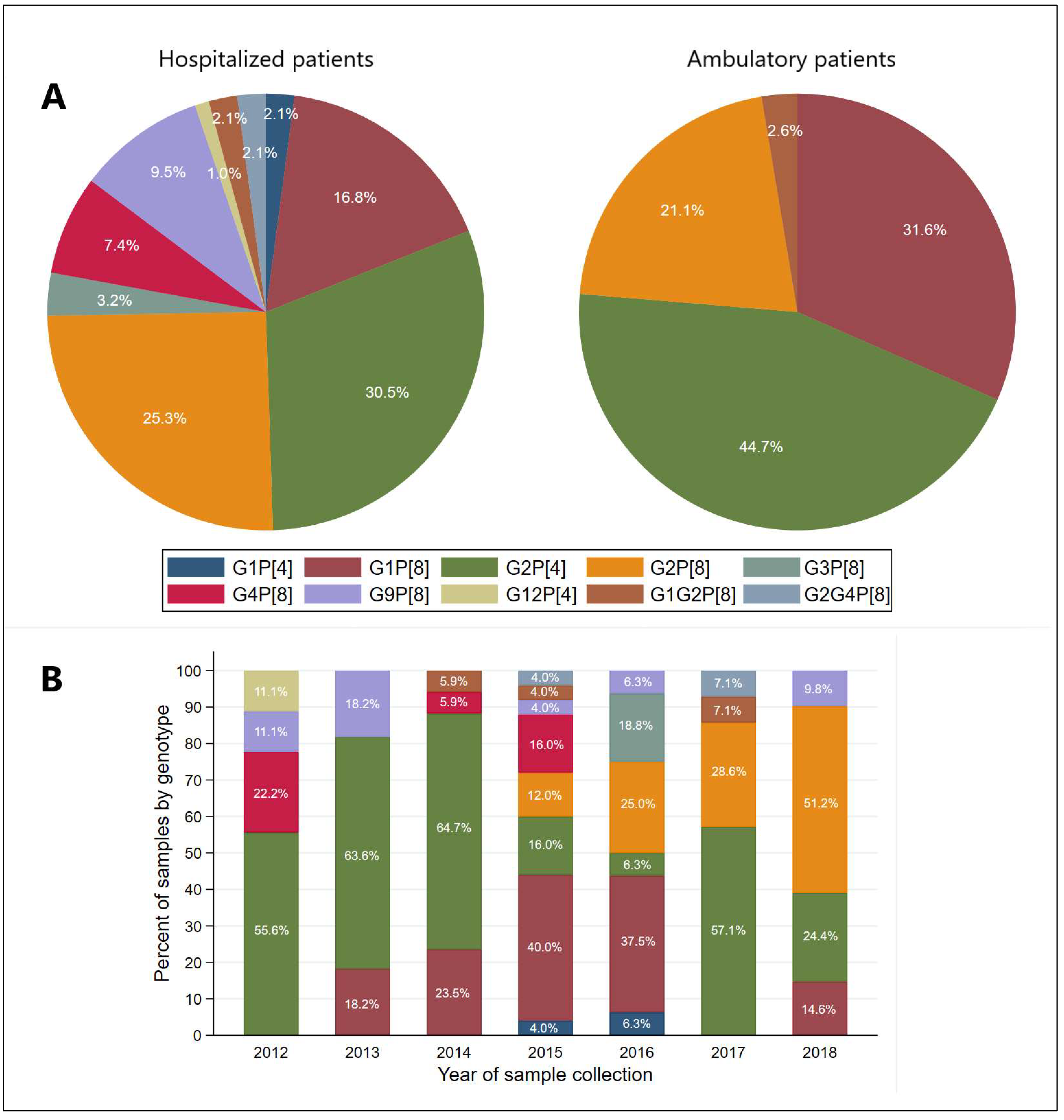
3.3. Distribution of G and P Genotypes of Group A Rotavirus
3.4. Phylogenetic Analysis and Comparison of the VP4 and VP7 Proteins of the RVA Strains with the Vaccine Strains
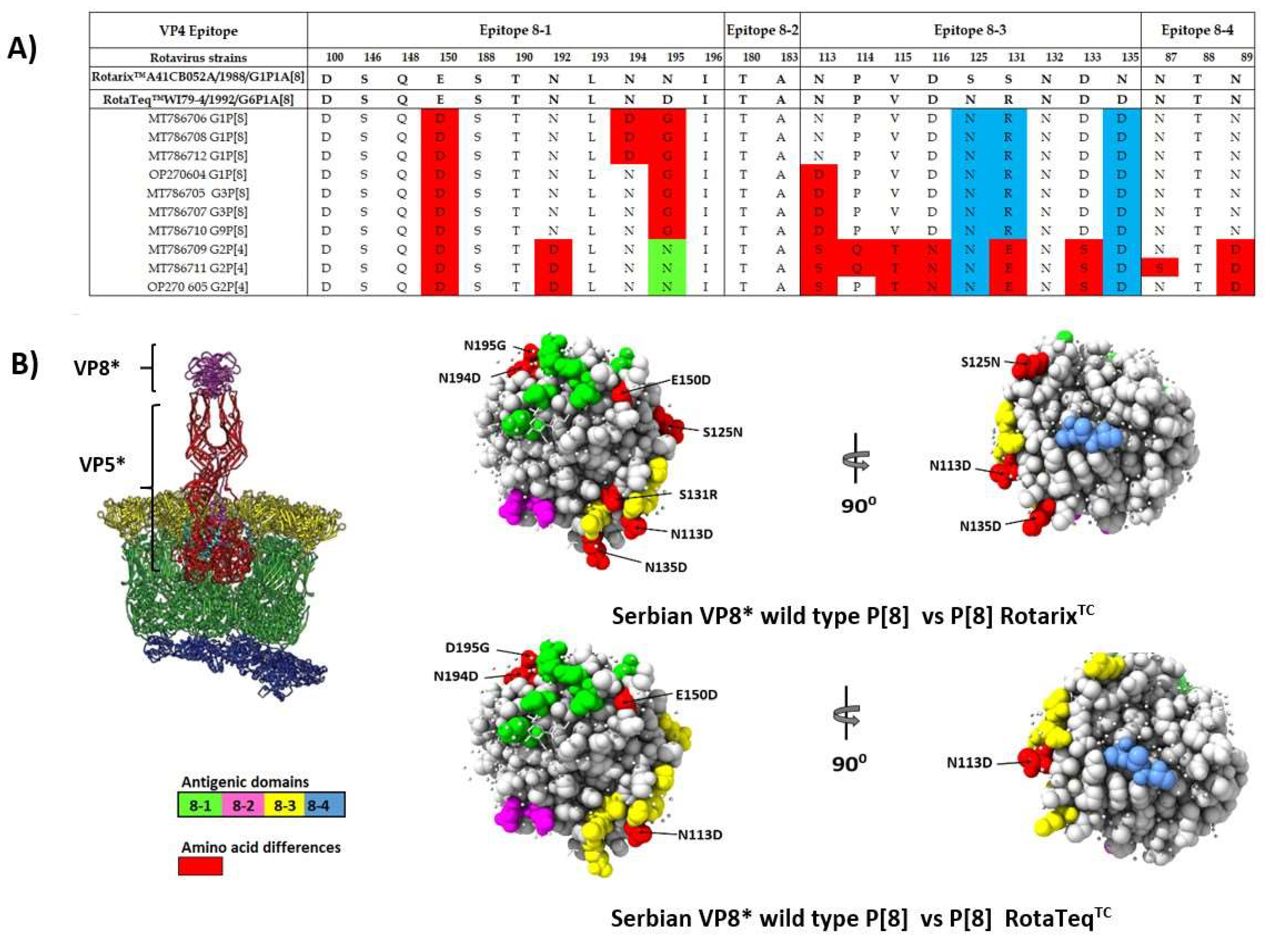
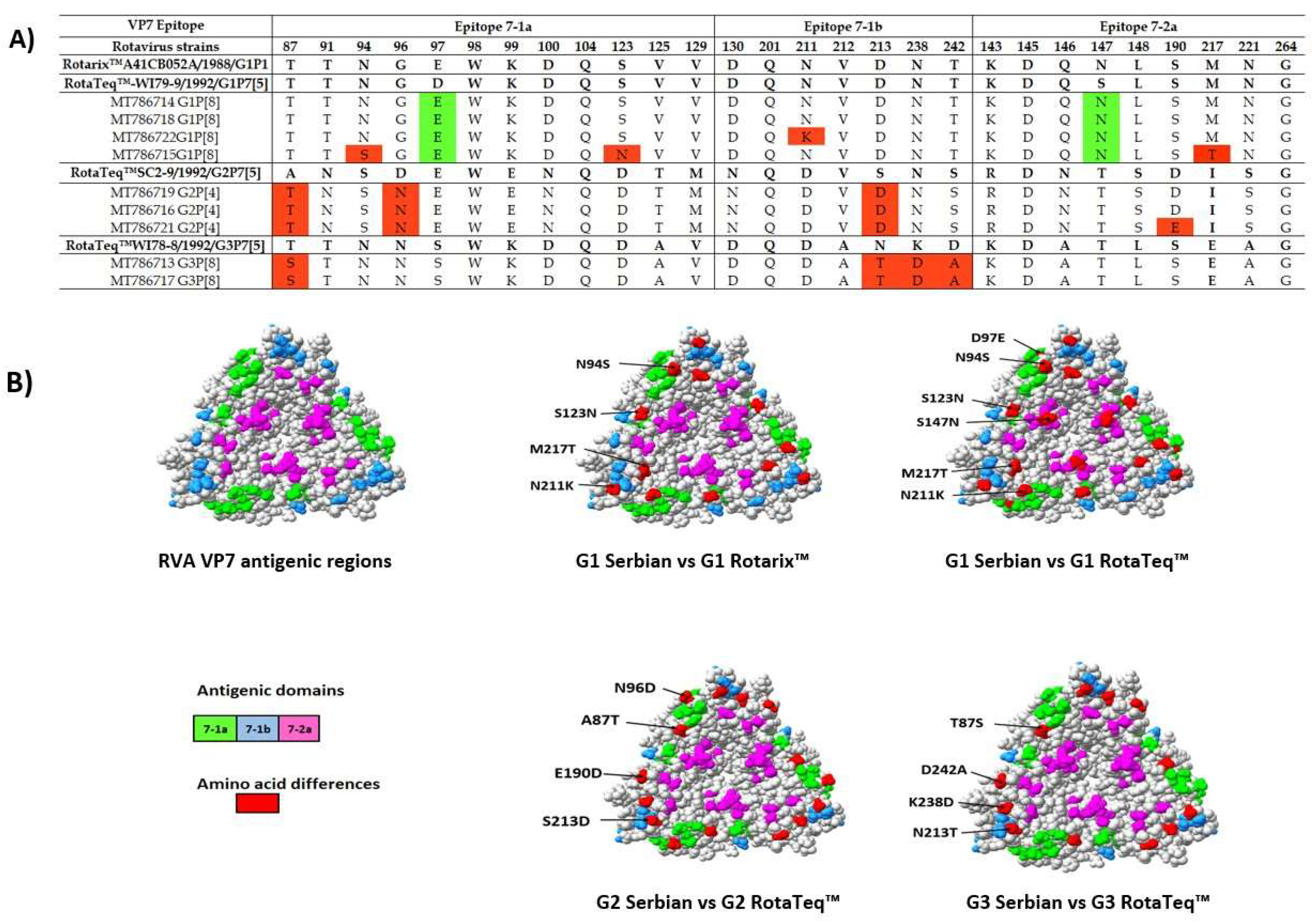
3.5. Comparison of the VP4 and VP7 Neutralizing Epitopes with Vaccine Strains
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Omatola, C.A.; Olaniran, A.O. Rotaviruses: From Pathogenesis to Disease Control—A Critical Review. Viruses 2022, 14, 875. [Google Scholar] [CrossRef] [PubMed]
- Karakusevic, A.; Devaney, P.; Enstone, A.; Kanibir, N.; Hartwig, S.; Carias, C.D.S. The burden of rotavirus-associated acute gastroenteritis in the elderly: Assessment of the epidemiology in the context of universal childhood vaccination programs. Expert Rev. Vaccines 2022, 21, 929–940. [Google Scholar] [CrossRef] [PubMed]
- Troeger, C.; Khalil, I.A.; Rao, P.C.; Cao, S.; Blacker, B.F.; Ahmed, T.; Armah, G.; Bines, J.E.; Brewer, T.G.; Colombara, D.V.; et al. Rotavirus Vaccination and the Global Burden of Rotavirus Diarrhea Among Children Younger Than 5 Years. JAMA Pediatr. 2018, 172, 958. [Google Scholar] [CrossRef] [PubMed]
