Cyclodepsipeptide Biosynthesis in Hypocreales Fungi and Sequence Divergence of The Non-Ribosomal Peptide Synthase Genes
Abstract
1. Introduction
2. Results and Discussion
2.1. Fungal Species Identification
2.2. Non-Ribosomal Peptide Synthetase Genes Divergence
2.3. In Vitro Cyclodepsipeptide Biosynthesis
3. Materials and Methods
3.1. Fungal Strains, Media and Growth Conditions
3.2. DNA Extraction, Molecular Identification, PCR Primers and DNA Sequencing
3.3. Sequence Analysis and Phylogeny Reconstruction
3.4. Mycotoxin Analyses
3.4.1. Chemicals
3.4.2. Extraction and purification
3.4.3. Liquid Chromatography Mass Spectrometry Analyses
3.4.4. Liquid Chromatography High-Resolution Mass Spectrometry (HRMS)
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jestoi, M. Emerging fusarium-mycotoxins fusaproliferin, beauvericin, enniatins, and moniliformin: A Review. Crit. Rev. Food Sci. Nutr. 2008, 48, 21–49. [Google Scholar] [CrossRef] [PubMed]
- Sivanathan, S.; Scherkenbeck, J. Cyclodepsipeptides: A rich source of biologically active compounds for drug research. Molecules 2014, 19, 12368–12420. [Google Scholar] [CrossRef]
- Urbaniak, M.; Stepien, L.; Uhlig, S. Evidence for naturally produced beauvericins containing N-Methyl-Tyrosine in hypocreales fungi. Toxins 2019, 11, 182. [Google Scholar] [CrossRef] [PubMed]
- Hornbogen, T.; Glinski, M.; Zocher, R. Biosynthesis of depsipeptide mycotoxins in fusarium. Eur. J. Plant. Pathol. 2002, 108, 713–718. [Google Scholar] [CrossRef]
- Stepien, L.; Jestoi, M.; Chelkowski, J. Cyclic hexadepsipeptides in wheat field samples and Esyn1 gene divergence among enniatin producing fusarium avenaceum strains. World Mycotoxin J. 2013, 6, 399–409. [Google Scholar] [CrossRef]
- Logrieco, A.; Moretti, A.; Castella, G.; Kostecki, M.; Golinski, P.; Ritieni, A.; Chelkowski, J. Beauvericin production by fusarium species. Appl. Environ. Microbiol. 1998, 64, 3084–3088. [Google Scholar] [CrossRef]
