Genomic, Antimicrobial, and Aphicidal Traits of Bacillus velezensis ATR2, and Its Biocontrol Potential against Ginger Rhizome Rot Disease Caused by Bacillus pumilus
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacteria, Fungi, and Aphids Used in Assays
2.2. Taxonomic Identification of Strain ATR2
2.3. Genome Sequencing, Assembly, Annotation, and Bioinformatics Analysis
2.4. UHPLC-QE-MS Analysis
2.5. Antimicrobial Assays of Extracellular Metabolites from Strain ATR2
2.6. Aphicidal Assays of Extracellular Metabolites from Strain ATR2
2.7. Purification and Mass Spectrometry Analysis of the Antagonistic Compound against B. pumilus GR8
2.8. Biocontrol Assays of Strain ATR2 against Ginger Rhizome Rot Disease
2.9. Accession Numbers
3. Results
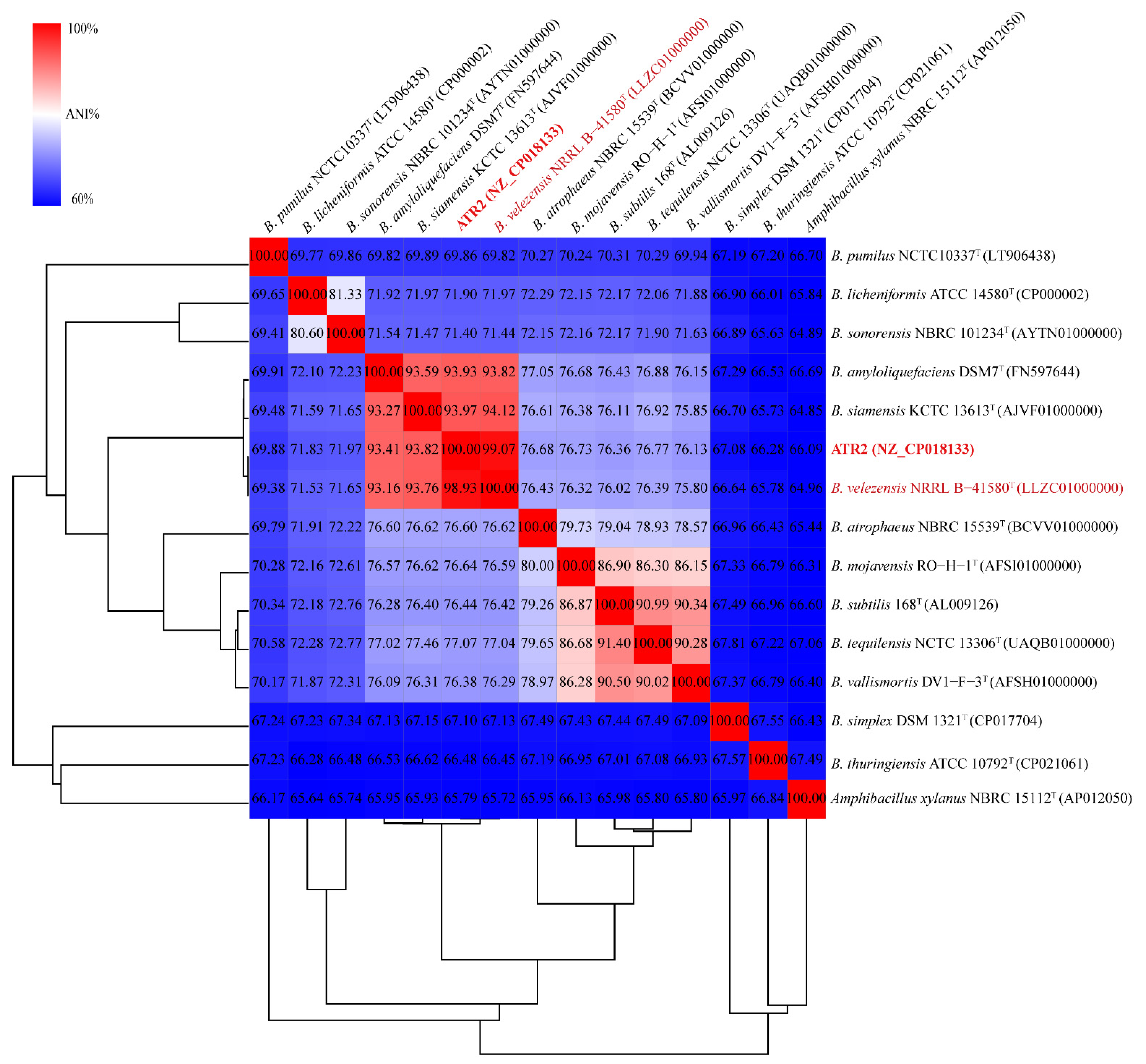
3.1. Strain Identification
3.2. Genomic Features of B. velezensis ATR2
3.3. Genes Involved in Plant Growth Promotion
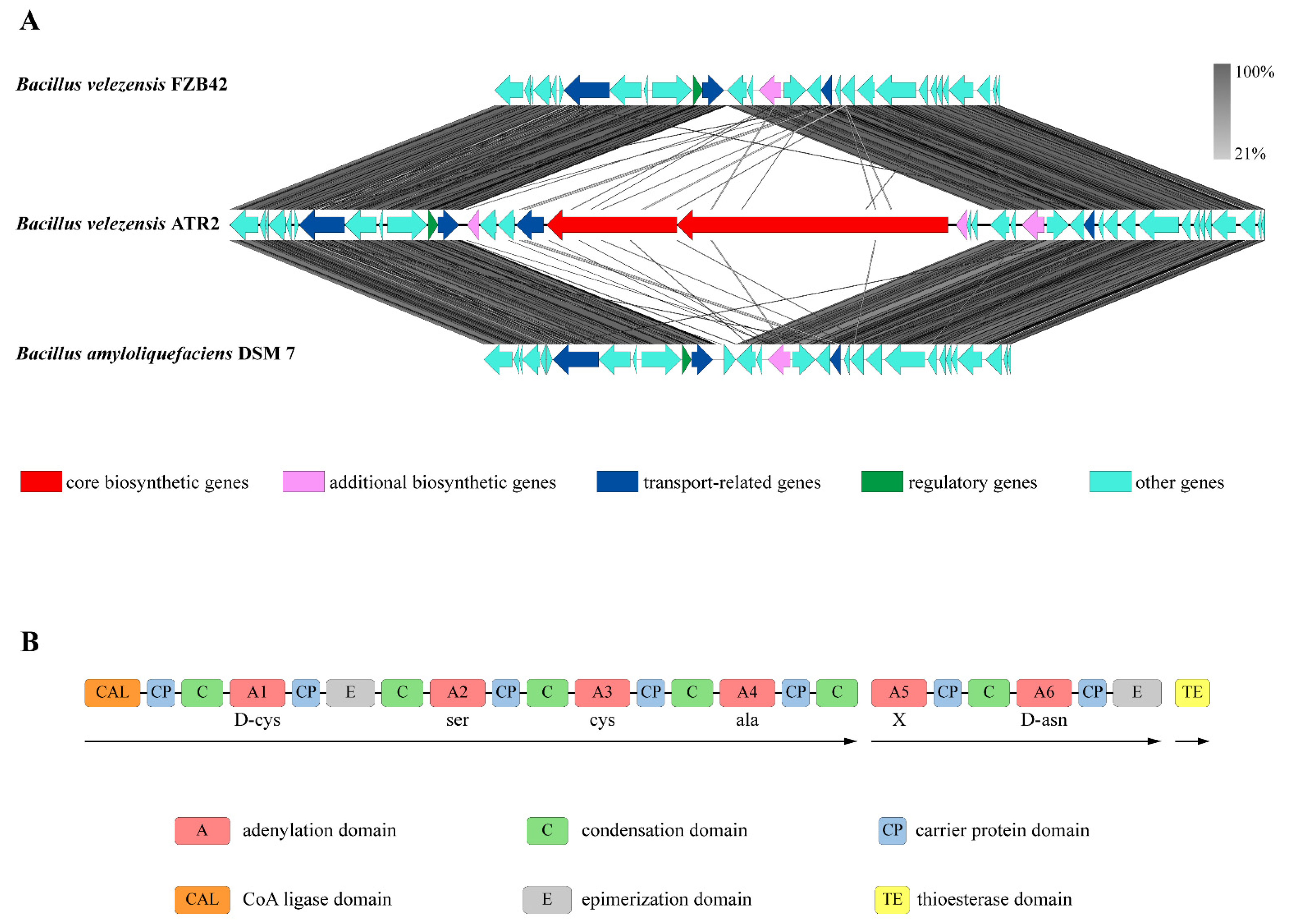
3.4. Genes Involved in Secondary Metabolite Synthesis
3.5. Antimicrobial Activities of Extracellular Metabolites from B. velezensis ATR2
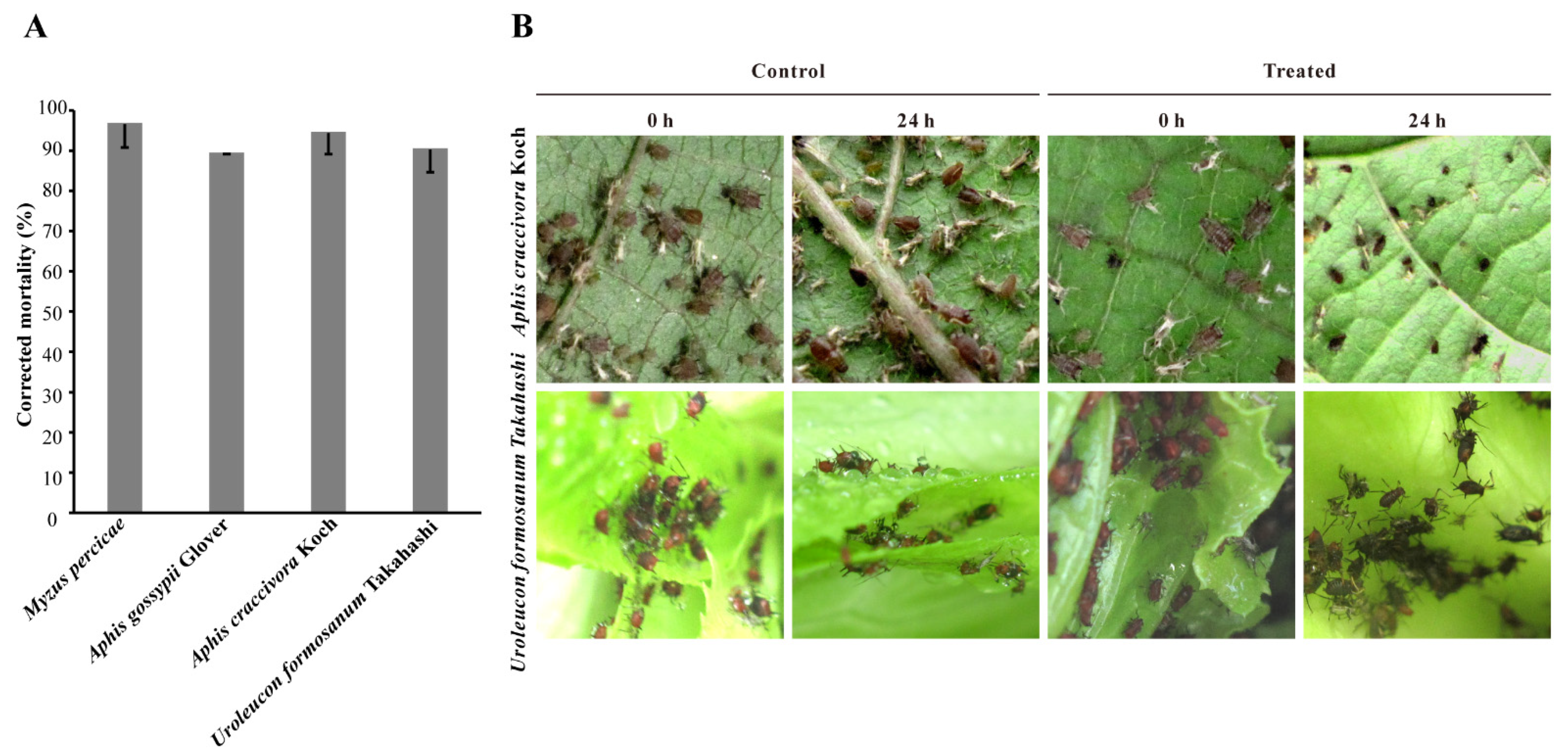
3.6. Aphicidal Activities of Extracellular Metabolites from B. velezensis ATR2
3.7. Identification of the Antagonistic Compound against B. pumilus GR8
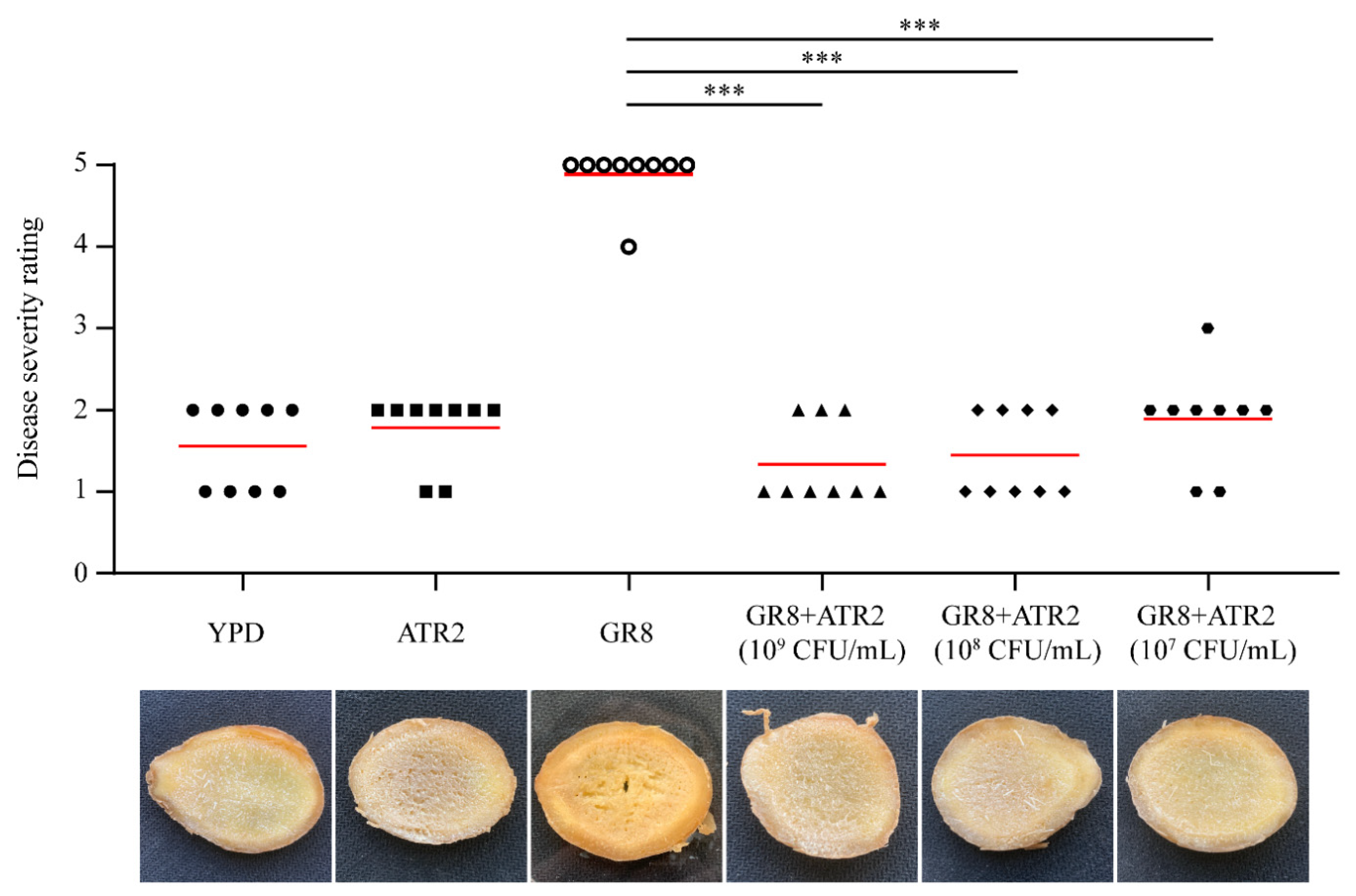
3.8. Biocontrol Efficacy of B. velezensis ATR2 against Ginger Rhizome Rot Disease
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Bodagh, M.N.; Maleki, I.; Hekmatdoost, A. Ginger in gastrointestinal disorders: A systematic review of clinical trials. Food Sci. Nutr. 2019, 7, 96–108. [Google Scholar] [CrossRef]
- Giacosa, A.; Morazzoni, P.; Bombardelli, E.; Riva, A.; Porro, G.B.; Rondanelli, M. Can nausea and vomiting be treated with gin-ger extract. Eur. Rev. Med. Pharm. 2015, 19, 1291–1296. [Google Scholar]
- Haniadka, R.; Saldanha, E.; Sunita, V.; Palatty, P.L.; Fayad, R.; Baliga, M.S. A review of the gastroprotective effects of ginger (Zingiber officinale Roscoe). Food Funct. 2013, 4, 845–855. [Google Scholar] [CrossRef] [PubMed]
- Kamada, T.; Kulip, J.; Tani, K.; Phan, C.-S.; Hatai, K.; Vairappan, C.S. Phillipsins A and B from Zingiber phillippsii Mood & Theilade in Borneo. Rec. Nat. Prod. 2018, 12, 317–322. [Google Scholar] [CrossRef]
- Kaur, I.P.; Deol, P.; Kondepudi, K.K.; Bishnoi, M. Anticancer Potential of Ginger: Mechanistic and Pharmaceutical Aspects. Curr. Pharm. Des. 2016, 22, 4160–4172. [Google Scholar] [CrossRef]
- Ohara, A.; Saito, F.; Matsuhisa, T. Screening of Antibacterial Activities of Edible Plants against Streptococcus mutans. Food Sci. Technol. Res. 2008, 14, 190–193. [Google Scholar] [CrossRef]
- Hayward, A.C. Characteristics of Pseudomonas solanacearum. J. Appl. Bacteriol. 1964, 27, 265–277. [Google Scholar] [CrossRef]
- Mathew, J.; Abraham, K.; Indrasenan, G.; Samuel, M. A New record of bacterial wilt of ginger incited by Pseudomonas solanacearum Smith, E.F. from India. Curr. Sci. India 1979, 48, 213–214. [Google Scholar]
- Koya, K.M.A. Role of rhizome maggot Mimegralla coeruleifrons macquart in rhizome rot of ginger. Entomon 1990, 15, 75–77. [Google Scholar]
- Li, Y.; Chi, L.D.; Mao, L.G.; Yan, D.D.; Wu, Z.F.; Ma, T.T.; Guo, M.X.; Wang, Q.X.; Ouyang, C.B.; Cao, A.C. First Report of Ginger Rhizome Rot Caused by Fusarium oxysporum in China. Plant Dis. 2014, 98, 282. [Google Scholar] [CrossRef]
- Wang, P.; Chung, C.; Lin, Y.; Yeh, Y. Use of polymerase chain reaction to detect the soft rot pathogen, Pythium myriotylum, in infected ginger rhizomes. Lett. Appl. Microbiol. 2003, 36, 116–120. [Google Scholar] [CrossRef]
- Peng, Q.; Yuan, Y.; Gao, M. Bacillus pumilus, a Novel Ginger Rhizome Rot Pathogen in China. Plant Dis. 2013, 97, 1308–1315. [Google Scholar] [CrossRef]
- Cai, X.-C.; Liu, C.-H.; Wang, B.-T.; Xue, Y.-R. Genomic and metabolic traits endow Bacillus velezensis CC09 with a potential biocontrol agent in control of wheat powdery mildew disease. Microbiol. Res. 2017, 196, 89–94. [Google Scholar] [CrossRef]
