Gre Factors Are Required for Biofilm Formation in Salmonella enterica Serovar Typhimurium by Targeting Transcription of the csgD Gene
Abstract
:1. Introduction
2. Materials and Methods
2.1. The Bacterial Strains and Culture Media
2.2. Genetic Manipulations
2.3. Biofilm Formation, Detection and Quantification
2.4. β-Galactosidase Assay
2.5. Cellulose Quantification
2.6. Rdar Morphotype
2.7. Immunodetection of CsgD
2.8. Expression Analysis by qPCR
2.9. Statistical Analysis
3. Results and Discussion
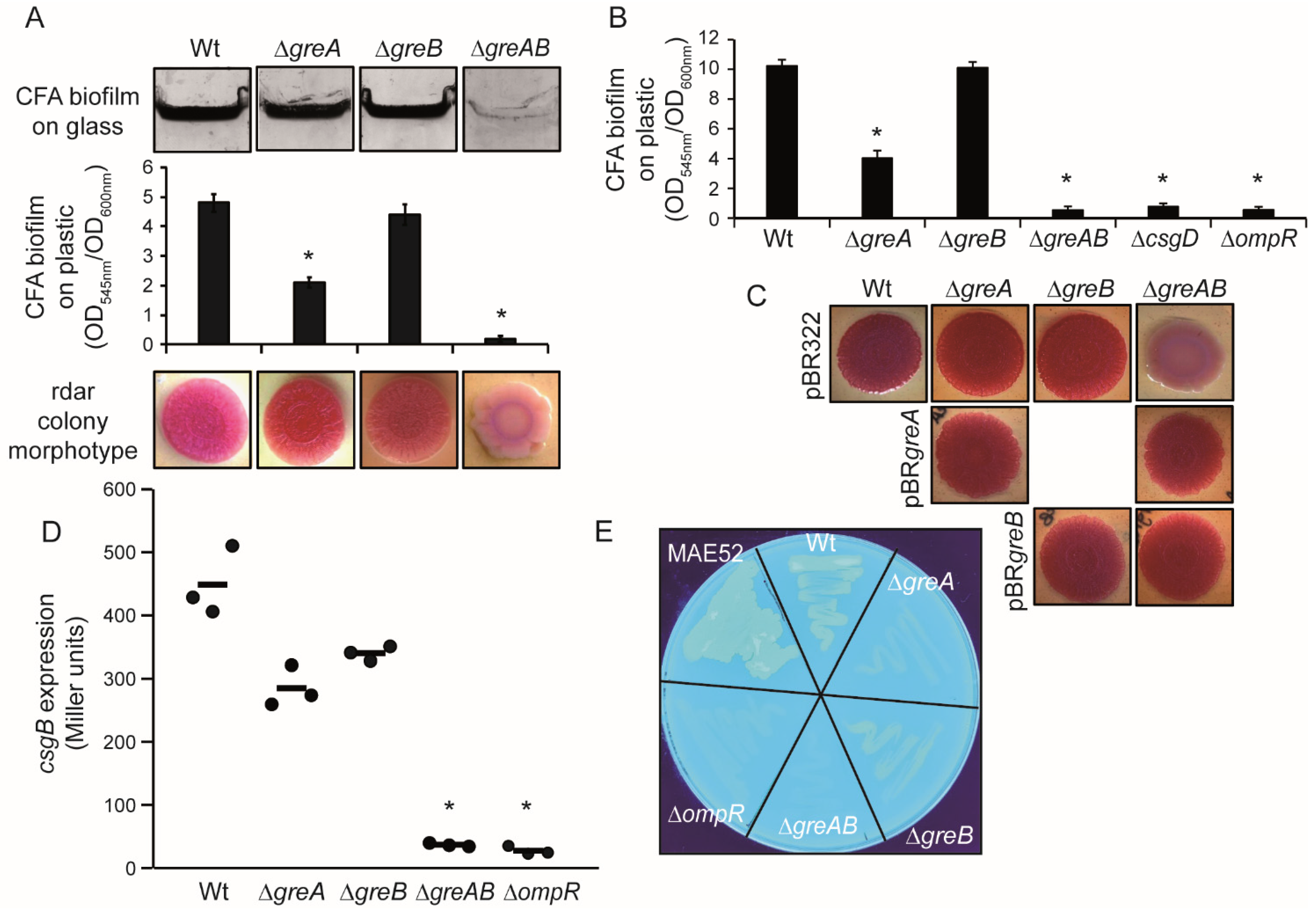
3.1. Rdar Biofilm Formation Is Impaired in S. Typhimurium When Deficient for the Transcription Elongation Factors GreA and GreB
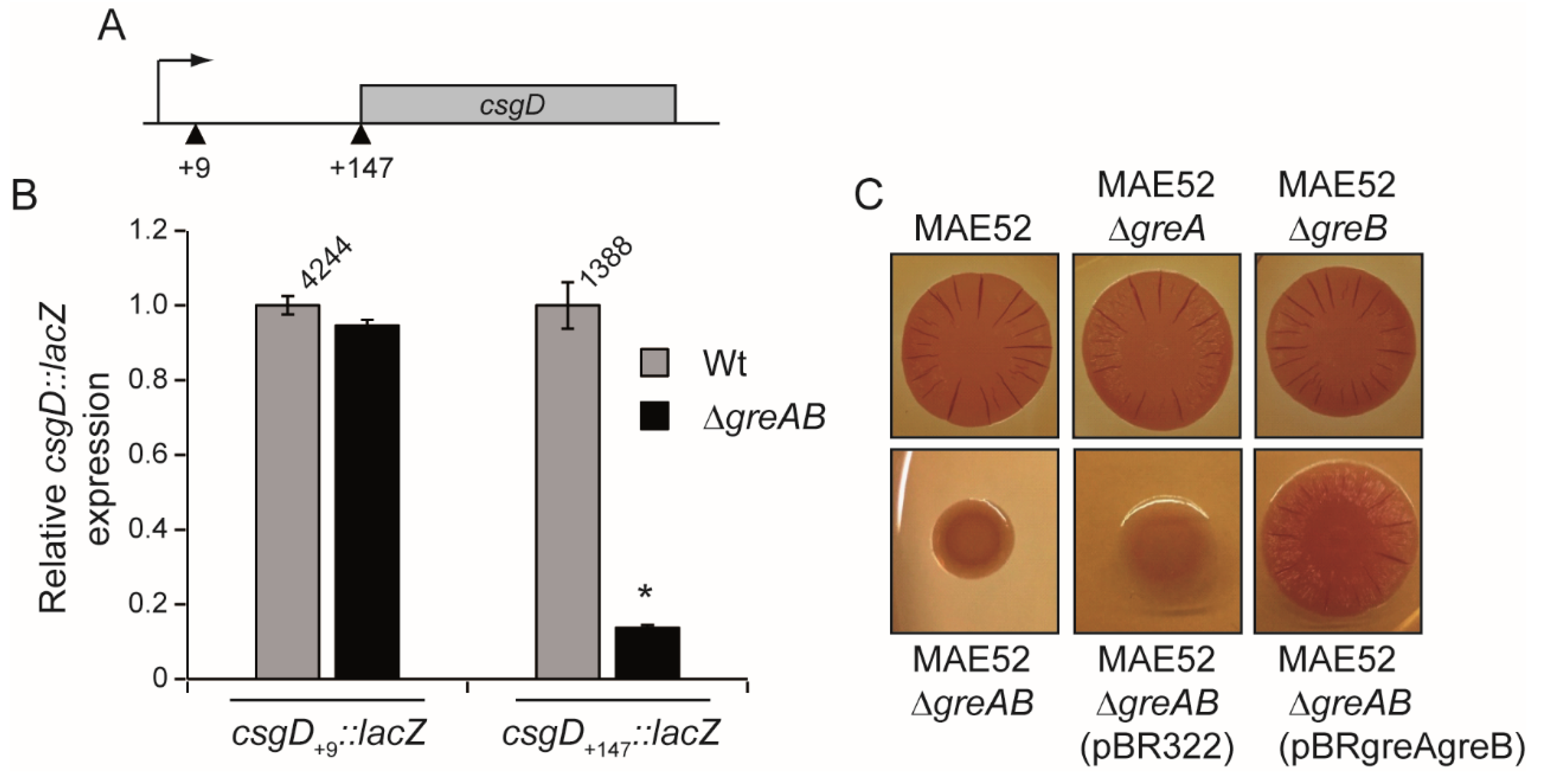
3.2. The Expression of CsgD, the Master Regulator of Biofilm Formation in S. Typhimurium Is Impaired in Strains Lacking the Gre Factors
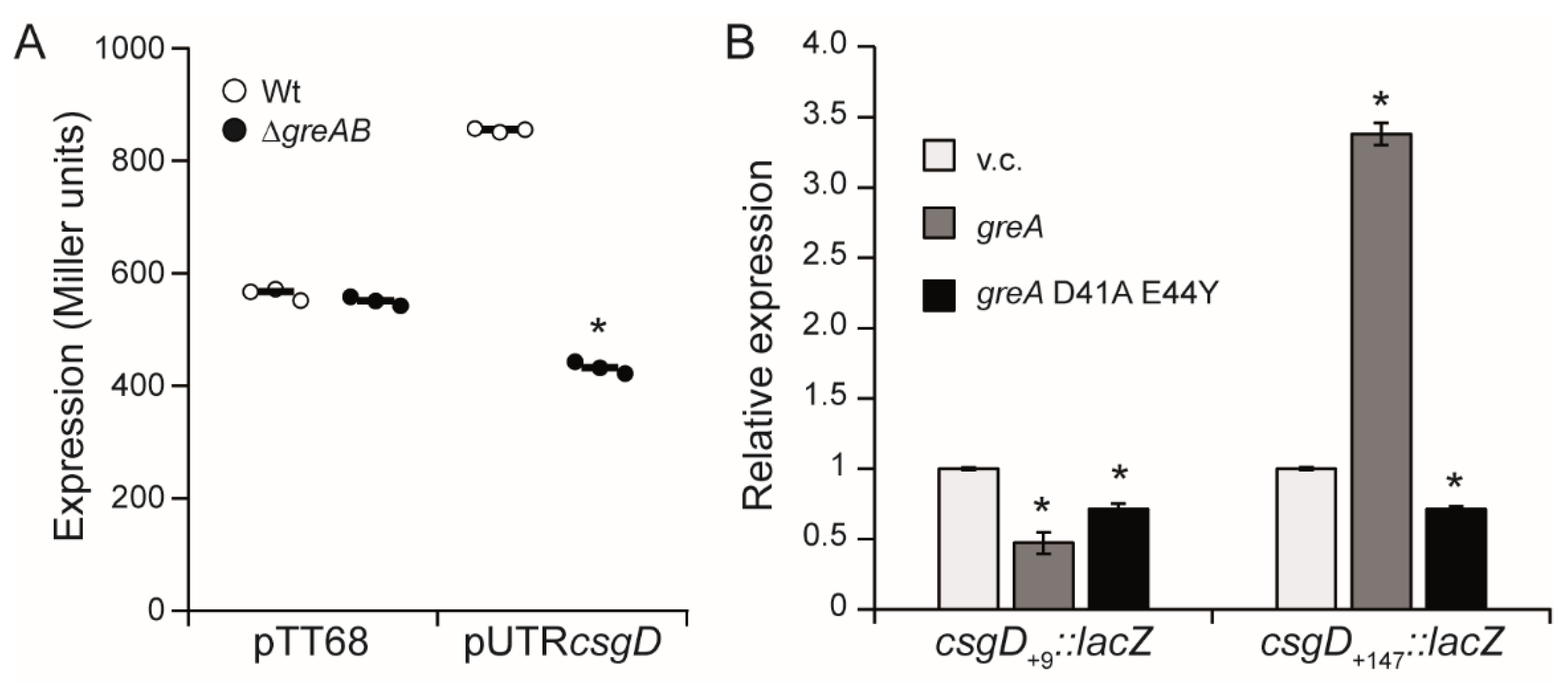
3.3. The 5′-UTR of the csgD Gene Is Required for Stimulation of csgD Expression by the Gre Factors
4. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Laptenko, O.; Lee, J.; Lomakin, I.; Borukhov, S. Transcript Cleavage Factors GreA and GreB Act as Transient Catalytic Components of RNA Polymerase. EMBO J. 2003, 22, 6322–6334. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Jiang, T.; Yu, B.; Wang, L.; Gao, C.; Ma, C.; Xu, P.; Ma, Y. Transcription Elongation Factor GreA Has Functional Chaperone Activity. PLoS ONE 2012, 7, e47521. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Yakhnin, A.V.; Fitzgerald, P.C.; Mclntosh, C.E.; Kashlev, M. Nascent RNA Sequencing Identifies a Widespread Sigma70-Dependent Pausing Regulated by Gre Factors in Bacteria. Nat Commun. 2021, 12, 906. [Google Scholar] [CrossRef] [PubMed]
