Differential Interaction between Invasive Thai Group B Streptococcus Sequence Type 283 and Caco-2 Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Isolates and Growth
2.2. Genomic DNA Extraction
2.3. Multilocus Sequence Typing (MLST)
2.4. Phylogenetic Analysis
2.5. Capsular Polysaccharide (CPS) Typing
2.6. Caco-2 Cell Culture
2.7. Quantitative Bacterial Interaction Assays
2.8. Cell Viability Testing by Trypan Blue Exclusion Test
2.9. Transepithelial Electrical Resistance (TEER) Measurement
2.10. Statistical Analysis
3. Results
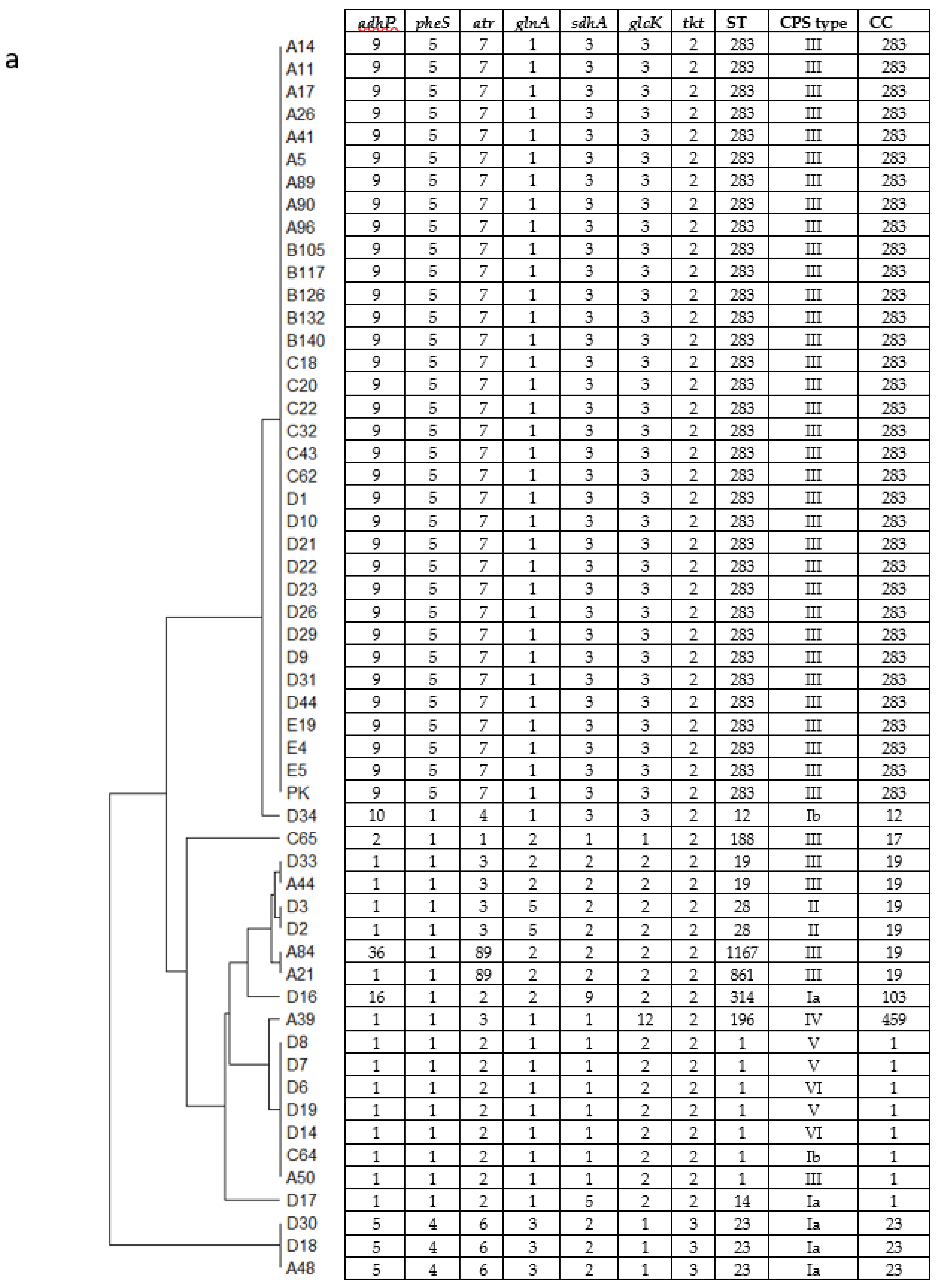
3.1. Genotyping of GBS Isolates and the Relationship with Patients’ Age
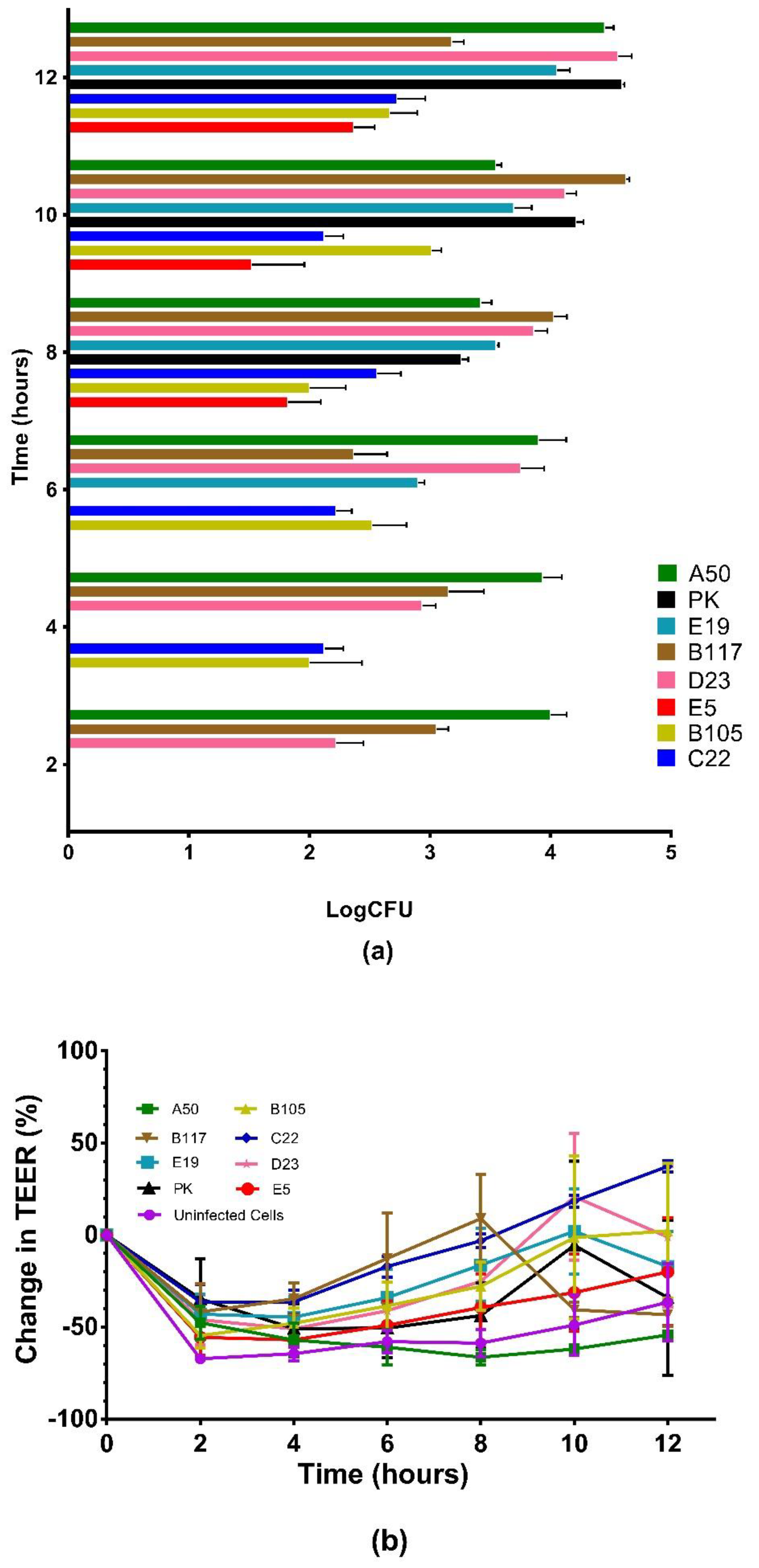
3.2. Comparison of Invasiveness among Selected CPSIII Isolates Using Caco-2 Cells
3.3. Intracellular Survival of the GBS Isolates in Caco-2 Cells
3.4. Translocation of the GBS Isolates across Polarized Caco-2 Cell Monolayers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- van Kassel, M.N.; Janssen, S.; Kofman, S.; Brouwer, M.C.; van de Beek, D.; Bijlsma, M.W. Prevalence of group B streptococcal colonization in the healthy non-pregnant population: A systematic review and meta-analysis. Clin. Microbiol. Infect. 2021, 27, 968–980. [Google Scholar] [CrossRef] [PubMed]
- Schuchat, A.; Wenger, J.D. Epidemiology of group B streptococcal disease. Risk factors, prevention strategies, and vaccine development. Epidemiol. Rev. 1994, 16, 374–402. [Google Scholar] [CrossRef] [PubMed]
- Edwards, M.S.; Baker, C.J. Group B streptococcal infections in elderly adults. Clin. Infect. Dis. 2005, 41, 839–847. [Google Scholar] [PubMed]
- Crespo-Ortiz Mdel, P.; Castaneda-Ramirez, C.R.; Recalde-Bolanos, M.; Velez-Londono, J.D. Emerging trends in invasive and noninvasive isolates of Streptococcus agalactiae in a Latin American hospital: A 17-year study. BMC Infect. Dis. 2014, 14, 428. [Google Scholar]
- Louthrenoo, W.; Kasitanon, N.; Wangkaew, S.; Hongsongkiat, S.; Sukitawut, W.; Wichainun, R. Streptococcus agalactiae: An emerging cause of septic arthritis. J. Clin. Rheumatol. 2014, 20, 74–78. [Google Scholar] [CrossRef]