- Dennehy, P.H. Rotavirus Infection. Adv. Pediatr. 2012, 59, 47–74. [Google Scholar] [CrossRef]
- Anderson, E.J.; Shippee, D.B.; Weinrobe, M.H.; Davila, M.D.; Katz, B.Z.; Reddy, S.; Cuyugan, M.G.K.P.; Lee, S.Y.; Simons, Y.M.; Yogev, R.; et al. Indirect Protection of Adults From Rotavirus by Pediatric Rotavirus Vaccination. Clin. Infect. Dis. 2013, 56, 755–760. [Google Scholar] [CrossRef]
- Naghavi, M.; Abajobir, A.A.; Abbafati, C.; Abbas, K.M.; Abd-Allah, F.; Abera, S.F.; Aboyans, V.; Adetokunboh, O.; Afshin, A.; Agrawal, A.; et al. Global, regional, and national age-sex specific mortality for 264 causes of death, 1980–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet 2017, 390, 1151–1210. [Google Scholar] [CrossRef]
- Caddy, S.; Papa, G.; Borodavka, A.; Desselberger, U. Rotavirus research: 2014–2020. Virus Res. 2021, 304, 198499. [Google Scholar] [CrossRef]
- Esona, M.D.; Gautam, R. Rotavirus. Clin. Lab. Med. 2015, 35, 363–391. [Google Scholar] [CrossRef]
- Donato, C.M.; Bines, J.E. Rotaviruses and Rotavirus Vaccines. Pathogens 2021, 10, 959. [Google Scholar] [CrossRef]