- García-Gutiérrez, L.; Zeriouh, H.; Romero, D.; Cubero, J.; de Vicente, A.; Pérez-García, A. The antagonistic strain Bacillus subtilis UMAF6639 also confers protection to melon plants against cucurbit powdery mildew by activation of jasmonate- and salicylic acid-dependent defence responses. Microb. Biotechnol. 2013, 6, 264–274. [Google Scholar] [CrossRef]
- Ghribi, D.; Abdelkefi-Mesrati, L.; Boukedi, H.; Elleuch, M.; Ellouze-Chaabouni, S.; Tounsi, S. The impact of the Bacillus subtilis SPB1 biosurfactant on the midgut histology of Spodoptera littoralis (Lepidoptera: Noctuidae) and determination of its putative receptor. J. Invertebr. Pathol. 2012, 109, 183–186. [Google Scholar] [CrossRef]
- Yun, D.C.; Yang, S.Y.; Kim, Y.C.; Kim, I.S.; Kim, Y.H. Identification of surfactin as an aphicidal metabolite produced by Bacillus amyloliquefaciens G1. J. Korean Soc. Appl. Biol. Chem. 2013, 56, 751–753. [Google Scholar] [CrossRef]
- Romero, D.; García, A.P.; Rivera, M.E.; Cazorla, F.M.; De Vicente, A. Isolation and evaluation of antagonistic bacteria towards the cucurbit powdery mildew fungus Podosphaera fusca. Appl. Microbiol. Biotechnol. 2003, 64, 263–269. [Google Scholar] [CrossRef]
- Arguelles-Arias, A.; Ongena, M.; Halimi, B.; Lara, Y.; Brans, A.; Joris, B.; Fickers, P. Bacillus amyloliquefaciens GA1 as a source of potent antibiotics and other secondary metabolites for biocontrol of plant pathogens. Microb. Cell Factories 2009, 8, 63. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-García, C.; Béjar, V.; Martínez-Checa, F.; Llamas, I.; Quesada, E. Bacillus velezensis sp. nov., a surfactant-producing bacterium isolated from the river Vélez in Málaga, southern Spain. Int. J. Syst. Evol. Microbiol. 2005, 55, 191–195. [Google Scholar] [CrossRef]
- Wang, L.-T.; Lee, F.-L.; Tai, C.-J.; Kuo, H.-P. Bacillus velezensis is a later heterotypic synonym of Bacillus amyloliquefaciens. Int. J. Syst. Evol. Microbiol. 2008, 58, 671–675. [Google Scholar] [CrossRef]
- Dunlap, C.A.; Kim, S.-J.; Kwon, S.-W.; Rooney, A.P. Bacillus velezensis is not a later heterotypic synonym of Bacillus amyloliquefaciens; Bacillus methylotrophicus, Bacillus amyloliquefaciens subsp. plantarum and ‘Bacillus oryzicola’ are later heterotypic synonyms of Bacillus velezensis based on phylogenomics. Int. J. Syst. Evol. Microbiol. 2016, 66, 1212–1217. [Google Scholar] [CrossRef] [PubMed]
- Rashid, H.-O.-R.; Kim, H.-J.; Yeom, S.-I.; Yu, H.-A.; Manir, M.; Moon, S.-S.; Kang, Y.J.; Chung, Y.R. Bacillus velezensis YC7010 Enhances Plant Defenses Against Brown Planthopper through Transcriptomic and Metabolic Changes in Rice. Front. Plant Sci. 2018, 9, 1904. [Google Scholar] [CrossRef]
- Al-Ali, A.; Deravel, J.; Krier, F.; Béchet, M.; Ongena, M.; Jacques, P. Biofilm formation is determinant in tomato rhizosphere colonization by Bacillus velezensis FZB42. Environ. Sci. Pollut. Res. 2018, 25, 29910–29920. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Shi, H.; Heng, J.; Wang, D.; Bian, K. Antimicrobial, plant growth-promoting and genomic properties of the peanut endophyte Bacillus velezensis LDO2. Microbiol. Res. 2019, 218, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.-H.; Liao, M.-J.; Wang, H.; Zheng, M.-Z.; Xu, J.-J.; Guo, J.-H. Bacillus velezensis, a potential and efficient biocontrol agent in control of pepper gray mold caused by Botrytis cinerea. Biol. Control 2018, 126, 147–157. [Google Scholar] [CrossRef]
- Gao, Z.; Zhang, B.; Liu, H.; Han, J.; Zhang, Y. Identification of endophytic Bacillus velezensis ZSY-1 strain and antifungal activity of its volatile compounds against Alternaria solani and Botrytis cinerea. Biol. Control 2017, 105, 27–39. [Google Scholar] [CrossRef]
- Huang, L.; Li, Q.-C.; Hou, Y.; Li, G.-Q.; Yang, J.-Y.; Li, D.-W.; Ye, J.-R. Bacillus velezensis strain HYEB5-6 as a potential biocontrol agent against anthracnose on Euonymus japonicus. Biocontrol Sci. Technol. 2017, 27, 636–653. [Google Scholar] [CrossRef]
- Chowdhury, S.P.; Dietel, K.; Rändler, M.; Schmid, M.; Junge, H.; Borriss, R.; Hartmann, A.; Grosch, R. Effects of Bacillus amyloliquefaciens FZB42 on Lettuce Growth and Health under Pathogen Pressure and Its Impact on the Rhizosphere Bacterial Community. PLoS ONE 2013, 8, e68818. [Google Scholar] [CrossRef]