- Fujita, H.; Nakamura, I.; Tsukimori, A.; Sato, A.; Ohkusu, K.; Matsumoto, T. Severe infective endocarditis in a healthy adult due to Streptococcus agalactiae. Int. J. Infect. Dis. 2015, 38, 43–45. [Google Scholar] [CrossRef]
- Rajendram, P.; Mar Kyaw, W.; Leo, Y.S.; Ho, H.; Chen, W.K.; Lin, R.; Pratim, P.; Badaruddin, H.; Ang, B.; Barkham, T.; et al. Group B Streptococcus Sequence Type 283 Disease Linked to Consumption of Raw Fish, Singapore. Emerg. Infect. Dis. 2016, 22, 1974–1977. [Google Scholar] [CrossRef]
- Barkham, T.; Zadoks, R.N.; Azmai, M.N.A.; Baker, S.; Bich, V.T.N.; Chalker, V.; Chau, M.L.; Dance, D.; Deepak, R.N.; van Doorn, H.R.; et al. One hypervirulent clone, sequence type 283, accounts for a large proportion of invasive Streptococcus agalactiae isolated from humans and diseased tilapia in Southeast Asia. PLoS Negl. Trop. Dis. 2019, 13, e0007421. [Google Scholar] [CrossRef]
- Kayansamruaj, P.; Soontara, C.; Unajak, S.; Dong, H.T.; Rodkhum, C.; Kondo, H.; Hirono, I.; Areechon, N. Comparative genomics inferred two distinct populations of piscine pathogenic Streptococcus agalactiae, serotype Ia ST7 and serotype III ST283, in Thailand and Vietnam. Genomics 2019, 111, 1657–1667. [Google Scholar] [CrossRef]
- Tan, S.; Lin, Y.; Foo, K.; Koh, H.F.; Tow, C.; Zhang, Y.; Ang, L.W.; Cui, L.; Badaruddin, H.; Ooi, P.L.; et al. Group B Streptococcus Serotype III Sequence Type 283 Bacteremia Associated with Consumption of Raw Fish, Singapore. Emerg. Infect. Dis. 2016, 22, 1970–1973. [Google Scholar] [CrossRef]
- Aiewsakun, P.; Ruangchai, W.; Thawornwattana, Y.; Jaemsai, B.; Mahasirimongkol, S.; Homkaew, A.; Suksomchit, P.; Dubbs, P.; Palittapongarnpim, P. Genomic epidemiology of Streptococcus agalactiae ST283 in Southeast Asia. Sci. Rep. 2022, 12, 4185. [Google Scholar] [CrossRef] [PubMed]
- Landwehr-Kenzel, S.; Henneke, P. Interaction of Streptococcus agalactiae and Cellular Innate Immunity in Colonization and Disease. Front. Immunol. 2014, 5, 519. [Google Scholar] [CrossRef] [PubMed]
- De Gaetano, G.V.; Lentini, G.; Galbo, R.; Coppolino, F.; Fama, A.; Teti, G.; Beninati, C. Invasion and trafficking of hypervirulent group B streptococci in polarized enterocytes. PLoS ONE 2021, 16, e0253242. [Google Scholar]