- Aberg, A.; Fernandez-Vazquez, J.; Cabrer-Panes, J.D.; Sanchez, A.; Balsalobre, C. Similar and Divergent Effects of PpGpp and DksA Deficiencies on Transcription in Escherichia coli. J. Bacteriol. 2009, 191, 3226–3236. [Google Scholar] [CrossRef] [PubMed]
- Åberg, A.; Shingler, V.; Balsalobre, C. Regulation of the fimB Promoter: A Case of Differential Regulation by PpGpp and DksA in vivo. Mol. Microbiol. 2008, 67, 1223–1241. [Google Scholar] [CrossRef] [PubMed]
- Stepanova, E.; Lee, J.; Ozerova, M.; Semenova, E.; Datsenko, K.; Wanner, B.L.; Severinov, K.; Borukhov, S. Analysis of Promoter Targets for Escherichia coli Transcription Elongation Factor GreA In Vivo and In Vitro. J. Bacteriol. 2007, 189, 8772–8785. [Google Scholar] [CrossRef]
- Gaviria-Cantin, T.; El Mouali, Y.; Le Guyon, S.; Römling, U.; Balsalobre, C. Gre Factors-Mediated Control of hilD Transcription Is Essential for the Invasion of Epithelial Cells by Salmonella enterica Serovar Typhimurium. PLoS Pathog. 2017, 13, e1006312. [Google Scholar] [CrossRef]
- Steenackers, H.; Hermans, K.; Vanderleyden, J.; De Keersmaecker, S.C.J. Salmonella Biofilms: An Overview on Occurrence, Structure, Regulation and Eradication. Food Res. Int. 2012, 45, 502–531. [Google Scholar] [CrossRef]
- Harrell, J.E.; Hahn, M.M.; D’Souza, S.J.; Vasicek, E.M.; Sandala, J.L.; Gunn, J.S.; McLachlan, J.B. Salmonella Biofilm Formation, Chronic Infection, and Immunity Within the Intestine and Hepatobiliary Tract. Front. Cell. Infect. Microbiol. 2021, 10, 910. [Google Scholar] [CrossRef]
- Römling, U.; Rohde, M. Flagella Modulate the Multicellular Behavior of Salmonella typhimurium on the Community Level. FEMS Microbiol. Lett. 1999, 180, 91–102. [Google Scholar] [CrossRef]
- Römling, U.; Sierralta, W.D.; Eriksson, K.; Normark, S. Multicellular and Aggregative Behaviour of Salmonella typhimurium Strains Is Controlled by Mutations in the AgfD Promoter. Mol. Microbiol. 1998, 28, 249–264. [Google Scholar] [CrossRef] [PubMed]