- Matthijnssens, J.; Ciarlet, M.; McDonald, S.M.; Attoui, H.; Bányai, K.; Brister, J.R.; Buesa, J.; Esona, M.D.; Estes, M.K.; Gentsch, J.R.; et al. Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG). Arch. Virol. 2011, 156, 1397–1413. [Google Scholar] [CrossRef] [PubMed]
- Crawford, S.E.; Ramani, S.; Tate, J.E.; Parashar, U.D.; Svensson, L.; Hagbom, M.; Franco, M.A.; Greenberg, H.B.; O’Ryan, M.; Kang, G.; et al. Rotavirus infection. Nat. Rev. Dis. Prim. 2017, 3, 17083. [Google Scholar] [CrossRef] [PubMed]
- Ministry of Health of the Republic of Serbia. Pravilnik o Programu Obavezne i Preporučene Imunizacije Stanovništva Protiv Određenih Zaraznih Bolesti, Službeni Glasnik Republike Srbije br. 65/2020; Ministry of Health of the Republic of Serbia: Belgrade, Serbia, 2020. (In Serbian)
- World Health Organization. Rotavirus: Vaccine Preventable Diseases Surveillance Standards; World Health Organization: Geneva, Switzerland, 2018. [Google Scholar]
- World Health Organization. Manual of Rotavirus Detection and Characterization Methods; World Health Organization: Geneva, Switzerland, 2009. [Google Scholar]