- Chowdhury, S.P.; Hartmann, A.; Gao, X.; Borriss, R. Biocontrol mechanism by root-associated Bacillus amyloliquefaciens FZB42—A review. Front. Microbiol. 2015, 6, 780. [Google Scholar] [CrossRef]
- Chowdhury, S.P.; Uhl, J.; Grosch, R.; Alquéres, S.; Pittroff, S.M.; Dietel, K.; Schmitt-Kopplin, P.; Borriss, R.; Hartmann, A. Cyclic Lipopeptides of Bacillus amyloliquefaciens subsp. plantarum Colonizing the Lettuce Rhizosphere Enhance Plant Defense Responses Toward the Bottom Rot Pathogen Rhizoctonia solani. Mol. Plant-Microbe Interact. 2015, 28, 984–995. [Google Scholar] [CrossRef]
- Gül, A.; Kidoglu, F.; Tüzel, Y.; Tüzel, I.H. Effects of nutrition and Bacillus amyloliquefaciens on tomato (Solanum lycopersicum, L.) growing in perlite. Span. J. Agric. Res. 2008, 6, 422. [Google Scholar] [CrossRef]
- Kang, X.; Zhang, W.; Cai, X.; Zhu, T.; Xue, Y.; Liu, C. Bacillus velezensis CC09: A Potential ‘Vaccine’ for Controlling Wheat Diseases. Mol. Plant-Microbe Interact. 2018, 31, 623–632. [Google Scholar] [CrossRef]
- Chen, L.; Gu, W.; Xu, H.-Y.; Yang, G.-L.; Shan, X.-F.; Chen, G.; Kang, Y.-H.; Wang, C.-F.; Qian, A.-D. Comparative genome analysis of Bacillus velezensis reveals a potential for degrading lignocellulosic biomass. 3 Biotech 2018, 8, 253. [Google Scholar] [CrossRef]
- Migunova, V.; Tomashevich, N.; Konrat, A.; Lychagina, S.; Dubyaga, V.; D’Addabbo, T.; Sasanelli, N.; Asaturova, A. Selection of Bacterial Strains for Control of Root-Knot Disease Caused by Meloidogyne incognita. Microorganisms 2021, 9, 1698. [Google Scholar] [CrossRef]
- Rashid, H.-O.-R.; Khan, A.; Hossain, M.T.; Chung, Y.R. Induction of Systemic Resistance against Aphids by Endophytic Bacillus velezensis YC7010 via Expressing PHYTOALEXIN DEFICIENT4 in Arabidopsis. Front. Plant Sci. 2017, 8, 211. [Google Scholar] [CrossRef]
- Susič, N.; Janežič, S.; Rupnik, M.; Stare, B.G. Whole Genome Sequencing and Comparative Genomics of Two Nematicidal Bacillus Strains Reveals a Wide Range of Possible Virulence Factors. G3 Genes|Genomes|Genetics 2020, 10, 881–890. [Google Scholar] [CrossRef]
- Logan, N.A.; Berkeley, R.C.W. Identification of Bacillus Strains Using the API System. Microbiology 1984, 130, 1871–1882. [Google Scholar] [CrossRef]
- Gao, M.; Li, R.; Dai, S.; Wu, Y.; Yi, D. Diversity of Bacillus thuringiensis strains from soil in China and their pesticidal activities. Biol. Control 2008, 44, 380–388. [Google Scholar] [CrossRef]
- Guardado-Valdivia, L.; Tovar-Pérez, E.; Chacón-López, A.; López-García, U.; Gutiérrez-Martínez, P.; Stoll, A.; Aguilera, S. Identification and characterization of a new Bacillus atrophaeus strain B5 as biocontrol agent of postharvest anthracnose disease in soursop (Annona muricata) and avocado (Persea americana). Microbiol. Res. 2018, 210, 26–32. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R.; Glöckner, F.O.; Peplies, J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef]
- Kelley, D.R.; Schatz, M.C.; Salzberg, S.L. Quake: Quality-aware detection and correction of sequencing errors. Genome Biol. 2010, 11, R116. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef]
- Roberts, R.J.; Vincze, T.; Posfai, J.; Macelis, D. REBASE—A database for DNA restriction and modification: Enzymes, genes and genomes. Nucleic Acids Res. 2015, 43, D298–D299. [Google Scholar] [CrossRef]
- Grissa, I.; Vergnaud, G.; Pourcel, C. CRISPRFinder: A web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007, 35, W52–W57. [Google Scholar] [CrossRef]
- Alikhan, N.-F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, T.; Shen, X.; Liu, J.; Zhao, D.; Sun, Y.; Wang, L.; Liu, Y.; Gong, X.; Liu, Y.; et al. Serum metabolomics for early diagnosis of esophageal squamous cell carcinoma by UHPLC-QTOF/MS. Metabolomics 2016, 12, 116. [Google Scholar] [CrossRef]
- Njimoh, D.L.; Assob, J.C.N.; Mokake, S.E.; Nyhalah, D.J.; Yinda, C.K.; Sandjon, B. Antimicrobial Activities of a Plethora of Medicinal Plant Extracts and Hydrolates against Human Pathogens and Their Potential to Reverse Antibiotic Resistance. Int. J. Microbiol. 2015, 2015, 547156. [Google Scholar] [CrossRef]
- Abbott, W.S. A Method of Computing the Effectiveness of an Insecticide. J. Econ. Èntomol. 1925, 18, 265–267. [Google Scholar] [CrossRef]