- Römling, U.; Rohde, M.; Olsén, A.; Normark, S.; Reinköster, J. agfD, the Checkpoint of Multicellular and Aggregative Behaviour in Salmonella typhimurium Regulates at Least Two Independent Pathways. Mol. Microbiol. 2000, 36, 10–23. [Google Scholar] [CrossRef] [PubMed]
- Ogasawara, H.; Ishizuka, T.; Hotta, S.; Aoki, M.; Shimada, T.; Ishihama, A. Novel Regulators of the csgD Gene Encoding the Master Regulator of Biofilm Formation in Escherichia coli K-12. Microbiology 2020, 166, 880–890. [Google Scholar] [CrossRef] [PubMed]
- Gerstel, U.; Römling, U. The csgD Promoter, a Control Unit for Biofilm Formation in Salmonella typhimurium. Res. Microbiol. 2003, 154, 659–667. [Google Scholar] [CrossRef] [PubMed]
- Andreassen, P.R.; Pettersen, J.S.; Szczerba, M.; Valentin-Hansen, P.; Møller-Jensen, J.; Jørgensen, M.G. SRNA-Dependent Control of Curli Biosynthesis in Escherichia coli: McaS Directs Endonucleolytic Cleavage of csgD mRNA. Nucleic Acids Res. 2018, 46, 6746–6760. [Google Scholar] [CrossRef] [PubMed]
- Bordeau, V.; Felden, B. Curli Synthesis and Biofilm Formation in Enteric Bacteria Are Controlled by a Dynamic Small RNA Module Made up of a Pseudoknot Assisted by an RNA Chaperone. Nucleic Acids Res. 2014, 42, 4682–4696. [Google Scholar] [CrossRef]
- Gerstel, U.; Kolb, A.; Römling, U. Regulatory Components at the CsgD Promoter--Additional Roles for OmpR and Integration Host Factor and Role of the 5’ Untranslated Region. FEMS Microbiol. Lett. 2006, 261, 109–117. [Google Scholar] [CrossRef]
- Holmqvist, E.; Reimegård, J.; Sterk, M.; Grantcharova, N.; Römling, U.; Wagner, E.G.H. Two Antisense RNAs Target the Transcriptional Regulator CsgD to Inhibit Curli Synthesis. EMBO J. 2010, 29, 1840–1850. [Google Scholar] [CrossRef]