- Engle, M.J.; Goetz, G.S.; Alpers, D.H. Caco-2 cells express a combination of colonocyte and enterocyte phenotypes. J. Cell. Physiol. 1998, 174, 362–369. [Google Scholar] [CrossRef]
- Soriani, M.; Santi, I.; Taddei, A.; Rappuoli, R.; Grandi, G.; Telford, J.L. Group B Streptococcus crosses human epithelial cells by a paracellular route. J. Infect. Dis. 2006, 193, 241–250. [Google Scholar] [CrossRef]
- de-Paris, F.; Machado, A.B.; Gheno, T.C.; Ascoli, B.M.; Oliveira, K.R.; Barth, A.L. Group B Streptococcus detection: Comparison of PCR assay and culture as a screening method for pregnant women. Braz. J. Infect. Dis. 2011, 15, 323–327. [Google Scholar] [CrossRef]
- Gaillard, J.L.; Finlay, B.B. Effect of cell polarization and differentiation on entry of Listeria monocytogenes into the enterocyte-like Caco-2 cell line. Infect. Immun. 1996, 64, 1299–1308. [Google Scholar] [CrossRef]
- Jones, N.; Bohnsack, J.F.; Takahashi, S.; Oliver, K.A.; Chan, M.S.; Kunst, F.; Glaser, P.; Rusniok, C.; Crook, D.W.; Harding, R.M.; et al. Multilocus sequence typing system for group B Streptococcus. J. Clin. Microbiol. 2003, 41, 2530–2536. [Google Scholar] [CrossRef]
- Poyart, C.; Tazi, A.; Reglier-Poupet, H.; Billoet, A.; Tavares, N.; Raymond, J.; Trieu-Cuot, P. Multiplex PCR assay for rapid and accurate capsular typing of group B streptococci. J. Clin. Microbiol. 2007, 45, 1985–1988. [Google Scholar] [CrossRef]
- Kim, K.P.; Loessner, M.J. Enterobacter sakazakii invasion in human intestinal Caco-2 cells requires the host cell cytoskeleton and is enhanced by disruption of tight junction. Infect. Immun. 2008, 76, 562–570. [Google Scholar] [CrossRef]
- Decleva, E.; Menegazzi, R.; Busetto, S.; Patriarca, P.; Dri, P. Common methodology is inadequate for studies on the microbicidal activity of neutrophils. J. Leukoc. Biol. 2006, 79, 87–94. [Google Scholar] [CrossRef]
- Kuhns, D.B.; Priel, D.A.L.; Chu, J.; Zarember, K.A. Isolation and Functional Analysis of Human Neutrophils. Curr. Protoc. Immunol. 2015, 111, 7.23.1–7.23.16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eyngor, M.; Chilmonczyk, S.; Zlotkin, A.; Manuali, E.; Lahav, D.; Ghittino, C.; Shapira, R.; Hurvitz, A.; Eldar, A. Transcytosis of Streptococcus iniae through skin epithelial barriers: An in vitro study. FEMS Microbiol. Lett. 2007, 277, 238–248. [Google Scholar] [CrossRef] [PubMed]