- Gong, Q.; Zhang, C.; Lu, F.; Zhao, H.; Bie, X.; Lu, Z. Identification of bacillomycin D from Bacillus subtilis fmbJ and its inhibition effects against Aspergillus flavus. Food Control 2014, 36, 8–14. [Google Scholar] [CrossRef]
- Ye, T.; Zhou, T.; Li, Q.; Xu, X.; Fan, X.; Zhang, L.; Chen, S. Cupriavidus sp. HN-2, a Novel Quorum Quenching Bacterial Isolate, is a Potent Biocontrol Agent against Xanthomonas campestris pv. campestris. Microorganisms 2019, 8, 45. [Google Scholar] [CrossRef]
- Nishijima, K.A.; Alvarez, A.M.; Hepperly, P.R.; Shintaku, M.H.; Keith, L.M.; Sato, D.M.; Bushe, B.C.; Armstrong, J.W.; Zee, F.T. Association of Enterobacter cloacae with Rhizome Rot of Edible Ginger in Hawaii. Plant Dis. 2004, 88, 1318–1327. [Google Scholar] [CrossRef] [PubMed]
- Chun, J.; Oren, A.; Ventosa, A.; Christensen, H.; Arahal, D.R.; Da Costa, M.S.; Rooney, A.P.; Yi, H.; Xu, X.-W.; De Meyer, S.; et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 2018, 68, 461–466. [Google Scholar] [CrossRef]
- Luo, C.; Liu, X.; Zhou, H.; Wang, X.; Chen, Z. Nonribosomal Peptide Synthase Gene Clusters for Lipopeptide Biosynthesis in Bacillus subtilis 916 and Their Phenotypic Functions. Appl. Environ. Microbiol. 2014, 81, 422–431. [Google Scholar] [CrossRef]
- Iyer, S.; Acharya, K.R. Tying the knot: The cystine signature and molecular-recognition processes of the vascular endothelial growth factor family of angiogenic cytokines. FEBS J. 2011, 278, 4304–4322. [Google Scholar] [CrossRef] [PubMed]
- Reva, O.N.; Larisa, S.A.; Mwakilili, A.D.; Tibuhwa, D.; Lyantagaye, S.; Chan, W.Y.; Lutz, S.; Ahrens, C.H.; Vater, J.; Borriss, R. Complete genome sequence and epigenetic profile of Bacillus velezensis UCMB5140 used for plant and crop protection in comparison with other plant-associated Bacillus strains. Appl. Microbiol. Biotechnol. 2020, 104, 7643–7656. [Google Scholar] [CrossRef]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef]
- Kurabachew, H.; Wydra, K. Characterization of plant growth promoting rhizobacteria and their potential as bioprotectant against tomato bacterial wilt caused by Ralstonia solanacearum. Biol. Control 2013, 67, 75–83. [Google Scholar] [CrossRef]
- Mácha, H.; Marešová, H.; Juříková, T.; Švecová, M.; Benada, O.; Škríba, A.; Baránek, M.; Novotný, Č.; Palyzová, A. Killing Effect of Bacillus velezensis FZB42 on a Xanthomonas campestris pv. Campestris (Xcc) Strain Newly Isolated from Cabbage Brassica oleracea Convar. Capitata (L.): A Metabolomic Study. Microorganisms 2021, 9, 1410. [Google Scholar] [CrossRef]
- Hu, Y.; Lu, P.; Wang, Y.; Ding, L.; Atkinson, S.; Chen, S. OmpR positively regulates urease expression to enhance acid survival of Yersinia pseudotuberculosis. Microbiology 2009, 155, 2522–2531. [Google Scholar] [CrossRef]
- Yuan, Y.; Zheng, D.; Hu, X.; Cai, Q.; Yuan, Z. Conjugative Transfer of Insecticidal Plasmid pHT73 from Bacillus thuringiensis to B. anthracis and Compatibility of This Plasmid with pXO1 and pXO2. Appl. Environ. Microbiol. 2010, 76, 468–473. [Google Scholar] [CrossRef][Green Version]
- Sagi, S.; Konduru, B.; Parida, M. Heterologous expression of Intimin and IpaB fusion protein in Lactococcus lactis and its mucosal delivery elicit protection against pathogenicity of Escherichia coli O157 and Shigella flexneri in a murine model. Int. Immunopharmacol. 2020, 85, 106617. [Google Scholar] [CrossRef]
- Casella, F.; Manenti, M.; Conca, C.; Repetti, V.; Longhi, P.; Lazzaroni, S.; Mercieri, A.; Furlan, R. Liver abscess caused by Klebsiella pneumoniae. Dig. Liver Dis. 2009, 41, 838. [Google Scholar] [CrossRef]
- Mazumder, R.N.; Salam, M.A.; Ali, M.; Bhattacharya, M.K. Reactive arthritis associated with Shigella dysenteriae type 1 infection. J. Diarrhoeal Dis. Res. 1997, 15, 21–24. [Google Scholar]
- Ellis, M.W.; Johnson, R.; Crawford, K.; Lanier, J.B.; Merrell, D.S. Molecular Characterization of a Catalase-Negative Methicillin-Susceptible Staphylococcus aureus subsp. aureus Strain Collected from a Patient with Cutaneous Abscess. J. Clin. Microbiol. 2014, 52, 344–346. [Google Scholar] [CrossRef]