- Maes, P.; Matthijnssens, J.; Rahman, M.; Van Ranst, M. RotaC: A web-based tool for the complete genome classification of group A rotaviruses. BMC Microbiol. 2009, 9, 238. [Google Scholar] [CrossRef] [PubMed]
- Settembre, E.C.; Chen, J.Z.; Dormitzer, P.R.; Grigorieff, N.; Harrison, S.C. Atomic model of an infectious rotavirus particle. EMBO J. 2011, 30, 408–416. [Google Scholar] [CrossRef] [PubMed]
- Aoki, S.T.; Settembre, E.C.; Trask, S.D.; Greenberg, H.B.; Harrison, S.C.; Dormitzer, P.R. Structure of Rotavirus Outer-Layer Protein VP7 Bound with a Neutralizing Fab. Science 2009, 324, 1444–1447. [Google Scholar] [CrossRef]
- Dormitzer, P.R. The rhesus rotavirus VP4 sialic acid binding domain has a galectin fold with a novel carbohydrate binding site. EMBO J. 2002, 21, 885–897. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef]
- Zeller, M.; Patton, J.T.; Heylen, E.; De Coster, S.; Ciarlet, M.; Van Ranst, M.; Matthijnssens, J. Genetic Analyses Reveal Differences in the VP7 and VP4 Antigenic Epitopes between Human Rotaviruses Circulating in Belgium and Rotaviruses in Rotarix and RotaTeq. J. Clin. Microbiol. 2012, 50, 966–976. [Google Scholar] [CrossRef]