- Römling, U. Characterization of the Rdar Morphotupe, a Multicellular Behaviour in Enterobacteriaceae. CMLS, Cell. Mol. Life. Sci. 2005, 62, 1234–1246. [Google Scholar] [CrossRef]
- Römling, U.; Bian, Z.; Hammar, M.; Sierralta, W.D.; Normark, S. Curli Fibers Are Highly Conserved between Salmonella typhimurium and Escherichia coli with Respect to Operon Structure and Regulation. J. Bacteriol. 1998, 180, 722–731. [Google Scholar] [CrossRef] [Green Version]
- Hoiseth, S.K.; Stocker, B.A. Aromatic-Dependent Salmonella typhimurium Are Non-Virulent and Effective as Live Vaccines. Nature 1981, 291, 238–239. [Google Scholar] [CrossRef] [PubMed]
- Vivero, A.; Banos, R.C.; Mariscotti, J.F.; Oliveros, J.C.; Garcia-del Portillo, F.; Juarez, A.; Madrid, C. Modulation of Horizontally Acquired Genes by the Hha-YdgT Proteins in Salmonella enterica Serovar Typhimurium. J. Bacteriol. 2008, 190, 1152–1156. [Google Scholar] [CrossRef] [PubMed]
- Datsenko, K.A.; Wanner, B.L. One-Step Inactivation of Chromosomal Genes in Escherichia coli K-12 Using PCR Products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef] [PubMed]
- Ellermeier, C.D.; Janakiraman, A.; Slauch, J.M. Construction of Targeted Single Copy Lac Fusions Using Lambda Red and FLP-Mediated Site-Specific Recombination in Bacteria. Gene 2002, 290, 153–161. [Google Scholar] [CrossRef]
- Sternberg, N.L.; Maurer, R. Bacteriophage-Mediated Generalized Transduction in Escherichia Coli and Salmonella Typhimurium. Methods Enzymol. 1991, 204, 18–43. [Google Scholar] [PubMed]
- Maloy, S.; Stewart, R. Genetic Analysis of Pathogenic Bacteria; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1996. [Google Scholar]