- Liu, Q.; Yi, J.; Liang, K.; Zhang, X. Salmonella Choleraesuis outer membrane vesicles: Proteomics and immunogenicity. J. Basic Microbiol. 2017, 57, 852–861. [Google Scholar] [CrossRef]
- Santos, R.; Zhang, S.; Tsolis, R.; Kingsley, R.; Adams, L.G.; Bäumler, A.J. Animal models of Salmonella infections: Enteritis versus typhoid fever. Microbes Infect. 2001, 3, 1335–1344. [Google Scholar] [CrossRef]
- Zhou, H.; Yao, Y.; Zhu, B.; Ren, D.; Yang, Q.; Fu, Y.; Yu, Y.; Zhou, J. Risk factors for acquisition and mortality of multidrug-resistant Acinetobacter baumannii bacteremia. Medicine 2019, 98, e14937. [Google Scholar] [CrossRef]
- Zhou, X.; Heyer, C.; Choi, Y.-E.; Mehrabi, R.; Xu, J.-R. The CID1 cyclin C-like gene is important for plant infection in Fusarium graminearum. Fungal Genet. Biol. 2010, 47, 143–151. [Google Scholar] [CrossRef]
- Tomioka, K.; Sawada, H.; Aoki, T.; Sato, T. Gray mold of pearl lupine caused by Botrytis cinerea. J. Gen. Plant Pathol. 2008, 74, 405–407. [Google Scholar] [CrossRef]
- Romão-Dumaresq, A.S.; Araújo, W.L.; Talbot, N.J.; Thornton, C.R. RNA Interference of Endochitinases in the Sugarcane Endophyte Trichoderma virens 223 Reduces Its Fitness as a Biocontrol Agent of Pineapple Disease. PLoS ONE 2012, 7, e47888. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Zhou, M.; Xiong, Z.; Peng, F.; Wei, W. The cAMP-PKA Signaling Pathway Regulates Pathogenicity, Hyphal Growth, Appressorial Formation, Conidiation, and Stress Tolerance in Colletotrichum higginsianum. Front. Microbiol. 2017, 8, 1416. [Google Scholar] [CrossRef]
- Starace, M.; Carpanese, M.A.; Alessandrini, A.; Piraccini, B.M.; Patrizi, A.; Neri, I. Tinea corporis incognito due to Microsporum Gypseum: Report of eight cases in children. Pediatr. Dermatol. 2021, 38, 652–654. [Google Scholar] [CrossRef]
- Peypoux, F.; Besson, F.; Michel, G.; Delcambe, L. Structure of Bacillomycin D, a New Antibiotic of the Iturin Group. JBIC J. Biol. Inorg. Chem. 2005, 118, 323–327. [Google Scholar] [CrossRef] [PubMed]
- Moyne, A.-L.; Shelby, R.; Cleveland, T.; Tuzun, S. Bacillomycin D: An iturin with antifungal activity against Aspergillus flavus. J. Appl. Microbiol. 2001, 90, 622–629. [Google Scholar] [CrossRef]
- Moyne, A.-L.; Cleveland, T.E.; Tuzun, S. Molecular characterization and analysis of the operon encoding the antifungal lipopeptide bacillomycin D. FEMS Microbiol. Lett. 2004, 234, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Russell, P.E. Fungicide resistance: Occurrence and management. J. Agric. Sci. 1995, 124, 317–323. [Google Scholar] [CrossRef]
- Yoo, J.-S.; Zheng, C.-J.; Lee, S.; Kwak, J.-H.; Kim, W.-G. Macrolactin N, a new peptide deformylase inhibitor produced by Bacillus subtilis. Bioorgan. Med. Chem. Lett. 2006, 16, 4889–4892. [Google Scholar] [CrossRef]
- Lee, S.J.; Cho, J.Y.; Cho, J.I.; Moon, J.H.; Park, K.D.; Lee, Y.J.; Park, K.H. Isolation and characterization of antimicrobial substance macrolactin a produced from Bacillus amyloliquefaciens CHO104 isolated from soil. J. Microbiol. Biotechnol. 2004, 14, 525–531. [Google Scholar]
- Kim, D.H.; Kim, H.K.; Kim, K.M.; Kim, C.K.; Jeong, M.H.; Ko, C.Y.; Moon, K.H.; Kang, J.S. Antibacterial activities of macrolactin a and 7-O-succinyl macrolactin a from Bacillus polyfermenticus KJS-2 against vancomycin-resistant enterococci and methicillin-resistant Staphylococcus aureus. Arch. Pharmacal Res. 2011, 34, 147–152. [Google Scholar] [CrossRef]
- Romero-Tabarez, M.; Jansen, R.; Sylla, M.; Lünsdorf, H.; Häußler, S.; Santosa, D.A.; Timmis, K.N.; Molinari, G. 7-O-Malonyl Macrolactin A, a New Macrolactin Antibiotic from Bacillus subtilis Active against Methicillin-Resistant Staphylococcus aureus, Vancomycin-Resistant Enterococci, and a Small-Colony Variant of Burkholderia cepacia. Antimicrob. Agents Chemother. 2006, 50, 1701–1709. [Google Scholar] [CrossRef]
- Patel, P.S.; Huang, S.; Fisher, S.; Pirnik, D.; Aklonis, C.; Dean, L.; Meyers, E.; Fernandes, P.; Mayerl, F. Bacillaene, a novel inhibitor of prokaryotic protein synthesis produced by Bacillus subtilis: Production, taxonomy, isolation, physico-chemical characteriza-tion and biological activity. J. Antibiot. 1995, 48, 997–1003. [Google Scholar] [CrossRef] [PubMed]