- Covarelli, L.; Beccari, G.; Prodi, A.; Generotti, S.; Etruschi, F.; Meca, G.; Juan, C.; Manes, J. Biosynthesis of beauvericin and enniatins in vitro by wheat fusarium species and natural grain contamination in an area of central Italy. Food Microbiol. 2015, 46, 618–626. [Google Scholar] [CrossRef]
- Weng, Q.; Zhang, X.; Chen, W.; Hu, Q. Secondary metabolites and the risks of isaria fumosorosea and isaria farinosa. Molecules 2019, 24, 664. [Google Scholar] [CrossRef]
- Luangsa-Ard, J.; Berkaew, P.; Ridkaew, R.; Hywel-Jones, N.L.; Isaka, M. A Beauvericin Hot spot in the genus isaria. Mycol. Res. 2009, 113, 1389–1395. [Google Scholar] [CrossRef]
- Bunyapaiboonsri, T.; Vongvilai, P.; Auncharoen, P.; Isaka, M. Cyclohexadepsipeptides from the filamentous fungus acremonium sp. Bcc 2629. Helv. Chim. Acta 2012, 95, 963–972. [Google Scholar] [CrossRef]
- Fukuda, T.; Arai, M.; Tomoda, H.; Omura, S. New beauvericins, potentiators of antifungal miconazole activity, produced by beauveria sp Fki-1366. II. structure elucidation. J. Antibiot. 2004, 57, 117–124. [Google Scholar] [CrossRef]
- Nilanonta, C.; Isaka, M.; Kittakoop, P.; Trakulnaleamsai, S.; Tanticharoen, M.; Thebtaranonth, Y. Precursor-directed biosynthesis of beauvericin analogs by the insect pathogenic fungus paecilomyces tenuipes Bcc 1614. Tetrahedron 2002, 58, 3355–3360. [Google Scholar] [CrossRef]
- Xu, Y.; Zhan, J.; Wijeratne, E.M.K.; Burns, A.M.; Gunatilaka, A.A.L.; Molnar, I. Cytotoxic and antihaptotactic beauvericin analogues from precursor-directed biosynthesis with the insect pathogen beauveria bassiana atcc 7159. J. Nat. Prod. 2007, 70, 1467–1471. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, L.; Skjerve, E.; Eriksen, G.S.; Uhlig, S. Cytotoxicity of enniatins a, A1, B, B1, B2 and B3 from fusarium avenaceum. Toxicon 2006, 47, 868–876. [Google Scholar] [CrossRef]
- Uhlig, S.; Ivanova, L. Determination of beauvericin and four other enniatins in grain by liquid chromatography-mass spectrometry. J. Chromatogr. A 2004, 1050, 173–178. [Google Scholar] [CrossRef]
- Song, H.H.; Lee, H.S.; Jeong, J.H.; Park, H.S.; Lee, C. Diversity in beauvericin and enniatins H, I, and Mk1688 by fusarium oxysporum isolated from potato. Int. J. Food Microbiol. 2008, 122, 296–301. [Google Scholar] [CrossRef] [PubMed]