- Paytubi, S.; Guirado, P.; Balsalobre, C.; Madrid, C. An Improved and Versatile Methodology to Quantify Biofilms Formed on Solid Surfaces and Exposed to the Air-Liquid Interphase. J. Microbiol. Methods 2014, 103. [Google Scholar] [CrossRef]
- Paytubi, S.; Cansado, C.; Madrid, C.; Balsalobre, C. Nutrient Composition Promotes Switching between Pellicle and Bottom Biofilm in Salmonella. Front. Microbiol. 2017, 8, 2160. [Google Scholar] [CrossRef]
- Miller, J.H. A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1992; ISBN 0879693495. [Google Scholar]
- Zafar, M.; Jahan, H.; Shafeeq, S.; Nimtz, M.; Jänsch, L.; Römling, U.; Iqbal Choudhary, M. Clarithromycin Exerts an Antibiofilm Effect against Salmonella enterica Serovar Typhimurium Rdar Biofilm Formation and Transforms the Physiology towards an Apparent Oxygen-Depleted Energy and Carbon Metabolism. Infect. Immun. 2020, 88, e00510-20. [Google Scholar] [CrossRef]
- Lamprokostopoulou, A.; Römling, U. Yin and Yang of Biofilm Formation and Cyclic Di-GMP Signaling of the Gastrointestinal Pathogen Salmonella enterica Serovar Typhimurium. J. Innate Immun. 2022, 14, 275–292. [Google Scholar] [CrossRef]
- Zakikhany, K.; Harrington, C.R.; Nimtz, M.; Hinton, J.C.D.; Römling, U. Unphosphorylated CsgD Controls Biofilm Formation in Salmonella enterica Serovar Typhimurium. Mol. Microbiol. 2010, 77, 771–786. [Google Scholar] [CrossRef]
- Vinella, D.; Potrykus, K.; Murphy, H.; Cashel, M. Effects on Growth by Changes of the Balance between GreA, GreB, and DksA Suggest Mutual Competition and Functional Redundancy in Escherichia coli. J. Bacteriol. 2012, 194, 261–273. [Google Scholar] [CrossRef] [PubMed]