- Zweerink, M.M.; Edison, A. Difficidin and oxydifficidin: Novel broad spectrum antibacterial antibiotics produced by Bacillus subtilis. III. Mode of action of difficidin. J. Antibiot. 1987, 40, 1692–1697. [Google Scholar] [CrossRef] [PubMed]
- Arima, K.; Kakinuma, A.; Tamura, G. Surfactin, a crystalline peptidelipid surfactant produced by Bacillussubtilis: Isolation, characterization and its inhibition of fibrin clot formation. Biochem. Biophys. Res. Commun. 1968, 31, 488–494. [Google Scholar] [CrossRef]
- Peypoux, F.; Bonmatin, J.M.; Wallach, J. Recent trends in the biochemistry of surfactin. Appl. Microbiol. Biotechnol. 1999, 51, 553–563. [Google Scholar] [CrossRef]
- Kracht, M.; Rokos, H.; Özel, M.; Kowall, M.; Pauli, G.; Vater, J. Antiviral and Hemolytic Activities of Surfactin Isoforms and Their Methyl Ester Derivatives. J. Antibiot. 1999, 52, 613–619. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhou, T.; He, D.; Li, X.-Z.; Wu, H.; Liu, W.; Gao, X. Functions of Lipopeptides Bacillomycin D and Fengycin in Antagonism of Bacillus amyloliquefaciens C06 towards Monilinia fructicola. J. Mol. Microbiol. Biotechnol. 2011, 20, 43–52. [Google Scholar] [CrossRef]
- Meena, K.R.; Kanwar, S.S. Lipopeptides as the Antifungal and Antibacterial Agents: Applications in Food Safety and Therapeutics. BioMed Res. Int. 2015, 2015, 473050. [Google Scholar] [CrossRef]
- Hu, L.B.; Shi, Z.Q.; Zhang, T.; Yang, Z.M. Fengycin antibiotics isolated from B-FS01 culture inhibit the growth of Fusarium moniliforme Sheldon ATCC 38932. FEMS Microbiol. Lett. 2007, 272, 91–98. [Google Scholar] [CrossRef]
- Ongena, M.; Jourdan, E.; Adam, A.; Paquot, M.; Brans, A.; Joris, B.; Arpigny, J.-L.; Thonart, P. Surfactin and fengycin lipopeptides of Bacillus subtilis as elicitors of induced systemic resistance in plants. Environ. Microbiol. 2007, 9, 1084–1090. [Google Scholar] [CrossRef] [PubMed]
- Loeffler, W.; Tschen, J.S.M.; Vanittanakom, N.; Kugler, M.; Knorpp, E.; Hsieh, T.F.; Wu, T.G. Antifungal Effects of Bacilysin and Fengymycin from Bacillus subtilis F-29-3 A Comparison with Activities of Other Bacillus Antibiotics. J. Phytopathol. 1986, 115, 204–213. [Google Scholar] [CrossRef]
- Kenig, M.; Abraham, E.P. Antimicrobial Activities and Antagonists of Bacilysin and Anticapsin. J. Gen. Microbiol. 1976, 94, 37–45. [Google Scholar] [CrossRef]
- Wu, L.; Wu, H.; Chen, L.; Xie, S.; Zang, H.; Borriss, R.; Gao, X. Bacilysin from Bacillus amyloliquefaciens FZB42 Has Specific Bactericidal Activity against Harmful Algal Bloom Species. Appl. Environ. Microbiol. 2014, 80, 7512–7520. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.H.; Koumoutsi, A.; Scholz, R.; Schneider, K.; Vater, J.; Süssmuth, R.; Piel, J.; Borriss, R. Genome analysis of Bacillus amyloliquefaciens FZB42 reveals its potential for biocontrol of plant pathogens. J. Biotechnol. 2009, 140, 27–37. [Google Scholar] [CrossRef]
- Chen, X.H.; Koumoutsi, A.; Scholz, R.; Eisenreich, A.; Schneider, K.; Heinemeyer, I.; Morgenstern, B.; Voss, B.; Hess, W.R.; Reva, O.; et al. Comparative analysis of the complete genome sequence of the plant growth–promoting bacterium Bacillus amyloliquefaciens FZB42. Nat. Biotechnol. 2007, 25, 1007–1014. [Google Scholar] [CrossRef]
- Chen, X.-H.; Vater, J.; Piel, J.; Franke, P.; Scholz, R.; Schneider, K.; Koumoutsi, A.; Hitzeroth, G.; Grammel, N.; Strittmatter, A.W.; et al. Structural and Functional Characterization of Three Polyketide Synthase Gene Clusters in Bacillus amyloliquefaciens FZB 42. J. Bacteriol. 2006, 188, 4024–4036. [Google Scholar] [CrossRef]
- Scholz, R.; Vater, J.; Budiharjo, A.; Wang, Z.; He, Y.; Dietel, K.; Schwecke, T.; Herfort, S.; Lasch, P.; Borriss, R. Amylocyclicin, a Novel Circular Bacteriocin Produced by Bacillus amyloliquefaciens FZB42. J. Bacteriol. 2014, 196, 1842–1852. [Google Scholar] [CrossRef] [PubMed]
- Scholz, R.; Molohon, K.J.; Nachtigall, J.; Vater, J.; Markley, A.L.; Süssmuth, R.D.; Mitchell, D.A.; Borriss, R. Plantazolicin, a Novel Microcin B17/Streptolysin S-Like Natural Product from Bacillus amyloliquefaciens FZB42. J. Bacteriol. 2011, 193, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Doroghazi, J.R.; Janga, S.C.; Zhang, J.K.; Circello, B.; Griffin, B.M.; Labeda, D.P.; Metcalf, W.W. Diversity and abundance of phosphonate biosynthetic genes in nature. Proc. Natl. Acad. Sci. USA 2013, 110, 20759–20764. [Google Scholar] [CrossRef]
