Abalone Viral Ganglioneuritis
Abstract
:1. Introduction
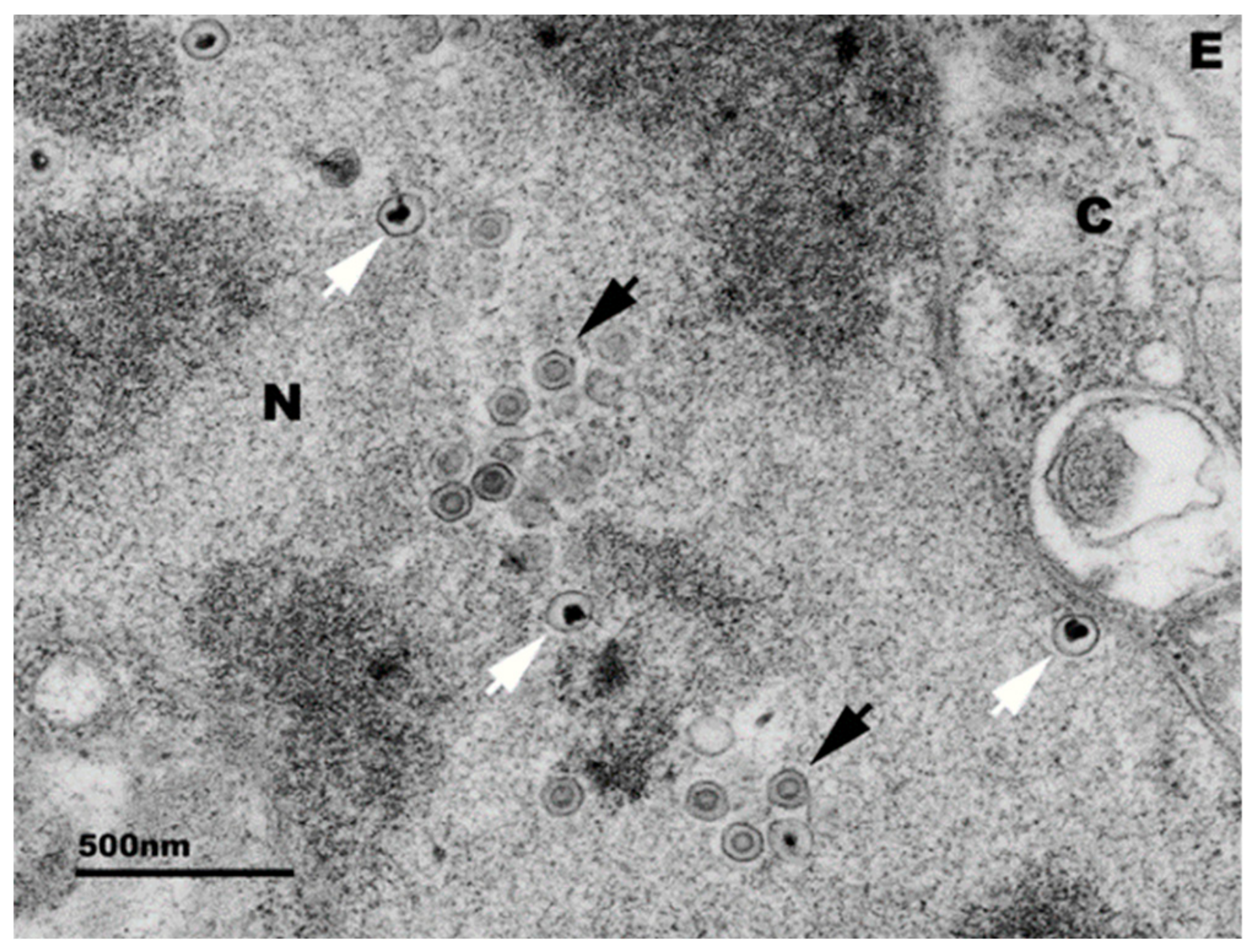
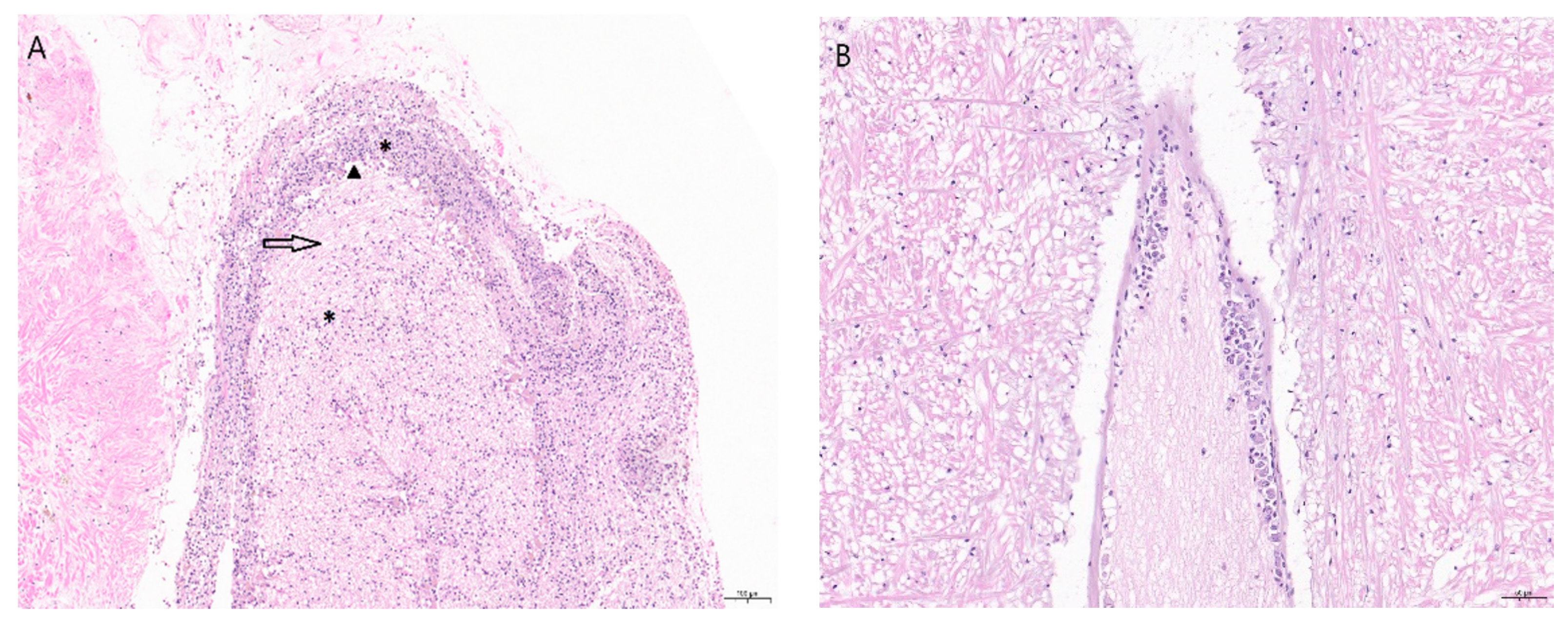
2. Clinical Signs and Pathology
3. Diagnostic Tests
4. Experimental Disease Model
5. Virus-Host Interaction
6. Molecular Vaccination/RNAi in Aquatic Invertebrates
7. Future of Abalone Production
Funding
Acknowledgments
Conflicts of Interest
References
- Nicolas, J.L.; Comps, M.; Cochennec, N. Herpes-like virus infecting Pacific oyster larvae, Crassostrea gigas. Bull. Eur. Assoc. Fish Pathol. 1992, 12, 11–13. [Google Scholar]
- Renault, T.; Le Deuff, R.M.; Cochennec, N.; Maffart, P. Herpesviruses associated with mortalities among Pacific oyster, Crassostrea gigas, in France—Comparative study. Rev. Med. Vet. 1994, 145, 735–742. [Google Scholar]
- Friedman, C.S.; Estes, R.M.; Stokes, N.A.; Burge, C.A.; Hargove, J.S.; Barber, B.J.; Elston, R.A.; Burreson, E.M.; Reece, K.S. Herpes virus in juvenile Pacific oysters Crassostrea gigas from Tomales Bay, California, coincides with summer mortality episodes. Dis. Aquat. Org. 2005, 63, 33–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hine, P.M.; Wesney, B.; Hay, B.E. Herpesvirus associated with mortalities among hatchery-reared larval Pacific oysters Crassostrea gigas. Dis. Aquat. Org. 1992, 12, 135–142. [Google Scholar] [CrossRef]