- Kamiloglu, S.; Sari, G.; Ozdal, T.; Capanoglu, E. Guidelines for cell viability assays. Food Front. 2020, 1, 332–349. [Google Scholar] [CrossRef]
- Srinivasan, B.; Kolli, A.R.; Esch, M.B.; Abaci, H.E.; Shuler, M.L.; Hickman, J.J. TEER measurement techniques for in vitro barrier model systems. J. Lab. Autom. 2015, 20, 107–126. [Google Scholar] [CrossRef]
- Shabayek, S.; Spellerberg, B. Group B Streptococcal Colonization, Molecular Characteristics, and Epidemiology. Front. Microbiol. 2018, 9, 437. [Google Scholar] [CrossRef]
- Dostal, A.; Gagnon, M.; Chassard, C.; Zimmermann, M.B.; O’Mahony, L.; Lacroix, C. Salmonella adhesion, invasion and cellular immune responses are differentially affected by iron concentrations in a combined in vitro gut fermentation-cell model. PLoS ONE 2014, 9, e93549. [Google Scholar] [CrossRef]
- Jaradat, Z.W.; Bhunia, A.K. Adhesion, invasion, and translocation characteristics of Listeria monocytogenes serotypes in Caco-2 cell and mouse models. Appl. Environ. Microbiol. 2003, 69, 3640–3645. [Google Scholar] [CrossRef]
- Deitch, E.A.; Haskel, Y.; Cruz, N.; Xu, D.; Kvietys, P.R. Caco-2 and IEC-18 intestinal epithelial cells exert bactericidal activity through an oxidant-dependent pathway. Shock 1995, 4, 345–350. [Google Scholar] [CrossRef]
- Wells, C.L.; van de Westerlo, E.M.; Jechorek, R.P.; Erlandsen, S.L. Intracellular survival of enteric bacteria in cultured human enterocytes. Shock 1996, 6, 27–34. [Google Scholar] [CrossRef]
- Ulluwishewa, D.; Anderson, R.C.; McNabb, W.C.; Moughan, P.J.; Wells, J.M.; Roy, N.C. Regulation of tight junction permeability by intestinal bacteria and dietary components. J. Nutr. 2011, 141, 769–776. [Google Scholar] [CrossRef] [PubMed]