- Chełkowski, J.; Ritieni, A.; Wiśniewska, H.; Mulè, G.; Logrieco, A. Occurrence of toxic hexadepsipeptides in preharvest maize ear rot infected by fusarium poae in Poland. J. Phytopathol. 2006, 155, 8–12. [Google Scholar] [CrossRef]
- Jow, G.M.; Chou, C.J.; Chen, B.F.; Tsai, J.H. Beauvericin induces cytotoxic effects in human acute lymphoblastic leukemia cells through cytochrome C release, caspase 3 activation: The causative role of calcium. Cancer Lett. 2004, 216, 165–173. [Google Scholar] [CrossRef]
- Shin, C.G.; An, D.G.; Song, H.H.; Lee, C. Beauvericin and enniatins H, I and Mk1688 are new potent inhibitors of human immunodeficiency virus type-1 integrase. J. Antibiot. 2009, 62, 687–690. [Google Scholar] [CrossRef]
- Dornetshuber, R.; Heffeter, P.; Kamyar, M.R.; Peterbauer, T.; Berger, W.; Lemmens-Gruber, R. Enniatin exerts p53-dependent cytostatic and p53-independent cytotoxic activities against human cancer cells. Chem. Res. Toxicol. 2007, 20, 465–473. [Google Scholar] [CrossRef]
- Watjen, W.; Debbab, A.; Hohlfeld, A.; Chovolou, Y.; Kampkotter, A.; Edrada, R.A.; Ebel, R.; Hakiki, A.; Mosaddak, M.; Totzke, F.; et al. Enniatins A1, B and B1 from an endophytic strain of fusarium tricinctum induce apoptotic cell death in H4IIE hepatoma cells accompanied by inhibition of ERK phosphorylation. Mol. Nutr. Food Res. 2009, 53, 431–440. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.F.; Tsai, M.C.; Jow, G.M. Induction of calcium influx from extracellular fluid by beauvericin in human leukemia cells. Biochem. Biophys. Res. Commun. 2006, 340, 134–139. [Google Scholar] [CrossRef] [PubMed]
- Kamyar, M.; Rawnduzi, P.; Studenik, C.R.; Kouri, K.; Lemmens-Gruber, R. Investigation of the electrophysiological properties of enniatins. Arch. Biochem. Biophys. 2004, 429, 215–223. [Google Scholar] [CrossRef]