- Cameron, A.; Crane, M. Final Report: OsHV-1 µVar International Workshop, Cairns, Queensland, Australia, 9–10 July 2011; AusVet Animal Health Services and Fisheries Research and Development Corporation: Canberra, Australia, 2011. [Google Scholar]
- Chang, P.H.; Kuo, S.T.; Lai, S.H.; Yang, H.S.; Ting, Y.Y.; Hsu, C.L.; Chen, H.C. Herpes-like virus infection causing mortality of cultured abalone Haliotis diversicolor supertexta in Taiwan. Dis. Aquat. Org. 2005, 65, 23–27. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Wu, W.; Yan, J.; Zhou, W.; Chen, X.; Ni, Z.; Chen, X. Investigation on an exterminate disease of Haliotis diversicolor aquatilis. Fujian J. Anim. Husb. 1999, 21, 4–5. [Google Scholar]
- Nie, Z.Q.; Wang, S.P. The status of abalone culture in China. J. Shellfish Res. 2004, 23, 941–945. [Google Scholar]
- Gu, L.; Qia, R.-J.; Yanga, R.; Hana, T.; Jianga, J.-Z.; Wanga, J.-W. The prevalence of abalone herpesvirus in two Haliotis species in South China during 2002–2013. Aquaculture 2019, 505, 18–26. [Google Scholar] [CrossRef]
- Bai, C.-M.; Li, Y.-N.; Chang, P.-H.; Jiang, J.-Z.; Xin, L.-S.; Li, C.; Wang, J.-Y.; Wang, C. Susceptibility of two abalone species, Haliotis diversicolor supertexta and Haliotis discus hannai, to Haliotid herpesvirus 1 infection. J. Invertebr. Pathol. 2019, 160, 26–32. [Google Scholar] [CrossRef]