- Nogales, J.; Campos, R.; BenAbdelkhalek, H.; Olivares, J.; Lluch, C.; Sanjuan, J. Rhizobium tropici Genes Involved in Free-Living Salt Tolerance Are Required for the Establishment of Efficient Nitrogen-Fixing Symbiosis with Phaseolus vulgaris. Mol. Plant. Microbe. Interact. 2002, 15, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Rhodius, V.A.; Suh, W.C.; Nonaka, G.; West, J.; Gross, C.A. Conserved and Variable Functions of the SigmaE Stress Response in Related Genomes. PLoS Biol. 2006, 4, e2. [Google Scholar] [CrossRef]
- Skovierova, H.; Rowley, G.; Rezuchova, B.; Homerova, D.; Lewis, C.; Roberts, M.; Kormanec, J. Identification of the SigmaE Regulon of Salmonella enterica Serovar Typhimurium. Microbiology 2006, 152, 1347–1359. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Overall, C.C.; Johnson, R.C.; Jones, M.B.; McDermott, J.E.; Heffron, F.; Adkins, J.N.; Cambronne, E.D. ChIP-Seq Analysis of the σE Regulon of Salmonella enterica Serovar Typhimurium Reveals New Genes Implicated in Heat Shock and Oxidative Stress Response. PLoS ONE 2015, 10, e0138466. [Google Scholar] [CrossRef] [PubMed]
- Gumber, S.; Taylor, D.L.; Marsh, I.B.; Whittington, R.J. Growth Pattern and Partial Proteome of Mycobacterium avium Subsp. Paratuberculosis during the Stress Response to Hypoxia and Nutrient Starvation. Vet. Microbiol. 2009, 133, 344–357. [Google Scholar] [CrossRef] [PubMed]
- Len, A.C.L.; Harty, D.W.S.; Jacques, N.A. Stress-Responsive Proteins Are Upregulated in Streptococcus mutans during Acid Tolerance. Microbiology 2004, 150, 1339–1351. [Google Scholar] [CrossRef]
- Campbell, G.R.O.; Sharypova, L.A.; Scheidle, H.; Jones, K.M.; Niehaus, K.; Becker, A.; Walker, G.C. Striking Complexity of Lipopolysaccharide Defects in a Collection of Sinorhizobium meliloti Mutants. J. Bacteriol. 2003, 185, 3853–3862. [Google Scholar] [CrossRef]
- Orlova, M.; Newlands, J.; Das, A.; Goldfarb, A.; Borukhov, S. Intrinsic Transcript Cleavage Activity of RNA Polymerase. Proc. Natl. Acad. Sci. USA 1995, 92, 4596–4600. [Google Scholar] [CrossRef]