- Fukuda, T.; Arai, M.; Yamaguchi, Y.; Masuma, R.; Tomoda, H.; Omura, S. New beauvericins, potentiators of antifungal miconazole activity, produced by beauveria sp FKI-1366. I. taxonomy, fermentation, Isolation and biological properties. J. Antibiot. 2004, 57, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.X.; Yan, K.Z.; Zhang, Y.; Huang, R.; Bian, J.; Zheng, C.S.; Sun, H.X.; Chen, Z.H.; Sun, N.; An, R.; et al. High-Throughput synergy screening identifies microbial metabolites as combination agents for the treatment of fungal infections. Proc. Natl. Acad. Sci. USA 2007, 104, 4606–4611. [Google Scholar] [CrossRef] [PubMed]
- Supothina, S.; Isaka, M.; Kirtikara, K.; Tanticharoen, M.; Thebtaranonth, Y. Enniatin production by the entomopathogenic fungus verticillium hemipterigenum BCC 1449. J. Antibiot. 2004, 57, 732–738. [Google Scholar] [CrossRef]
- Lin, H.I.; Lee, Y.J.; Chen, B.F.; Tsai, M.C.; Lu, J.L.; Chou, C.J.; Jow, G.M. Involvement of Bcl-2 family, cytochrome C and caspase 3 in induction of apoptosis by beauvericin in human non-small cell lung Cancer cells. Cancer Lett. 2005, 230, 248–259. [Google Scholar] [CrossRef]
- Galvez, L.; Urbaniak, M.; Waskiewicz, A.; Stepien, L.; Palmero, D. fusarium proliferatum–Causal agent of garlic bulb rot in Spain: Genetic variability and mycotoxin production. Food Microbiol. 2017, 67, 41–48. [Google Scholar] [CrossRef]
- Jestoi, M.; Rokka, M.; Yli-Mattila, T.; Parikka, P.; Rizzo, A.; Peltonen, K. Presence and concentrations of the fusarium-related mycotoxins beauvericin, enniatins and moniliformin in finnish grain samples. Food Addit. Contam. 2004, 21, 794–802. [Google Scholar] [CrossRef]
- Logrieco, A.; Rizzo, A.; Ferracane, R.; Ritieni, A. Occurrence of beauvericin and enniatins in wheat affected by fusarium avenaceum head blight. Appl. Environ. Microbiol. 2002, 68, 82–85. [Google Scholar] [CrossRef]
- Bushley, K.E.; Turgeon, B.G. Phylogenomics reveals subfamilies of fungal nonribosomal peptide synthetases and their evolutionary relationships. BMC Evol. Biol. 2010, 10, 26. [Google Scholar] [CrossRef]