- Hooper, C.; Hardy-Smith, P.; Handlinger, J. Ganglioneuritis causing high mortalities in farmed Australian abalone [Haliotis laevigata and Haliotis rubra]. Aust. Vet. J. 2007, 85, 188–193. [Google Scholar] [CrossRef]
- Ellard, K.; Pyecroft, S.; Handlinger, J.; Andrewartha, R. Findings of disease investigations following the recent detection of AVG in Tasmania. In Proceedings of the Fourth National FRDC Aquatic Animal Health Scientific Conference, Cairns, Australia, 22–24 July 2009. [Google Scholar]
- Cowley, J.A.; Corbeil, S.; Chen, H.; Wong, F.; Moody, N.J.; Ellard, K.; Fegan, M.; Savin, K.; Warner, S.; Crane, M.S.-J. Sequence variations amongst abalone herpes-like virus [AbHV] strains provide insights into its origins in Victoria and Tasmania. In Proceedings of the First FRDC Australasian Scientific Conference on Aquatic Animal Health, Cairn, Australia, 5–8 July 2011. [Google Scholar]
- Savin, K.W.; Cocks, B.G.; Wong, F.; Sawbridge, T.; Cogan, N.; Savage, D.; Warner, S. A neurotropic herpesvirus infecting the gastropod, abalone, shares ancestry with oyster herpesvirus and a herpesvirus associated with the amphioxus genome. Virol. J. 2010, 7, 308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savin, K.; Davison, A. Create Genus Aurivirus in the Family Malacoherpesviridae, Order Herpesvirales. 2011. Available online: https://talk.ictvonline.org/taxonomy/ (accessed on 1 September 2020).