- Tcheremenskaia, O.; Marucci, G.; De Petris, S.; Ruggeri, F.M.; Dovecar, D.; Sternak, S.L.; Matyasova, I.; Dhimolea, M.K.; Mladenova, Z.; Fiore, L. Molecular Epidemiology of Rotavirus in Central and Southeastern Europe. J. Clin. Microbiol. 2007, 45, 2197–2204. [Google Scholar] [CrossRef][Green Version]
- Giri, S.; Hemavathy, R.P.; Arumugam, R.; Sherchand, J.B.; Thu, H.M.; Galagoda, G.; Myat, T.W.; Abeysinghe, N.; Gunasekara, M.; Janakan, N.; et al. Molecular epidemiology of rotaviruses in the south-east Asian region from 2009 to 2015. Vaccine 2018, 36, 7851–7855. [Google Scholar] [CrossRef]
- Ouermi, D.; Soubeiga, D.; Nadembega, W.M.C.; Sawadogo, P.M.; Zohoncon, T.M.; Obiri-Yebo, D.; Djigma, F.W.; Nordgren, J.; Simpore, J. Molecular Epidemiology of Rotavirus in Children under Five in Africa (2006–2016): A Systematic Review. Pak. J. Biol. Sci. 2017, 20, 59–69. [Google Scholar] [CrossRef]
- Food and Drug Administration Viruses. Bad Bug Book, Foodborne Pathogenic Microorganisms and Natural Toxins; Food and Drug Administration: Silver Spring, MD, USA, 2012; pp. 167–187.