- Gallo, A.; Ferrara, M.; Perrone, G. Phylogenetic study of polyketide synthases and nonribosomal peptide synthetases involved in the biosynthesis of mycotoxins. Toxins 2013, 5, 717–742. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Orozco, R.; Wijeratne, E.M.; Gunatilaka, A.A.; Stock, S.P.; Molnar, I. Biosynthesis of the cyclooligomer depsipeptide beauvericin, a virulence factor of the entomopathogenic fungus beauveria bassiana. Chem. Biol. 2008, 15, 898–907. [Google Scholar] [CrossRef]
- Zhang, T.; Zhuo, Y.; Jia, X.; Liu, J.; Gao, H.; Song, F.; Liu, M.; Zhang, L. Cloning and characterization of the gene cluster required for beauvericin biosynthesis in fusarium proliferatum. Sci. China Life Sci. 2013, 56, 628–637. [Google Scholar] [CrossRef]
- Zocher, R.; Keller, U.; Kleinkauf, H. Enniatin synthetase, a novel type of multifunctional enzyme catalyzing depsipeptide synthesis in fusarium oxysporum. Biochemistry 1982, 21, 43–48. [Google Scholar] [CrossRef]
- Billich, A.; Zocher, R. Constitutive expression of enniatin synthetase during fermentative growth of fusarium scirpi. Appl. Environ. Microbiol. 1988, 54, 2504–2509. [Google Scholar] [CrossRef] [PubMed]
- Hornbogen, T.; Riechers, S.P.; Prinz, B.; Schultchen, J.; Lang, C.; Schmidt, S.; Mugge, C.; Turkanovic, S.; Sussmuth, R.D.; Tauberger, E.; et al. Functional characterization of the recombinant N-Methyltransferase domain from the multienzyme enniatin synthetase. ChemBioChem 2007, 8, 1048–1054. [Google Scholar] [CrossRef] [PubMed]
- Glinski, M.; Urbanke, C.; Hornbogen, T.; Zocher, R. Enniatin synthetase is a monomer with extended structure: Evidence for an intramolecular reaction mechanism. Arch. Microbiol. 2002, 178, 267–273. [Google Scholar] [CrossRef]
- Zocher, R.; Keller, U.; Kleinkauf, H. Mechanism of depsipeptide formation catalyzed by enniatin synthetase. Biochem. Biophys. Res. Commun. 1983, 110, 292–299. [Google Scholar] [CrossRef]
- Haese, A.; Schubert, M.; Herrmann, M.; Zocher, R. Molecular characterization of the enniatin synthetase gene encoding a multifunctional enzyme catalysing N-Methyldepsipeptide formation in fusarium scirpi. Mol. Microbiol. 1993, 7, 905–914. [Google Scholar] [CrossRef]