- Metcalf, W.W.; van der Donk, W.A. Biosynthesis of Phosphonic and Phosphinic Acid Natural Products. Annu. Rev. Biochem. 2009, 78, 65–94. [Google Scholar] [CrossRef] [PubMed]
- Seto, H.; Kuzuyama, T. Bioactive natural products with carbon–phosphorus bonds and their biosynthesis. Nat. Prod. Rep. 1999, 16, 589–596. [Google Scholar] [CrossRef] [PubMed]
- Bais, H.P.; Fall, R.; Vivanco, J.M. Biocontrol of Bacillus subtilis against Infection of Arabidopsis Roots by Pseudomonas syringae Is Facilitated by Biofilm Formation and Surfactin Production. Plant Physiol. 2004, 134, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Dong, W.; Li, S.; Lu, X.; Wang, P.; Zhang, X.; Wang, Y.; Ma, P. Fengycin produced by Bacillus subtilis NCD-2 plays a major role in biocontrol of cotton seedling damping-off disease. Microbiol. Res. 2014, 169, 533–540. [Google Scholar] [CrossRef]
- Wu, T.; Chen, M.; Zhou, L.; Lu, F.; Bie, X.; Lu, Z. Bacillomycin D effectively controls growth of Malassezia globosa by disrupting the cell membrane. Appl. Microbiol. Biotechnol. 2020, 104, 3529–3540. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Ishihara, A.; Nakajima, H. Isolation of anteiso-C17, iso-C17, iso-C16, and iso-C15 Bacillomycin D from Bacillus amyloliquefaciens SD-32 and Their Antifungal Activities against Plant Pathogens. J. Agric. Food Chem. 2014, 62, 1469–1476. [Google Scholar] [CrossRef] [PubMed]
- Dutta, R.; Banerjee, A.; Behere, G.T.; Devi, K.J.; Chandra, S.; Ngachan, V. PCR detection of Ralstonia solanacearum: A new ap-proach for rapid detection of bacterium from infected plants. Indian J. Agric. Sci. 2015, 85, 1007–1011. [Google Scholar]
- Liu, M.J.; Zhao, Z.; Wang, C. First Report of Rhizome Rot on Ginger (Zingiber officinale) Caused by Enterobacter cloacae in Shandong Province, China. Plant Dis. 2021, 105, 210. [Google Scholar] [CrossRef] [PubMed]


| Gene and Gene Cluster | Position | Product | Description |
|---|---|---|---|
| phyC | 2,149,490–2,150,641 | 3-Phytase | Phytase synthesis |
| trpC | 2,265,499–2,266,251 | Indole-3-glycerol phosphate synthase | IAA synthesis |
| bdhA | 660,137–661,177 | Butanediol dehydrogenase | 2,3-butanediol synthesis |
| ilvN | 2,764,326–2,764,844 | Acetolactate synthase small subunit | 3-hydroxy-2-butanone synthesis |
| ilvB | 2,764,841–2,766,565 | Acetolactate synthase large subunit | |
| alsD | 3,559,880–3,560,647 | alpha-acetolactate decarboxylase | |
| alsS | 3,560,708–3,562,420 | Acetolactate synthase | |
| treP | 820,991–822,406 | Trehalose permease IIC protein | Trehalose synthesis |
| treR | 824,189–824,905 | Trehalose operon repressor | |
| speE | 3,685,891–3,686,721 | Spermidine synthase | Spermidine synthesis |
| speB | 3,684,960–3,685,832 | Agmatinase | |
| dhbABCEF | 3,110,521–3,129,932 | Bacillibactin | Iron transport and utilization |
| nirD | 351,031–351,351 | Nitrite reductase small subunit | Nitrogen transport and utilization |
| nasD | 351,372–353,789 | Nitrite reductase large subunit | |
| nasC | 353,909–356,041 | Assimilatory nitrate reductase catalytic subunit | |
| narI | 3,668,425–3,669,096 | Nitrate reductase subunit gamma | |
| narJ | 3,669,093–3,669,650 | Nitrate reductase molybdenum cofactor assembly chaperone | |
| narH | 3,669,676–3,671,139 | Nitrate reductase subunit beta | |
| narG | 3,671,129–3,674,815 | Nitrate reductase subunit alpha | |
| khtT | 1,027,931–1,028,428 | Potassium: proton antiporter subunit KhtT | Potassium transport and utilization |
| ktrC | 1,448,732–1,449,397 | Ktr system potassium transporter KtrC | |
| ktrA | 3,028,064–3,028,732 | Potassium uptake protein KtrA | |
| corA | 847,676–848,635 | Magnesium and cobalt transport protein CorA | Magnesium transport and utilization |
| mgtE | 1,338,402–1,339,757 | Magnesium transporter MgtE |
| Metabolite | Genes and Gene Clusters | Genbank Accession | Main Function | Strains (Genetic Similarity) | |||
|---|---|---|---|---|---|---|---|
| B. velezensis ATR2 | B. velezensis FZB42 | B. amyloliquefaciens DSM 7T | B. subtilis 168T | ||||