- Van Damme, P.; Giaquinto, C.; Huet, F.; Gothefors, L.; Maxwell, M.; Van der Wielen, M. Multicenter Prospective Study of the Burden of Rotavirus Acute Gastroenteritis in Europe, 2004–2005: The REVEAL Study. J. Infect. Dis. 2007, 195, S4–S16. [Google Scholar] [CrossRef]
- Saánchez-Fauquier, A.; Wilhelmi, I.; Colomina, J.; Cubero, E.; Roman, E. Diversity of Group A Human Rotavirus Types Circulating over a 4-Year Period in Madrid, Spain. J. Clin. Microbiol. 2004, 42, 1609–1613. [Google Scholar] [CrossRef][Green Version]
- O’Halloran, F.; Lynch, M.; Cryan, B.; O’Shea, H.; Fanning, S. Molecular Characterization of Rotavirus in Ireland: Detection of Novel Strains Circulating in the Population. J. Clin. Microbiol. 2000, 38, 3370–3374. [Google Scholar] [CrossRef] [PubMed]
- De Donno, A.; Grassi, T.; Bagordo, F.; Idolo, A.; Cavallaro, A.; Gabutti, G. Emergence of unusual human rotavirus strains in Salento, Italy, during 2006–2007. BMC Infect. Dis. 2009, 9, 43. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cilla, G.; Montes, M.; Gomariz, M.; Alkorta, M.; Iturzaeta, A.; Perez-Yarza, E.G.; Perez-Trallero, E. Rotavirus genotypes in children in the Basque Country (North of Spain): Rapid and intense emergence of the G12[P8] genotype. Epidemiol. Infect. 2013, 141, 868–874. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hungerford, D.; Vivancos, R. In-season and out-of-season variation of rotavirus genotype distribution and age of infection across 12 European countries before the introduction of routine vaccination, 2007/08 to 2012/13. Eurosurveillance 2016, 21, 30106. [Google Scholar] [CrossRef]
- Zeller, M.; Rahman, M.; Heylen, E.; De Coster, S.; De Vos, S.; Arijs, I.; Novo, L.; Verstappen, N.; Van Ranst, M.; Matthijnssens, J. Rotavirus incidence and genotype distribution before and after national rotavirus vaccine introduction in Belgium. Vaccine 2010, 28, 7507–7513. [Google Scholar] [CrossRef] [PubMed]
- Hungerford, D.; Allen, D.J.; Nawaz, S.; Collins, S.; Ladhani, S.; Vivancos, R.; Iturriza-Gómara, M. Impact of rotavirus vaccination on rotavirus genotype distribution and diversity in England, September 2006 to August 2016. Eurosurveillance 2019, 24, 1700774. [Google Scholar] [CrossRef]
- Bonura, F.; Mangiaracina, L.; Filizzolo, C.; Bonura, C.; Martella, V.; Ciarlet, M.; Giammanco, G.M.; De Grazia, S. Impact of Vaccination on Rotavirus Genotype Diversity: A Nearly Two-Decade-Long Epidemiological Study before and after Rotavirus Vaccine Introduction in Sicily, Italy. Pathogens 2022, 11, 424. [Google Scholar] [CrossRef]
- Yandle, Z.; Coughlan, S.; Dean, J.; Tuite, G.; Conroy, A.; De Gascun, C.F. Group A Rotavirus Detection and Genotype Distribution before and after Introduction of a National Immunisation Programme in Ireland: 2015–2019. Pathogens 2020, 9, 449. [Google Scholar] [CrossRef]
- Kaplon, J.; Grangier, N.; Pillet, S.; Minoui-Tran, A.; Vabret, A.; Wilhelm, N.; Prieur, N.; Lazrek, M.; Alain, S.; Mekki, Y.; et al. Predominance of G9P[8] rotavirus strains throughout France, 2014–2017. Clin. Microbiol. Infect. 2018, 24, 660.e1–660.e4. [Google Scholar] [CrossRef] [PubMed]