- Stoll, B.J.; Hansen, N.I.; Sanchez, P.J.; Faix, R.G.; Poindexter, B.B.; Van Meurs, K.P.; Bizzarro, M.J.; Goldberg, R.N.; Frantz, I.D., 3rd; Hale, E.C.; et al. Early onset neonatal sepsis: The burden of group B Streptococcal and E. coli disease continues. Pediatrics 2011, 127, 817–826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jamrozy, D.; Bijlsma, M.W.; de Goffau, M.C.; van de Beek, D.; Kuijpers, T.W.; Parkhill, J.; van der Ende, A.; Bentley, S.D. Increasing incidence of group B Streptococcus neonatal infections in The Netherlands is associated with clonal expansion of CC17 and CC23. Sci. Rep. 2020, 10, 9539. [Google Scholar] [CrossRef]
- Thatrimontrichai, A.; Khunnarakpong, N.; Tantichanthakarun, P.; Techato, C.; Nurak, A.; Sukumal, R.; Pasee, S.; Sujjanunt, T.; Rujeerapaiboon, N. Neonatal group B Streptococcus sepsis: A multicenter study in Thailand. Southeast Asian J. Trop. Med. Public Health 2017, 48, 1063–1071. [Google Scholar]
- Kovavisarach, E.; Jarupisarnlert, P.; Kanjanaharuetai, S. The accuracy of late antenatal screening cultures in predicting intrapartum group B streptococcal colonization. J. Med. Assoc. Thai. 2008, 91, 1796–1800. [Google Scholar]
- Tor-Udom, S.; Tor-Udom, P.; Hiriote, W. The prevalence of Streptococcus agalactiae (group B) colonization in pregnant women at Thammasat Hospital. J. Med. Assoc. Thai. 2006, 89, 411–414. [Google Scholar]
- Thinkhamrop, J.; Limpongsanurak, S.; Festin, M.R.; Daly, S.; Schuchat, A.; Lumbiganon, P.; Zell, E.; Chipato, T.; Win, A.A.; Perilla, M.J.; et al. Infections in international pregnancy study: Performance of the optical immunoassay test for detection of group B Streptococcus. J. Clin. Microbiol. 2003, 41, 5288–5290. [Google Scholar] [CrossRef]
- Al-Taiar, A.; Hammoud, M.S.; Cuiqing, L.; Lee, J.K.; Lui, K.M.; Nakwan, N.; Isaacs, D. Neonatal infections in China, Malaysia, Hong Kong and Thailand. Arch. Dis. Child Fetal Neonatal Ed. 2013, 98, F249–F255. [Google Scholar] [CrossRef] [PubMed]
- Madrid, L.; Seale, A.C.; Kohli-Lynch, M.; Edmond, K.M.; Lawn, J.E.; Heath, P.T.; Madhi, S.A.; Baker, C.J.; Bartlett, L.; Cutland, C.; et al. Infant Group B Streptococcal Disease Incidence and Serotypes Worldwide: Systematic Review and Meta-analyses. Clin. Infect. Dis. 2017, 65 (Suppl. 2), S160–S172. [Google Scholar] [CrossRef]
- Navarro-Torne, A.; Curcio, D.; Moisi, J.C.; Jodar, L. Burden of invasive group B Streptococcus disease in non-pregnant adults: A systematic review and meta-analysis. PLoS ONE 2021, 16, e0258030. [Google Scholar] [CrossRef]