- Peeters, H.; Zocher, R.; Kleinkauf, H. Synthesis of beauvericin by a multifunctional enzyme. J. Antibiot. 1988, 41, 352–359. [Google Scholar] [CrossRef]
- Stepien, L.; Waskiewicz, A. Sequence divergence of the enniatin synthase gene in relation to production of beauvericin and enniatins in fusarium Species. Toxins 2013, 5, 537–555. [Google Scholar] [CrossRef]
- Gallou, A.; Serna-Dominguez, M.G.; Berlanga-Padilla, A.M.; Ayala-Zermeno, M.A.; Mellin-Rosas, M.A.; Montesinos-Matias, R.; Arredondo-Bernal, H.C. Species clarification of isaria isolates used as biocontrol agents against diaphorina citri (hemiptera: Liviidae) in Mexico. Fungal Biol. 2016, 120, 414–423. [Google Scholar] [CrossRef]
- Zhang, J.; Fu, B.; Lin, Q.; Riley, I.T.; Ding, S.; Chen, L.; Cui, J.; Yang, L.; Li, H. Colonization of beauveria bassiana 08f04 in root-zone soil and its biocontrol of cereal cyst nematode (heterodera filipjevi). PLoS ONE 2020, 15, e0232770. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.J.; Qiu, L.; Chu, Z.J.; Ying, S.H.; Feng, M.G. The connection of protein O-mannosyltransferase family to the biocontrol potential of beauveria bassiana, a fungal entomopathogen. Glycobiology 2014, 24, 638–648. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yang, H.; Qin, C.S.; Chen, Y.M.; Zhang, G.Y.; Dong, L.H.; Wan, S.Q. Persistence of metarhizium (hypocreales: Clavicipitaceae) and beauveria bassiana (hypocreales: Clavicipitaceae) in Tobacco Soils and Potential as Biocontrol Agents of Spodoptera litura (lepidoptera: Noctuidae). Environ. Entomol. 2019, 48, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Bhadauria, B.P.; Singh, P.K.; Shailesh, P.; Zaidi, N.W.; Singh, U.S. Characterization and biocontrol potential of entomopathogenic fungus, beauveria bassiana isolates against spilarctia obliqua. J. Environ. Biol. 2013, 34, 917–921. [Google Scholar] [PubMed]
- Batta, Y.A. Biocontrol of almond bark beetle (scolytus amygdali geurin-meneville, coleoptera: Scolytidae) using beauveria bassiana (Bals.) vuill. (deuteromycotina: Hyphomycetes). J. Appl. Microbiol. 2007, 103, 1406–1414. [Google Scholar] [CrossRef] [PubMed]