- Matthijnssens, J.; Nakagomi, O.; Kirkwood, C.D.; Ciarlet, M.; Desselberger, U.; van Ranst, M. Group A rotavirus universal mass vaccination: How and to what extent will selective pressure influence prevalence of rotavirus genotypes? Expert Rev. Vaccines 2012, 11, 1347–1354. [Google Scholar] [CrossRef] [PubMed]
- Gentsch, J.R.; Laird, A.R.; Bielfelt, B.; Griffin, D.D.; Bányai, K.; Ramachandran, M.; Jain, V.; Cunliffe, N.A.; Nakagomi, O.; Kirkwood, C.D.; et al. Serotype Diversity and Reassortment between Human and Animal Rotavirus Strains: Implications for Rotavirus Vaccine Programs. J. Infect. Dis. 2005, 192, S146–S159. [Google Scholar] [CrossRef]
- Maunula, L.; von Bonsdorff, C.-H. Frequent Reassortments May Explain the Genetic Heterogeneity of Rotaviruses: Analysis of Finnish Rotavirus Strains. J. Virol. 2002, 76, 11793–11800. [Google Scholar] [CrossRef]
- Poelaert, D.; Pereira, P.; Gardner, R.; Standaert, B.; Benninghoff, B. A review of recommendations for rotavirus vaccination in Europe: Arguments for change. Vaccine 2018, 36, 2243–2253. [Google Scholar] [CrossRef] [PubMed]
- Soares-Weiser, K.; Bergman, H.; Henschke, N.; Pitan, F.; Cunliffe, N. Vaccines for preventing rotavirus diarrhoea: Vaccines in use. Cochrane Database Syst. Rev. 2019, 1–266. [Google Scholar] [CrossRef]
- Luchs, A.; Timenetsky, M.d.C.S.T. Group A rotavirus gastroenteritis: Post-vaccine era, genotypes and zoonotic transmission. Einstein 2016, 14, 278–287. [Google Scholar] [CrossRef] [PubMed]
- Rasebotsa, S.; Mwangi, P.N.; Mogotsi, M.T.; Sabiu, S.; Magagula, N.B.; Rakau, K.; Uwimana, J.; Mutesa, L.; Muganga, N.; Murenzi, D.; et al. Whole genome and in-silico analyses of G1P[8] rotavirus strains from pre- and post-vaccination periods in Rwanda. Sci. Rep. 2020, 10, 13460. [Google Scholar] [CrossRef]
- Maringa, W.M.; Simwaka, J.; Mwangi, P.N.; Mpabalwani, E.M.; Mwenda, J.M.; Mphahlele, M.J.; Seheri, M.L.; Nyaga, M.M. Whole Genome Analysis of Human Rotaviruses Reveals Single Gene Reassortant Rotavirus Strains in Zambia. Viruses 2021, 13, 1872. [Google Scholar] [CrossRef] [PubMed]
- Motamedi-Rad, M.; Farahmand, M.; Arashkia, A.; Jalilvand, S.; Shoja, Z. VP7 and VP4 genotypes of rotaviruses cocirculating in Iran, 2015 to 2017: Comparison with cogent sequences of Rotarix and RotaTeq vaccine strains before their use for universal mass vaccination. J. Med. Virol. 2020, 92, 1110–1123. [Google Scholar] [CrossRef]
- Harastani, H.H.; Reslan, L.; Sabra, A.; Ali, Z.; Hammadi, M.; Ghanem, S.; Hajar, F.; Matar, G.M.; Dbaibo, G.S.; Zaraket, H. Genetic Diversity of Human Rotavirus A Among Hospitalized Children Under-5 Years in Lebanon. Front. Immunol. 2020, 11, 317. [Google Scholar] [CrossRef] [PubMed]
- Degiuseppe, J.I.; Parra, G.I.; Stupka, J.A. Genetic Diversity of G3 Rotavirus Strains Circulating in Argentina during 1998–2012 Assessed by Full Genome Analyses. PLoS ONE 2014, 9, e110341. [Google Scholar] [CrossRef] [PubMed]
- Kirkwood, C.; Masendycz, P.J.; Coulson, B.S. Characteristics and Location of Cross-Reactive and Serotype-Specific Neutralization Sites on VP7 of Human G Type 9 Rotaviruses. Virology 1993, 196, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Betts, M.J.; Russell, R.B. Amino-Acid Properties and Consequences of Substitutions. In Bioinformatics for Geneticists; John Wiley & Sons, Ltd.: Chichester, UK; pp. 311–342.