- Manning, S.D.; Springman, A.C.; Lehotzky, E.; Lewis, M.A.; Whittam, T.S.; Davies, H.D. Multilocus sequence types associated with neonatal group B streptococcal sepsis and meningitis in Canada. J. Clin. Microbiol. 2009, 47, 1143–1148. [Google Scholar] [CrossRef] [PubMed]
- Poyart, C.; Reglier-Poupet, H.; Tazi, A.; Billoet, A.; Dmytruk, N.; Bidet, P.; Bingen, E.; Raymond, J.; Trieu-Cuot, P. Invasive group B streptococcal infections in infants, France. Emerg. Infect. Dis. 2008, 14, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Kao, Y.; Tsai, M.H.; Lai, M.Y.; Chu, S.M.; Huang, H.R.; Chiang, M.C.; Fu, R.H.; Lu, J.J.; Hsu, J.F. Emerging serotype III sequence type 17 group B Streptococcus invasive infection in infants: The clinical characteristics and impacts on outcomes. BMC Infect. Dis. 2019, 19, 538. [Google Scholar] [CrossRef] [PubMed]
- Flores, A.R.; Galloway-Pena, J.; Sahasrabhojane, P.; Saldana, M.; Yao, H.; Su, X.; Ajami, N.J.; Holder, M.E.; Petrosino, J.F.; Thompson, E.; et al. Sequence type 1 group B Streptococcus, an emerging cause of invasive disease in adults, evolves by small genetic changes. Proc. Natl. Acad. Sci. USA 2015, 112, 6431–6436. [Google Scholar] [CrossRef]
- Teatero, S.; Athey, T.B.; Van Caeseele, P.; Horsman, G.; Alexander, D.C.; Melano, R.G.; Li, A.; Flores, A.R.; Shelburne, S.A., 3rd; McGeer, A.; et al. Emergence of Serotype IV Group B Streptococcus Adult Invasive Disease in Manitoba and Saskatchewan, Canada, Is Driven by Clonal Sequence Type 459 Strains. J. Clin. Microbiol. 2015, 53, 2919–2926. [Google Scholar] [CrossRef] [Green Version]
- Kalimuddin, S.; Chen, S.L.; Lim, C.T.K.; Koh, T.H.; Tan, T.Y.; Kam, M.; Wong, C.W.; Mehershahi, K.S.; Chau, M.L.; Ng, L.C.; et al. 2015 Epidemic of Severe Streptococcus agalactiae Sequence Type 283 Infections in Singapore Associated With the Consumption of Raw Freshwater Fish: A Detailed Analysis of Clinical, Epidemiological, and Bacterial Sequencing Data. Clin. Infect. Dis. 2017, 64 (Suppl. 2), S145–S152. [Google Scholar] [CrossRef]
- Farley, M.M. Group B streptococcal disease in nonpregnant adults. Clin. Infect. Dis. 2001, 33, 556–561. [Google Scholar] [CrossRef]