- Tomczyk, L.; Stepien, L.; Urbaniak, M.; Szablewski, T.; Cegielska-Radziejewska, R.; Stuper-Szablewska, K. Characterisation of the mycobiota on the shell surface of table eggs acquired from different egg-laying hen breeding systems. Toxins 2018, 10, 293. [Google Scholar] [CrossRef]
- Kozlowska, E.; Hoc, N.; Sycz, J.; Urbaniak, M.; Dymarska, M.; Grzeszczuk, J.; Kostrzewa-Suslow, E.; Stepien, L.; Plaskowska, E.; Janeczko, T. Biotransformation of steroids by entomopathogenic strains of isaria farinosa. Microb. Cell Factories 2018, 17, 71. [Google Scholar] [CrossRef]
- Kozlowska, E.; Urbaniak, M.; Hoc, N.; Grzeszczuk, J.; Dymarska, M.; Stepien, L.; Plaskowska, E.; Kostrzewa-Suslow, E.; Janeczko, T. Cascade biotransformation of dehydroepiandrosterone (DHEA) by beauveria Species. Sci. Rep. 2018, 8, 13449. [Google Scholar] [CrossRef] [PubMed]
- Stepien, L.; Koczyk, G.; Waskiewicz, A. Diversity of fusarium species and mycotoxins contaminating pineapple. J. Appl. Genet. 2013, 54, 367–380. [Google Scholar] [CrossRef]
- Stepien, L.; Waskiewicz, A.; Urbaniak, M. Wildly growing asparagus (Asparagus officinalis L.) hosts pathogenic fusarium species and accumulates their mycotoxins. Microb. Ecol. 2016, 71, 927–937. [Google Scholar] [CrossRef]
- Von Bargen, S.; Martinez, O.; Schadock, I.; Eisold, A.M.; Gossmann, M.; Buttner, C. Genetic variability of phytopathogenic fusarium proliferatum associated with crown rot in asparagus officinalis. J. Phytopathol. 2009, 157, 446–456. [Google Scholar] [CrossRef]
- Xu, L.; Li, Y.; Biggins, J.B.; Bowman, B.R.; Verdine, G.L.; Gloer, J.B.; Alspaugh, J.A.; Bills, G.F. Identification of cyclosporin C from amphichorda felina using a cryptococcus neoformans differential temperature sensitivity assay. Appl. Microbiol. Biotechnol. 2018, 102, 2337–2350. [Google Scholar] [CrossRef] [PubMed]
- Kepler, R.M.; Luangsa-Ard, J.J.; Hywel-Jones, N.L.; Quandt, C.A.; Sung, G.H.; Rehner, S.A.; Aime, M.C.; Henkel, T.W.; Sanjuan, T.; Zare, R.; et al. A phylogenetically-based nomenclature for cordycipitaceae (hypocreales). IMA Fungus 2017, 8, 335–353. [Google Scholar] [CrossRef]