- FAO. Statistics—Introduction; Fisheries and Aquaculture Department, Food and Agriculture Organization of the United Nations: Rome, Italy, 2020; Available online: http://www.fao.org/fishery/statistics/en (accessed on 1 September 2020).
- Cook, P.A. Recent Trends in Worldwide Abalone Production. J. Shellfish Res. 2016, 35, 581–583. [Google Scholar] [CrossRef]
- Mobsby, D.; Steven, H.A.; Curtotti, R. Australian Fisheries and Aquaculture Outlook 2020; Department of Agriculture Wat Env ABARES: Canberra, Australia, 2020; p. 4. [Google Scholar]
- Wang, J.; Guo, Z.; Feng, J.; Liu, G.; Xu, L.; Chen, B.; Pan, J. Virus infection in cultured abalone, Haliotis diversicolor Reeve in Guangdong Province, China. J. Shellfish Res. 2004, 23, 1163–1168. [Google Scholar]
- Zhang, Z.; Wang, J.; Su, Y.; Yan, Q.; Chi, X.; Zhou, H.; Zhou, Y. Pathogeny and Histopathology of the Epidemic Disease in Haliotis diversicolor supertexta. J. Xiamen Univ. [Nat. Sci.] 2001, 40, 949–956. [Google Scholar]
- Chen, I.-W.; Chang, P.-H.; Chen, M.-S.; Renault, T.; Chen, M.-M.; Kuo, S.-T.; Cheng, C.-H. Exploring the chronic mortality affecting abalone in Taiwan: Differentiation of abalone herpesvirus-associated acute infection from chronic mortality by PCR and in situ hybridization and histopathology. Taiwan Vet. J. 2016, 42, 1–9. [Google Scholar] [CrossRef]
- Whitley, R.J. Advances in the management of herpesvirus infections—Introduction. Semin. Dermatol. 1996, 15 (Suppl. 1), 1–3. [Google Scholar]
- Chen, M.H.; Kuo, S.T.; Renault, T.; Friedman, C.S.; Chang, P.H. Development of a polymerase chain reaction for the detection of abalone herpesvirus infection based on the DNA polymerase gene. J. Virol. Methods 2012, 185, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Corbeil, S.; Colling, A.; Williams, L.M.; Wong, F.Y.K.; Savin, K.; Warner, S.; Murdoch, B.; Cogan, N.O.I.; Sawbridge, T.I.; Fegan, M.; et al. Development and validation of a TaqMan® PCR assay for the Australian abalone herpes-like virus. Dis. Aquat. Org. 2010, 92, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Caraguel, C.G.B.; Ellard, K.; Moody, N.J.G.; Corbeil, S.; Williams, L.M.; Mohr, P.G.; Cummins, D.M.; Hoad, J.; Slater, J.; Crane, M.S.-J. Diagnostic test accuracy when screening for Haliotid herpesvirus 1 [AbHV] in apparently healthy populations of Australian abalone Haliotis spp. Dis. Aquat. Org. 2019, 136, 199–207. [Google Scholar] [CrossRef] [Green Version]
- OIE. 2.4.1 Infection with Abalone Herpesvirus. 2019. Available online: https://www.oie.int/index.php?id=2439&L=0&htmfile=chapitre_abalone_herpesvirus.htm (accessed on 1 September 2020).
- Chen, M.H.; Kuo, S.T.; Renault, T.; Chang, P.H. The development of a loop-mediated isothermal amplification for rapid and sensitive detection of abalone herpesvirus DNA. J. Virol. Methods 2014, 196, 199–203. [Google Scholar] [CrossRef] [Green Version]
- Corbeil, S.; Williams, L.M.; McColl, K.A.; Mohammad, I.; Hyatt, A.D.; Crameri, S.G.; Fegan, M.; Crane, M.S.-J. Abalone viral ganglioneuritis: Establishment of an experimental immersion challenge system and kinetics of detection of the herpes viral infection in Australian abalone. Virus Res. 2012, 165, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Corbeil, S.; Williams, L.M.; Bergfeld, J.; Crane, M. St-J. Abalone herpesvirus stability in sea water and susceptibility to chemical disinfectants. Aquaculture 2012, 326–329, 20–26. [Google Scholar] [CrossRef]
- Corbeil, S.; Williams, L.M.; Kube, P.; King, H.; Elliot, N.; Crane, M.S.-J. Fisheries Research Development and Corporation Project—Final Report—011/003—Investigation into the Genetic Basis of Resistance to Infection by Abalone Herpes-Like Virus. 2013. Final Report Available upon Request: Project 2011-003. Available online: http://frdc.com.au (accessed on 1 September 2020).