- Foxman, B.; Gillespie, B.W.; Manning, S.D.; Marrs, C.F. Risk factors for group B streptococcal colonization: Potential for different transmission systems by capsular type. Ann. Epidemiol. 2007, 17, 854–862. [Google Scholar] [CrossRef]
- Carl, M.A.; Ndao, I.M.; Springman, A.C.; Manning, S.D.; Johnson, J.R.; Johnston, B.D.; Burnham, C.A.; Weinstock, E.S.; Weinstock, G.M.; Wylie, T.N.; et al. Sepsis from the gut: The enteric habitat of bacteria that cause late-onset neonatal bloodstream infections. Clin. Infect. Dis. 2014, 58, 1211–1218. [Google Scholar] [CrossRef]
- Baron, M.J.; Bolduc, G.R.; Goldberg, M.B.; Auperin, T.C.; Madoff, L.C. Alpha C protein of group B Streptococcus binds host cell surface glycosaminoglycan and enters cells by an actin-dependent mechanism. J. Biol. Chem. 2004, 279, 24714–24723. [Google Scholar] [CrossRef]
- Rubens, C.E.; Smith, S.; Hulse, M.; Chi, E.Y.; van Belle, G. Respiratory epithelial cell invasion by group B streptococci. Infect. Immun. 1992, 60, 5157–5163. [Google Scholar] [CrossRef] [PubMed]
- Korir, M.L.; Laut, C.; Rogers, L.M.; Plemmons, J.A.; Aronoff, D.M.; Manning, S.D. Differing mechanisms of surviving phagosomal stress among group B Streptococcus strains of varying genotypes. Virulence 2017, 8, 924–937. [Google Scholar] [CrossRef] [PubMed]
- Marouni, M.J.; Barzilai, A.; Keller, N.; Rubinstein, E.; Sela, S. Intracellular survival of persistent group A streptococci in cultured epithelial cells. Int. J. Med. Microbiol. 2004, 294, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Balkovetz, D.F.; Katz, J. Bacterial invasion by a paracellular route: Divide and conquer. Microbes Infect. 2003, 5, 613–619. [Google Scholar] [CrossRef]
- Kerneis, S.; Chauviere, G.; Darfeuille-Michaud, A.; Aubel, D.; Coconnier, M.H.; Joly, B.; Servin, A.L. Expression of receptors for enterotoxigenic Escherichia coli during enterocytic differentiation of human polarized intestinal epithelial cells in culture. Infect. Immun. 1992, 60, 2572–2580. [Google Scholar] [CrossRef] [Green Version]




| Patients’ Age | Total No. of Patients | No. of Patients (ST/CPS of GBS) | |||||
|---|---|---|---|---|---|---|---|