- Agapov, A.; Esyunina, D.; Kulbachinskiy, A. Gre-Family Factors Modulate DNA Damage Sensing by Deinococcus radiodurans RNA Polymerase. RNA Biol. 2019, 16, 1711–1720. [Google Scholar] [CrossRef]
- Kaberdin, V.R.; Bläsi, U. Translation Initiation and the Fate of Bacterial MRNAs. FEMS Microbiol. Rev. 2006, 30, 967–979. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gaviria-Cantin, T.; Vargas, A.F.; Mouali, Y.E.; Jiménez, C.J.; Cimdins-Ahne, A.; Madrid, C.; Römling, U.; Balsalobre, C. Gre Factors Are Required for Biofilm Formation in Salmonella enterica Serovar Typhimurium by Targeting Transcription of the csgD Gene. Microorganisms 2022, 10, 1921. https://doi.org/10.3390/microorganisms10101921
Gaviria-Cantin T, Vargas AF, Mouali YE, Jiménez CJ, Cimdins-Ahne A, Madrid C, Römling U, Balsalobre C. Gre Factors Are Required for Biofilm Formation in Salmonella enterica Serovar Typhimurium by Targeting Transcription of the csgD Gene. Microorganisms. 2022; 10(10):1921. https://doi.org/10.3390/microorganisms10101921
Chicago/Turabian StyleGaviria-Cantin, Tania, Andrés Felipe Vargas, Youssef El Mouali, Carlos Jonay Jiménez, Annika Cimdins-Ahne, Cristina Madrid, Ute Römling, and Carlos Balsalobre. 2022. "Gre Factors Are Required for Biofilm Formation in Salmonella enterica Serovar Typhimurium by Targeting Transcription of the csgD Gene" Microorganisms 10, no. 10: 1921. https://doi.org/10.3390/microorganisms10101921
APA StyleGaviria-Cantin, T., Vargas, A. F., Mouali, Y. E., Jiménez, C. J., Cimdins-Ahne, A., Madrid, C., Römling, U., & Balsalobre, C. (2022). Gre Factors Are Required for Biofilm Formation in Salmonella enterica Serovar Typhimurium by Targeting Transcription of the csgD Gene. Microorganisms, 10(10), 1921. https://doi.org/10.3390/microorganisms10101921