- Crane, M.S.-J.; Corbeil, S.; William, L.M.; McColl, K.A.; Gannon, V. Evaluation of Abalone Viral Ganglioneuritis Resistance Among Wild Abalone Populations Along the Victorian Coast of Australia Source. J. Shellfish Res. 2013, 32, 67–72. [Google Scholar] [CrossRef]
- Corbeil, S.; William, L.M.; McColl, K.A.; Crane, M.S.-J. Australian abalone [Haliotis laevigata, H. rubra and H. conicorpora] are susceptible to infection by multiple abalone herpesvirus genotypes. Dis. Aquat. Org. 2016, 119, 101–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corbeil, S.; McColl, K.A.; William, L.M.; Slater, J.; Crane, M.S.-J. Innate resistance of New Zealand paua to abalone viral ganglioneuritis. J. Invertebr. Pathol. 2017, 146, 31–35. [Google Scholar] [CrossRef]
- Neave, M.J.; Corbeil, S.; McColl, K.A.; Crane, M.S.-J. Investigating the natural resistance of Haliotis iris [blackfoot pāua] to abalone viral ganglioneuritis using whole transcriptome analysis. Dis. Aquat. Org. 2019, 135, 107–119. [Google Scholar] [CrossRef]
- Hamid, R.; Khan, M.A.; Ahmad, M.; Ahmad, M.M.; Abdin, M.Z.; Musarrat, J.; Javed, S. Chitinases: An update. J. Pharm. Bioallied Sci. 2013, 5, 21–29. [Google Scholar]
- Sarker, S.; Ebrahimie, E.; Corbeil, S.; Helbig, K. In Proceedings of the abalone viral ganglioneuritis resistant päua [Haliotis iris] displays a highly divergent tissue transcriptome profile in respect to the susceptible Australian Haliotis rubra × H. In laevigata hybrid abalone. In Proceedings of the Australasian Virology Society Meeting, Queenstown, New Zealand, 2–5 December 2019. [Google Scholar]
- Cheng, T.C. Some cellular mechanisms governing self and nonself recognition and pathogenicity in vertebrates and invertebrates relative to protistan parasites. Aquaculture 1987, 67, 1–14. [Google Scholar] [CrossRef]
- Green, T.J.; Speck, P. Antiviral Defense and Innate Immune Memory in the Oyster. Viruses 2018, 10, 133. [Google Scholar] [CrossRef] [Green Version]
- Escoubas, J.M.; Gourbal, B.; Duval, D.; Green, T.J.; Charrière, G.M.; Destoumieux-Garzón, D.; Montagnani, C. Immunity in molluscs. In Encyclopedia of Immunobiology; Ratcliffe, M.J.H., Ed.; Academic Press: Oxford, UK, 2016; pp. 417–436. [Google Scholar]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef]
- Fritz, J.H.; Girardin, S.; Philpott, D.J. Innate immune defense through RNA interference. Sci. STKE 2006, 339, pe27. [Google Scholar] [CrossRef]
- Owens, L.; Malham, S. Review of the RNA Interference pathway in molluscs including some possibilities for use in bivalves in aquaculture. J. Mar. Sci. Eng. 2015, 3, 87–99. [Google Scholar] [CrossRef] [Green Version]
- Green, T.J.; Speck, P.; Geng, L.; Raftos, D.; Beard, M.R.; Helbig, K.J. Oyster viperin retains direct antiviral activity and its transcription occurs via a signalling pathway involving a heat-stable haemolymph protein. J. Gen. Virol. 2015, 96, 3587–3597. [Google Scholar] [CrossRef] [PubMed]