| Sterile Body Site | Non-Sterile Body Site | ||||||
| CSF | Blood | Joint Fluid | Pus | Urine | Genital Area (Swab) | ||
| Neonates (0–89 d) | 4 | 1 (ST283/III) 1 (ST861/III) | 1 (ST28/II) 1 (ST19/III) | 0 | 0 | 0 | 0 |
| Adolescents (10–19 y) | 2 | 0 | 1 (ST283/III) | 0 | 0 | 0 | 1 (ST1167/III) |
| Young adult (20–39 y) | 18 | 2 (ST283/III) | 9 (ST283/III) 1 (ST314/Ia) | 3 (ST283/III) | 1 (ST1/V) | 1 (ST23/Ia) | 1 (ST196/IV) |
| Middle aged adults (40–59 y) | 14 | 1 (ST283/III) | 4 (ST283/III) 2 (ST1/V) | 2 (ST283/III) 1 (ST188/III) 1 (ST28/II) 1 (ST1/Ib) | 1 (ST1/VI) | 1 (ST19/III) | 0 |
| Elderly (≥60 y) | 17 | 2 (ST283/III) | 4 (ST283/III) 1 (ST1/III) 1 (ST1/VI) | 4 (ST283/III) | 2 (ST23/Ia) 1 (ST14/Ia) | 1 (ST12/Ib) | 1 (ST283/III) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boonsilp, S.; Nealiga, M.J.; Wangchuk, K.; Homkaew, A.; Wongsuk, T.; Thuncharoon, H.; Suksomchit, P.; Wasipraphai, D.; Chaturongakul, S.; Dubbs, P. Differential Interaction between Invasive Thai Group B Streptococcus Sequence Type 283 and Caco-2 Cells. Microorganisms 2022, 10, 1917. https://doi.org/10.3390/microorganisms10101917
Boonsilp S, Nealiga MJ, Wangchuk K, Homkaew A, Wongsuk T, Thuncharoon H, Suksomchit P, Wasipraphai D, Chaturongakul S, Dubbs P. Differential Interaction between Invasive Thai Group B Streptococcus Sequence Type 283 and Caco-2 Cells. Microorganisms. 2022; 10(10):1917. https://doi.org/10.3390/microorganisms10101917
Chicago/Turabian StyleBoonsilp, Siriphan, Marea Jikka Nealiga, Kinley Wangchuk, Anchalee Homkaew, Thanwa Wongsuk, Huttaya Thuncharoon, Paveesuda Suksomchit, Daranee Wasipraphai, Soraya Chaturongakul, and Padungsri Dubbs. 2022. "Differential Interaction between Invasive Thai Group B Streptococcus Sequence Type 283 and Caco-2 Cells" Microorganisms 10, no. 10: 1917. https://doi.org/10.3390/microorganisms10101917
APA StyleBoonsilp, S., Nealiga, M. J., Wangchuk, K., Homkaew, A., Wongsuk, T., Thuncharoon, H., Suksomchit, P., Wasipraphai, D., Chaturongakul, S., & Dubbs, P. (2022). Differential Interaction between Invasive Thai Group B Streptococcus Sequence Type 283 and Caco-2 Cells. Microorganisms, 10(10), 1917. https://doi.org/10.3390/microorganisms10101917