| Sfp-dependent nonribosomal synthesis | |||||||
| Surfactin | srfABCD | AJ575642.1 | Antibacterial, ISR | 82% | 91% | 82% | 82% |
| Bacillomycin | bmyABCD | CP000560.1 | Antifungal | 100% | 100% | 93% | ND |
| Fengycin | fenABCDE | CP000560.1 | Antifungal, ISR | 100% | 100% | ND | 100% |
| Bacillibactin | dhbABCEF | AL009126.3 | Siderophore | 100% | 100% | 100% | 100% |
| Bacillaene | baeBCDEGHIJLMNRS | AJ634060.2 | Antibacterial | 100% | 100% | 100% | 100% |
| Macrolactin | pks2ABCDEFGHI | AJ634061.2 | Antibacterial | 100% | 100% | ND | ND |
| Difficidin | difABCDEFGHIJKLMNO | AJ634062.2 | Antibacterial | 100% | 100% | ND | ND |
| Sfp-independent nonribosomal synthesis | |||||||
| Bacilysin | bacABCDE, ywfAG | CP000560.1 | Antimicrobial | 100% | 100% | 100% | 100% |
| Ribosomal synthesis of bacteriocins and modified peptides | |||||||
| Plantazolicin | ptnABCDEFGHIJKL | FN668567.1 | Antibacterial | 91% | 91% | ND | ND |
| Amylocyclicin | acnABCDEF | CP000560.1 | Antibacterial | 98% | 100% | 93% | ND |
| Unknown metabolite | |||||||
| Putative lipopeptide | / | / | Unknown | D a | ND | ND | ND |
| Putative phosphonate | / | / | Unknown | D | ND | ND | ND |
| Microorganism | Description | Source or Reference | Diameter of Inhibition Zone (mm) a | Inhibition Ratio (%) |
|---|---|---|---|---|
| Bacillus pumilus GR8 | Pathogen of ginger rhizome rot | [12] | 27 ± 0.67 | / |
| Ralstonia solanacearum (Smith) | Pathogen of tomato bacterial wilt | [59] | 33.89 ± 0.19 | / |
| Xanthomonas campestris pv. campestris | Pathogen of crucifers black rot | [60] | 35.11 ± 0.19 | / |
| Enterobacter cloacae | Pathogen of rhizome rot of ginger | [53] | 19 ± 0 | / |
| Yersinia pseudotuberculosis YpⅢ | Pathogen of gastroenteritis | [61] | 24.78 ± 0.38 | / |
| Bacillus anthracis | Pathogen of anthrax | [62] | 20 ± 0.67 | / |
| Escherichia coli EHEC O157:H7 | Pathogen of diarrhea | [63] | 20 ± 0 | / |
| Klebsiella pneumoniae | Pathogen of liver abscess | [64] | 16 ± 0 | / |
| Shigella dysenteriae | Pathogen of bacillary dysentery | [65] | 20.56 ± 0.51 | / |
| Staphylococcus aureus subsp. aureus | Pathogen of cutaneous abscess | [66] | 15.11 ± 0.19 | / |
| Salmonella Choleraesuis | Pathogen of swine paratyphoid | [67] | 17.78 ± 0.38 | / |
| Salmonella Typhimurium | Pathogen of enteritis | [68] | 16.33 ± 0.33 | / |
| Acinetobacter baumannii | Pathogen of bloodstream infection | [69] | 14.89 ± 0.19 | / |
| Fusarium graminearum Sehw | Pathogen of wheat head scab | [70] | / | 75.42 ± 0.72 |
| Botrytis cinerea | Pathogen of gray mold | [71] | / | 80.83 ± 0.72 |
| Ceratocystis paradoxa (Dode) Moreau | Pathogen of pineapple disease of sugarcane | [72] | / | 91.67 ± 0.72 |
| Colletotrichum higginsianum Sacc | Pathogen of crucifer anthracnose disease | [73] | / | 81.67 ± 0.72 |
| Microsporum gypseum | Pathogen of tinea corporis | [74] | / | 52.5 ± 1.25 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, L.; Fu, Y.; Deng, S.; Wu, Y.; Gao, M. Genomic, Antimicrobial, and Aphicidal Traits of Bacillus velezensis ATR2, and Its Biocontrol Potential against Ginger Rhizome Rot Disease Caused by Bacillus pumilus. Microorganisms 2022, 10, 63. https://doi.org/10.3390/microorganisms10010063
Liang L, Fu Y, Deng S, Wu Y, Gao M. Genomic, Antimicrobial, and Aphicidal Traits of Bacillus velezensis ATR2, and Its Biocontrol Potential against Ginger Rhizome Rot Disease Caused by Bacillus pumilus. Microorganisms. 2022; 10(1):63. https://doi.org/10.3390/microorganisms10010063
Chicago/Turabian StyleLiang, Leiqin, Yajuan Fu, Sangsang Deng, Yan Wu, and Meiying Gao. 2022. "Genomic, Antimicrobial, and Aphicidal Traits of Bacillus velezensis ATR2, and Its Biocontrol Potential against Ginger Rhizome Rot Disease Caused by Bacillus pumilus" Microorganisms 10, no. 1: 63. https://doi.org/10.3390/microorganisms10010063
APA StyleLiang, L., Fu, Y., Deng, S., Wu, Y., & Gao, M. (2022). Genomic, Antimicrobial, and Aphicidal Traits of Bacillus velezensis ATR2, and Its Biocontrol Potential against Ginger Rhizome Rot Disease Caused by Bacillus pumilus. Microorganisms, 10(1), 63. https://doi.org/10.3390/microorganisms10010063