- Feng, D.; Qi, L.; Hong, Y. RNA Interference by Ingested dsRNA-Expressing Bacteria to Study Shell Biosynthesis and Pigmentation in Crassostrea gigas. Mar. Biotechnol. 2019, 21, 526–536. [Google Scholar] [CrossRef] [PubMed]
- Robalino, J.; Browdy, C.L.; Prior, S.; Metz, A.; Parnell, P.; Gross, P.S.; Warr, G.W. Induction of Antiviral Immunity by Double-Stranded RNA in a Marine Invertebrate. J. Virol. 2004, 78, 10442–10448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carrera-Naipil, C.; Valenzuela-Munoz, V.; Valdés, J.A.; Molina, A.; Gallardo-Escárate, C. RNA interference in Haliotis rufescens myostatin evidences upregulation of insulin signaling pathway. Agri Gene 2016, 1, 93–99. [Google Scholar] [CrossRef]
- He, C.; Yue, H.; Xu, L.; Liu, Y.; Song, Y.; Tang, C.; Yin, C. siRNA release kinetics from polymeric nanoparticles correlate with RNAi efficiency and inflammation therapy via oral delivery. Acta Biomater. 2020, 103, 213–222. [Google Scholar] [CrossRef]
- Lafont, M.; Petton, B.; Vergnes, A.; Pauletto, M.; Segarra, A. Long-lasting antiviral innate immune priming in the Lophotrochozoan Pacific oyster, Crassostrea gigas. Sci. Rep. 2017, 7, 13143. [Google Scholar] [CrossRef]
- Green, T.J.; Helbig, K.; Speck, P.; Raftos, D.A. Primed for success: Oyster parents treated with poly[I:C] produce offspring with enhanced protection against Ostreid herpesvirus type I infection. Mol. Immunol. 2016, 78, 113–120. [Google Scholar] [CrossRef]
- Green, T.J.; Montagnani, C. Poly I:C induces a protective antiviral immune response in the Pacific oyster [Crassostrea gigas] against subsequent challenge with Ostreid herpesvirus [OsHV-1 μvar]. Fish Shellfish Immunol. 2013, 35, 382–388. [Google Scholar] [CrossRef]
- Tetreau, G.; Dhinaut, J.; Gourbal, B.; Moret, Y. Trans-generational Immune Priming in Invertebrates: Current Knowledge and Future Prospects. Front. Immunol. 2019, 10, 1938. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Test | Analytical Sensitivity Viral Gene Copies (v.g.c.) | Characteristics |
|---|---|---|
| cPCR [23] | 2000 v.g.c. | DNA sequencing verifies target sequence and variation. |
| TaqMan ORF49 [25] | 20 v.g.c. | Fast, sensitive, specific. |
| TaqMan ORF66[25] | 20 v.g.c. | Fast, sensitive, specific. |
| TaqMan ORF77 [25] | 20 v.g.c. | Fast, sensitive, specific. |
| In situ hybridization [26] | Not evaluated | Visualization of DNA in infected tissues. |
| LAMP [27] | 20 v.g.c. | Fast, sensitive, specific, low cost, and field application. |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Corbeil, S. Abalone Viral Ganglioneuritis. Pathogens 2020, 9, 720. https://doi.org/10.3390/pathogens9090720
Corbeil S. Abalone Viral Ganglioneuritis. Pathogens. 2020; 9(9):720. https://doi.org/10.3390/pathogens9090720
Chicago/Turabian StyleCorbeil, Serge. 2020. "Abalone Viral Ganglioneuritis" Pathogens 9, no. 9: 720. https://doi.org/10.3390/pathogens9090720
APA StyleCorbeil, S. (2020). Abalone Viral Ganglioneuritis. Pathogens, 9(9), 720. https://doi.org/10.3390/pathogens9090720