- Liu, X.; Wang, M.; Li, S.; Li, J.; Xiao, J.; Li, H.; Zhang, Q.; Kong, X.; Wang, H.; Li, D.; et al. Genomic and evolutionary characteristics of G9P[8], the dominant group a rotavirus in China (2016–2018). Front. Microbiol. 2022, 13, 997957. [Google Scholar] [CrossRef]



| Total, Positive/Tested (%) | Hospitalized Patients, Positive/Tested (%) | Ambulatory Patients, Positive/Tested (%) | p-Value 1 | |
|---|---|---|---|---|
| Total | 538/1853 (29.0) | 486/1376 (35.3) | 52/477 (10.9) | <0.001 |
| Sex, n (%) | ||||
| Male | 292/995 (29.3) | 265/733 (36.2) | 27/262 (10.3) | <0.001 |
| Female | 246/858 (28.7) | 221/643 (34.4) | 25/215 (11.6) | <0.001 |
| Age category (years), n (%) | ||||
| <1 | 62/209 (29.7) | 56/182 (30.8) | 6/27 (22.2) | 0.364 |
| 1 | 164/394 (41.6) | 153/337 (45.4) | 11/57 (19.3) | <0.001 |
| 2 | 83/209 (39.7) | 70/154 (45.5) | 13/55 (23.6) | 0.005 |
| 3–5 | 87/256 (34.0) | 83/205 (40.5) | 4/51 (7.8) | <0.001 |
| 6–10 | 44/189 (23.3) | 40/128 (31.3) | 4/61 (6.6) | <0.001 |
| 11–15 | 20/128 (15.6) | 16/91 (17.6) | 4/37 (10.8) | 0.428 |
| 16–20 | 15/110 (13.6) | 14/89 (15.7) | 1/21 (4.8) | 0.294 |
| 21–30 | 19/102 (18.6) | 18/71 (25.4) | 1/31 (3.2) | 0.011 |
| 31–40 | 12/73 (16.4) | 11/37 (29.7) | 1/36 (2.8) | 0.003 |
| 41–50 | 6/42 (14.3) | 5/21 (23.8) | 1/21 (4.8) | 0.184 |
| >50 | 26/141 (18.4) | 20/61 (32.8) | 6/80 (7.5) | <0.001 |
| G Genotypes | P Genotypes, n (%) | ||
|---|---|---|---|
| P[4] | P[8] | Total | |
| G1 | 2 (4.1) | 28 (33.3) | 30 (22.6) |
| G2 | 46 (93.9) | 32 (38.1) | 78 (58.6) |
| G3 | 0 | 3 (3.6) | 3 (2.2) |
| G4 | 0 | 7 (8.3) | 7 (5.3) |
| G9 | 0 | 9 (10.7) | 9 (6.8) |
| G12 | 1 (2.0) | 0 | 1 (0.7) |
| G-mixed | 0 | 5 (6.0) | 5 (3.8) |
| Total | 49 (100) | 84 (100) | 133 (100) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Patić, A.; Vuković, V.; Kovačević, G.; Petrović, V.; Ristić, M.; Djilas, M.; Knežević, P.; Pustahija, T.; Štrbac, M.; Djekić Malbaša, J.; et al. Detection and Molecular Characterization of Rotavirus Infections in Children and Adults with Gastroenteritis from Vojvodina, Serbia. Microorganisms 2022, 10, 2050. https://doi.org/10.3390/microorganisms10102050
Patić A, Vuković V, Kovačević G, Petrović V, Ristić M, Djilas M, Knežević P, Pustahija T, Štrbac M, Djekić Malbaša J, et al. Detection and Molecular Characterization of Rotavirus Infections in Children and Adults with Gastroenteritis from Vojvodina, Serbia. Microorganisms. 2022; 10(10):2050. https://doi.org/10.3390/microorganisms10102050
Chicago/Turabian StylePatić, Aleksandra, Vladimir Vuković, Gordana Kovačević, Vladimir Petrović, Mioljub Ristić, Milan Djilas, Petar Knežević, Tatjana Pustahija, Mirjana Štrbac, Jelena Djekić Malbaša, and et al. 2022. "Detection and Molecular Characterization of Rotavirus Infections in Children and Adults with Gastroenteritis from Vojvodina, Serbia" Microorganisms 10, no. 10: 2050. https://doi.org/10.3390/microorganisms10102050
APA StylePatić, A., Vuković, V., Kovačević, G., Petrović, V., Ristić, M., Djilas, M., Knežević, P., Pustahija, T., Štrbac, M., Djekić Malbaša, J., Rajčević, S., & Hrnjaković Cvjetković, I. (2022). Detection and Molecular Characterization of Rotavirus Infections in Children and Adults with Gastroenteritis from Vojvodina, Serbia. Microorganisms, 10(10), 2050. https://doi.org/10.3390/microorganisms10102050