- Mongkolsamrit, S.; Noisripoom, W.; Thanakitpipattana, D.; Wutikhun, T.; Spatafora, J.W.; Luangsa-Ard, J. Disentangling cryptic species with isaria-like morphs in cordycipitaceae. Mycologia 2018, 110, 230–257. [Google Scholar] [CrossRef]
- Xu, L.J.; Liu, Y.S.; Zhou, L.G.; Wu, J.Y. Enhanced beauvericin production with in situ adsorption in mycelial liquid culture of fusarium redolens Dzf2. Process. Biochem. 2009, 44, 1063–1067. [Google Scholar] [CrossRef]
- Decleer, M.; Landschoot, S.; De Saeger, S.; Rajkovic, A.; Audenaert, K. Impact of fungicides and weather on cyclodepsipeptide-producing fusarium spp. and beauvericin and enniatin levels in wheat grains. J. Sci. Food Agric. 2019, 99, 253–262. [Google Scholar] [CrossRef]
- Song, H.H.; Ahn, J.H.; Lim, Y.H.; Lee, C. Analysis of beauvericin and unusual enniatins co-produced by fusarium oxysporum Fb1501 (Kfcc 11363p). J. Microbiol. Biotechn. 2006, 16, 1111–1119. [Google Scholar]
- Gorczyca, A.; Oleksy, A.; Gala-Czekaj, D.; Urbaniak, M.; Laskowska, M.; Waskiewicz, A.; Stepien, L. fusarium head blight incidence and mycotoxin accumulation in three durum wheat cultivars in relation to sowing date and density. Sci. Nat. 2018, 105. [Google Scholar] [CrossRef] [PubMed]
- Carbone, I.; Kohn, L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetic. PCR Protoc. Guide Methods Appl. 1990, 5, 315–322. [Google Scholar]
- Gouy, M.; Guindon, S.; Gascuel, O. Seaview version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010, 27, 221–224. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. Iq-Tree: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Stanciu, O.; Juan, C.; Miere, D.; Loghin, F.; Manes, J. Analysis of enniatins and beauvericin by LC-MS/MS in wheat-based products. CyTA–J. Food 2017, 15, 433–440. [Google Scholar] [CrossRef]
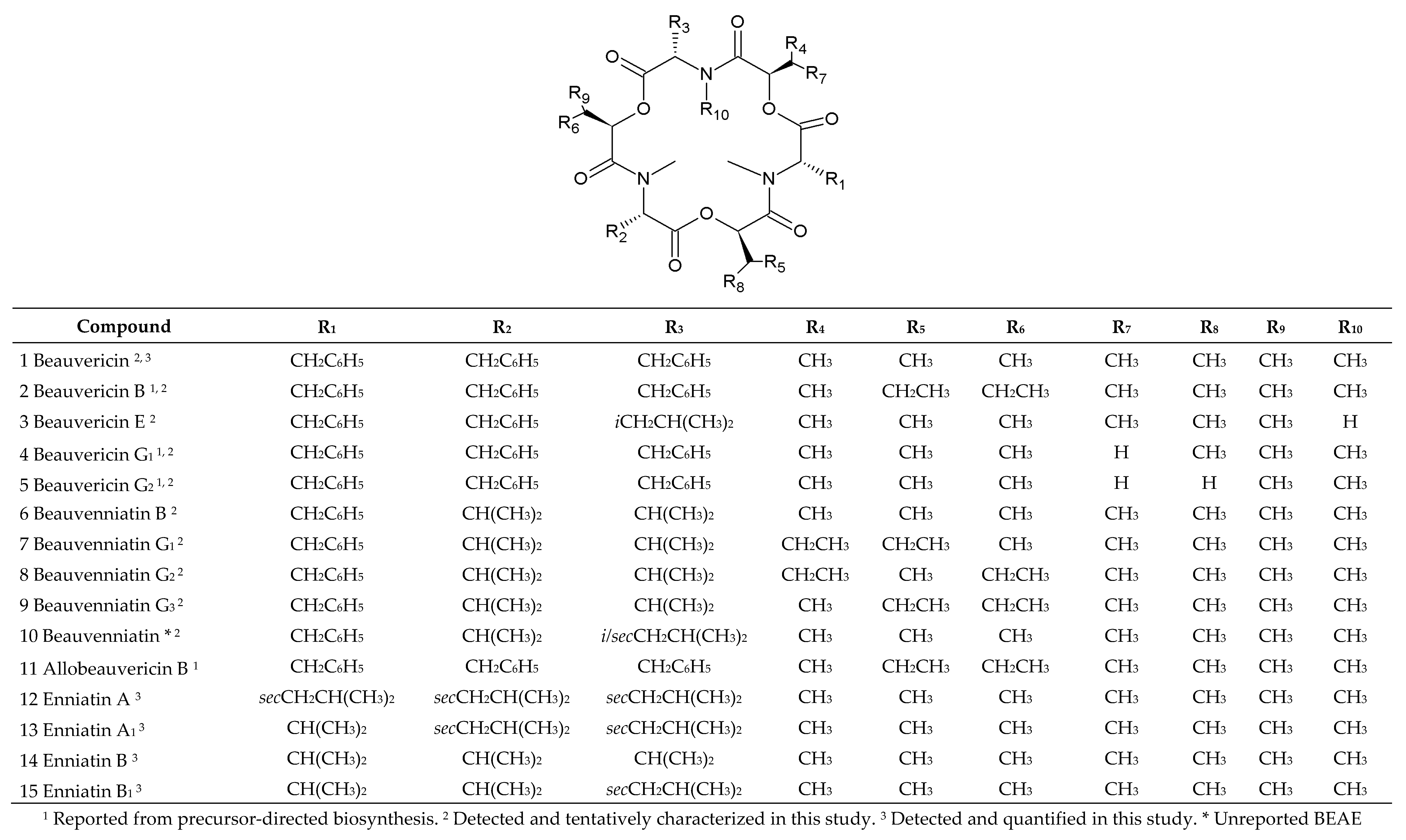
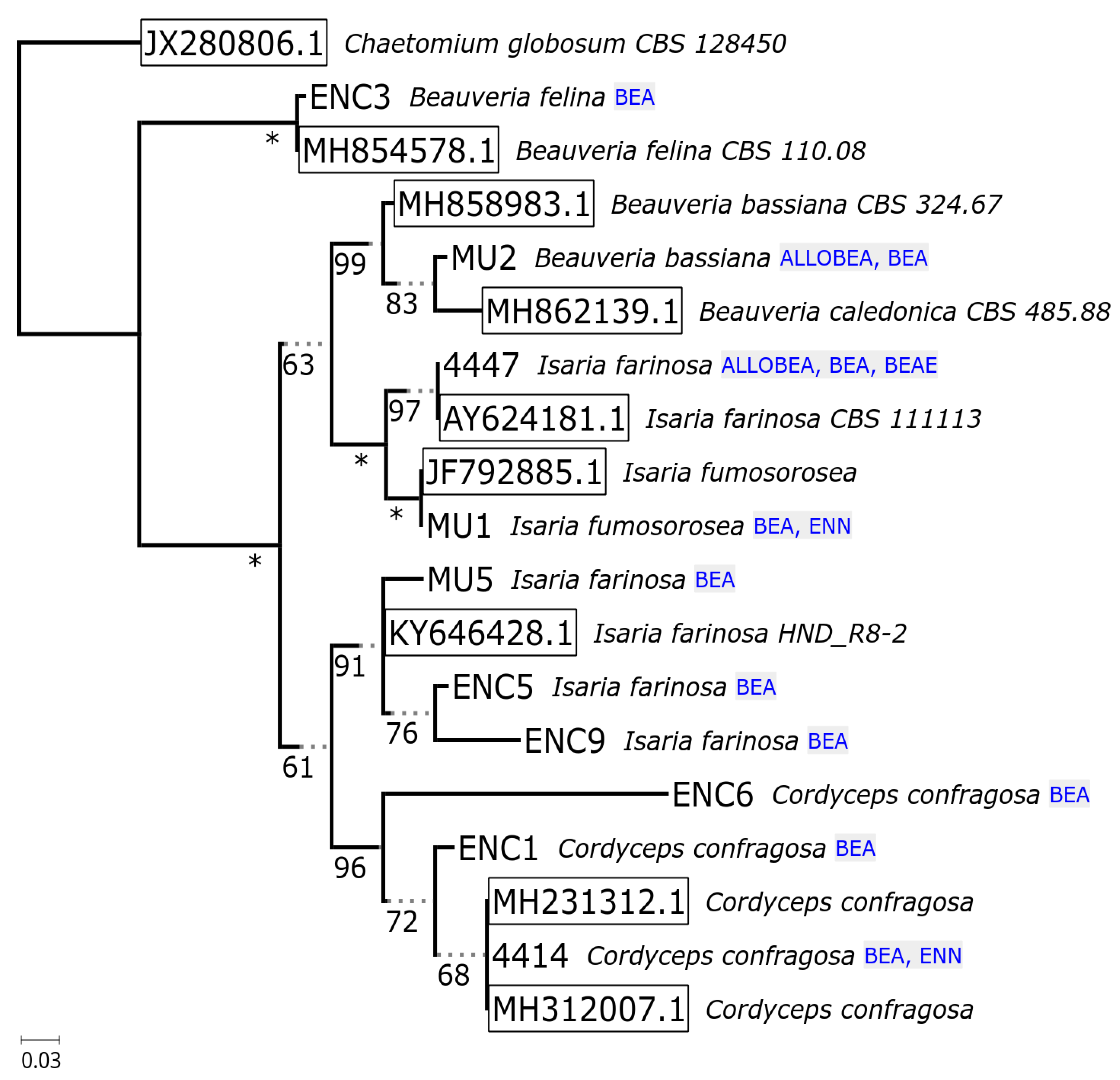
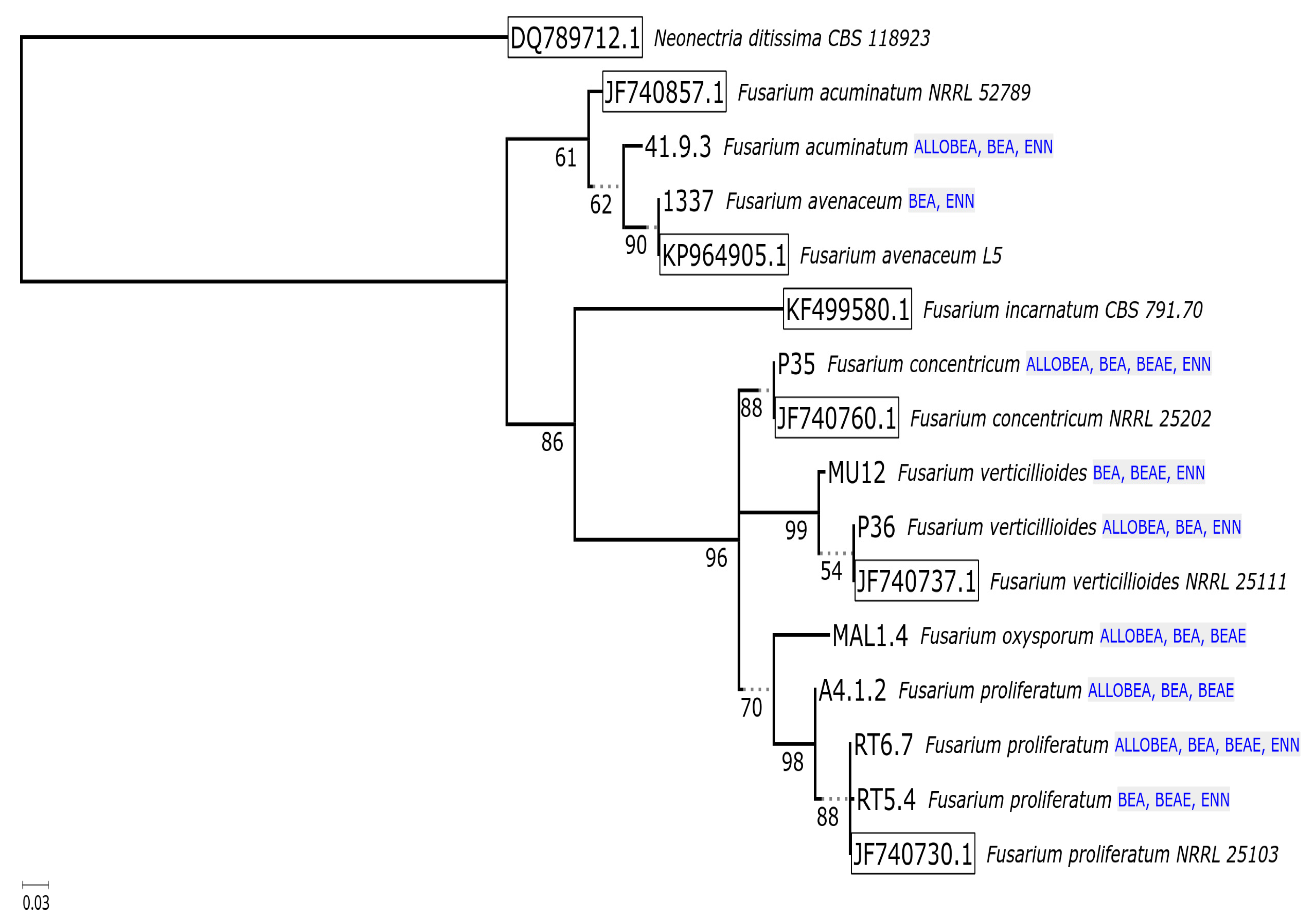
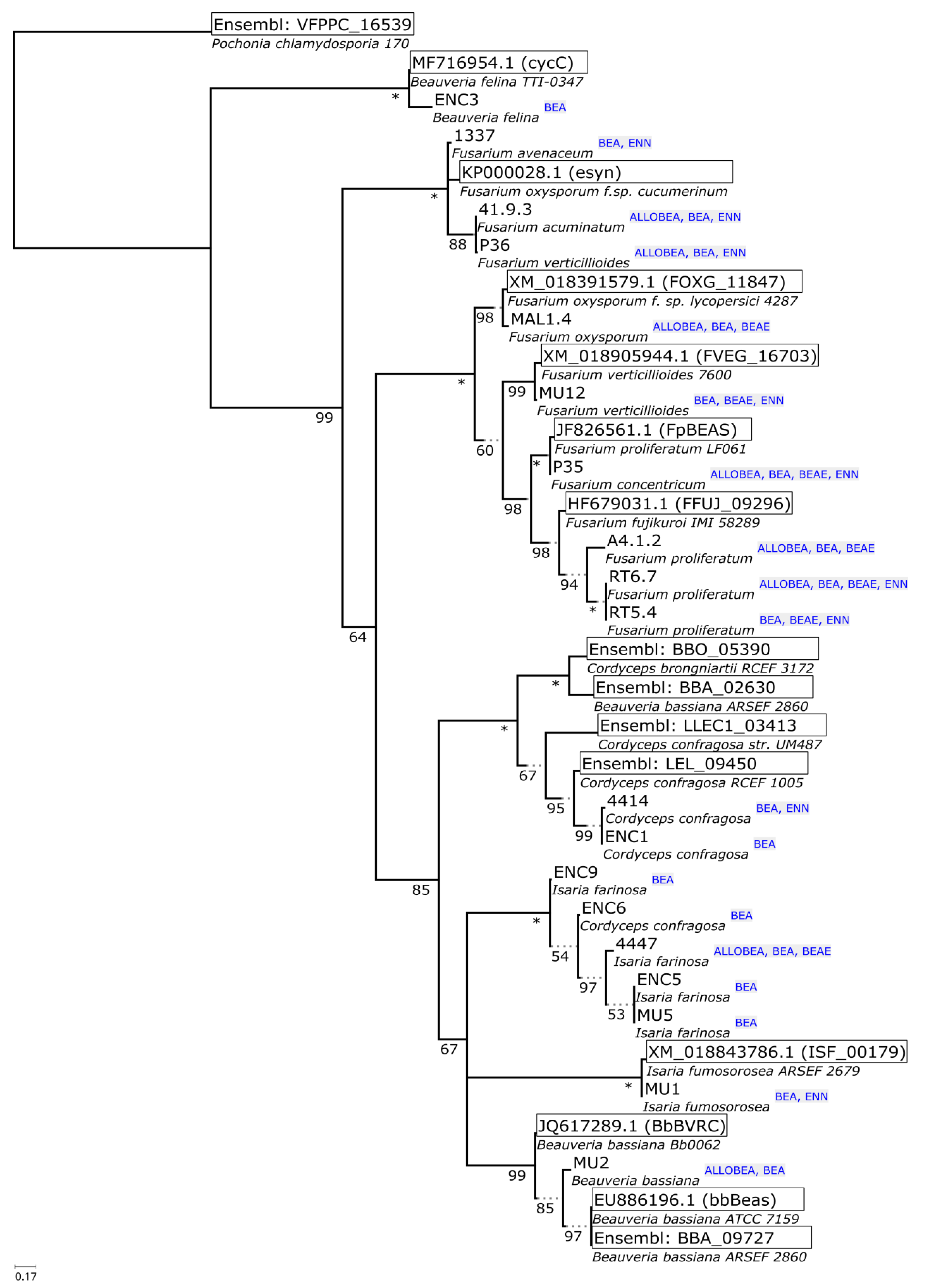
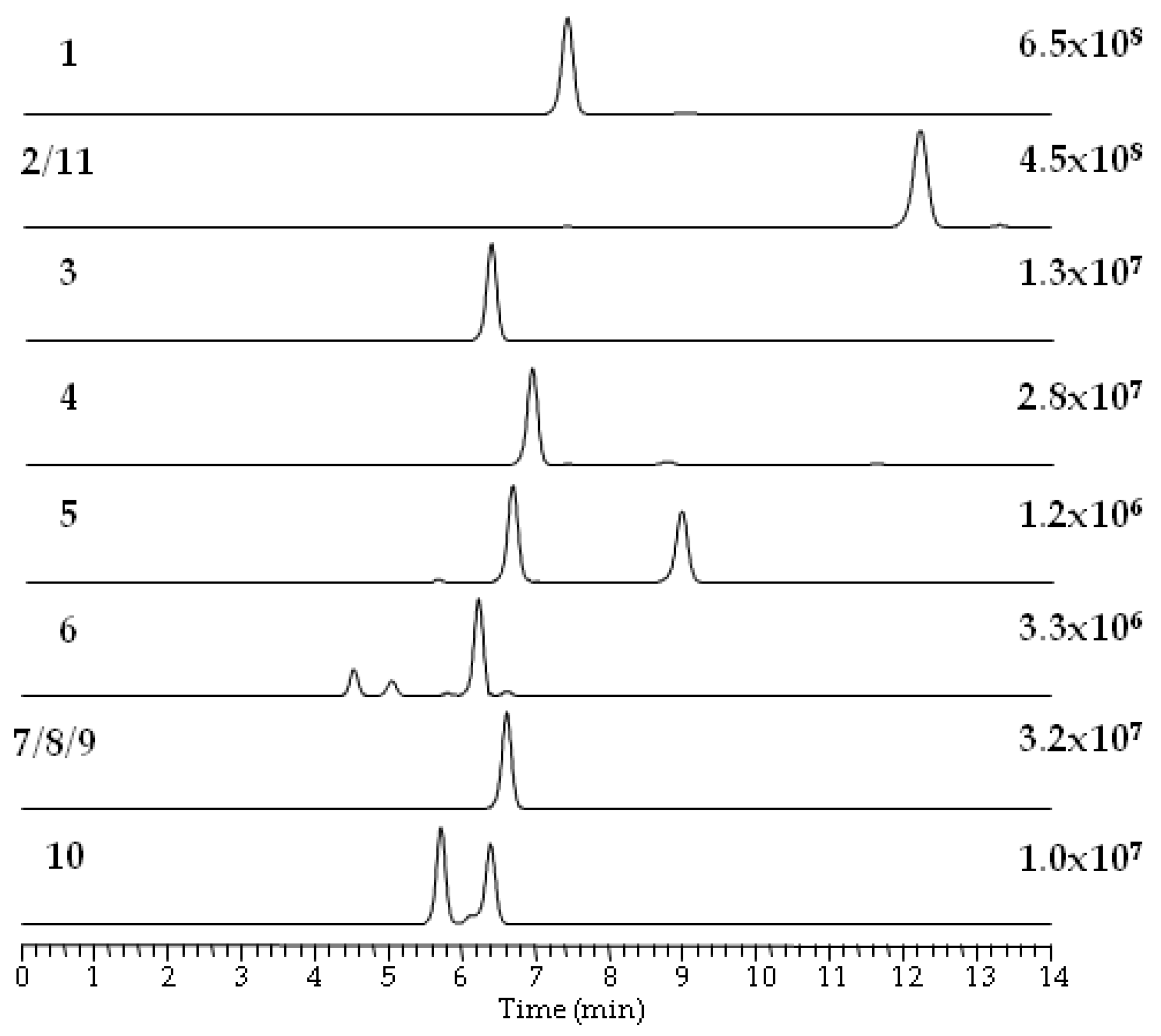
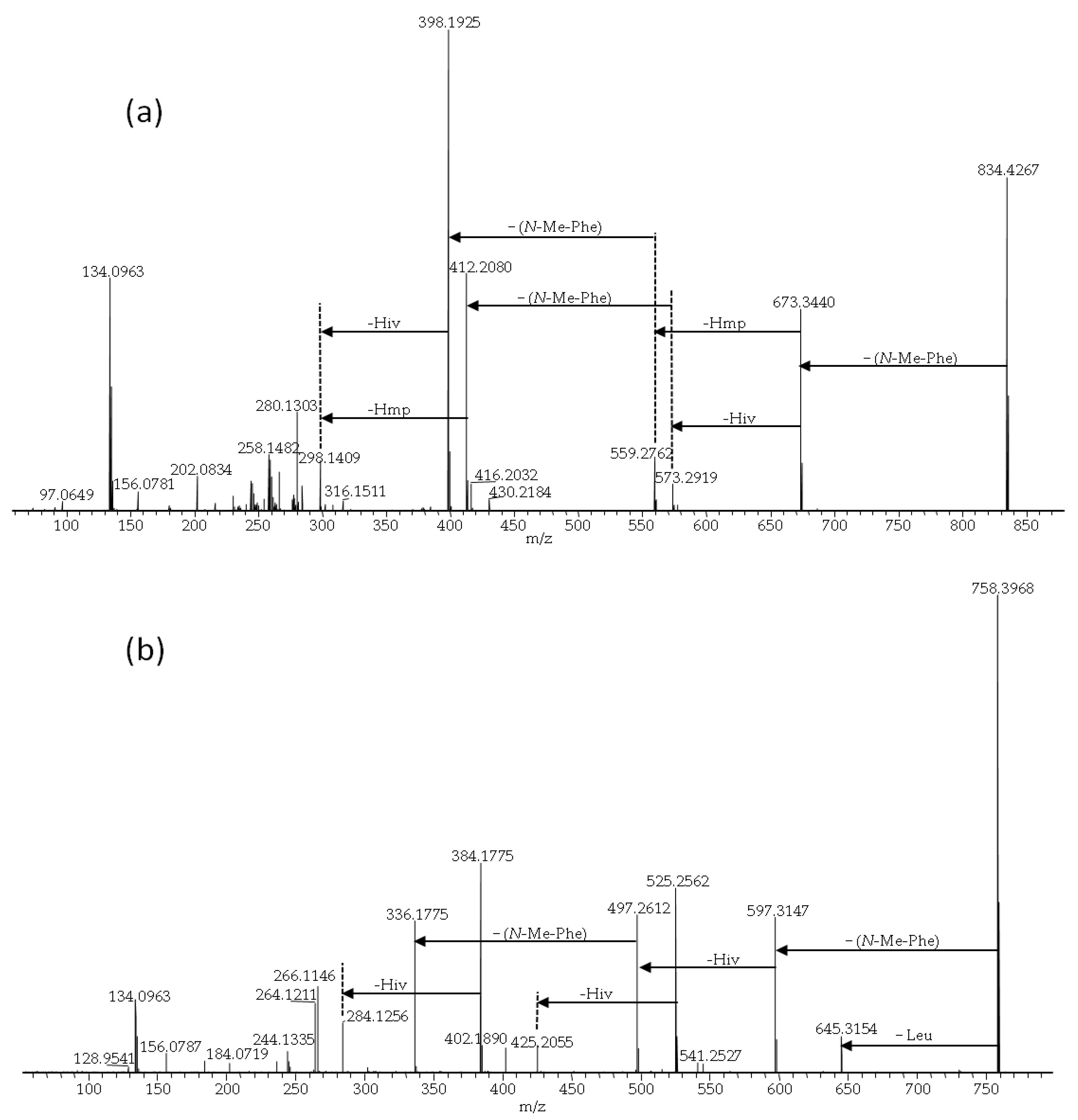
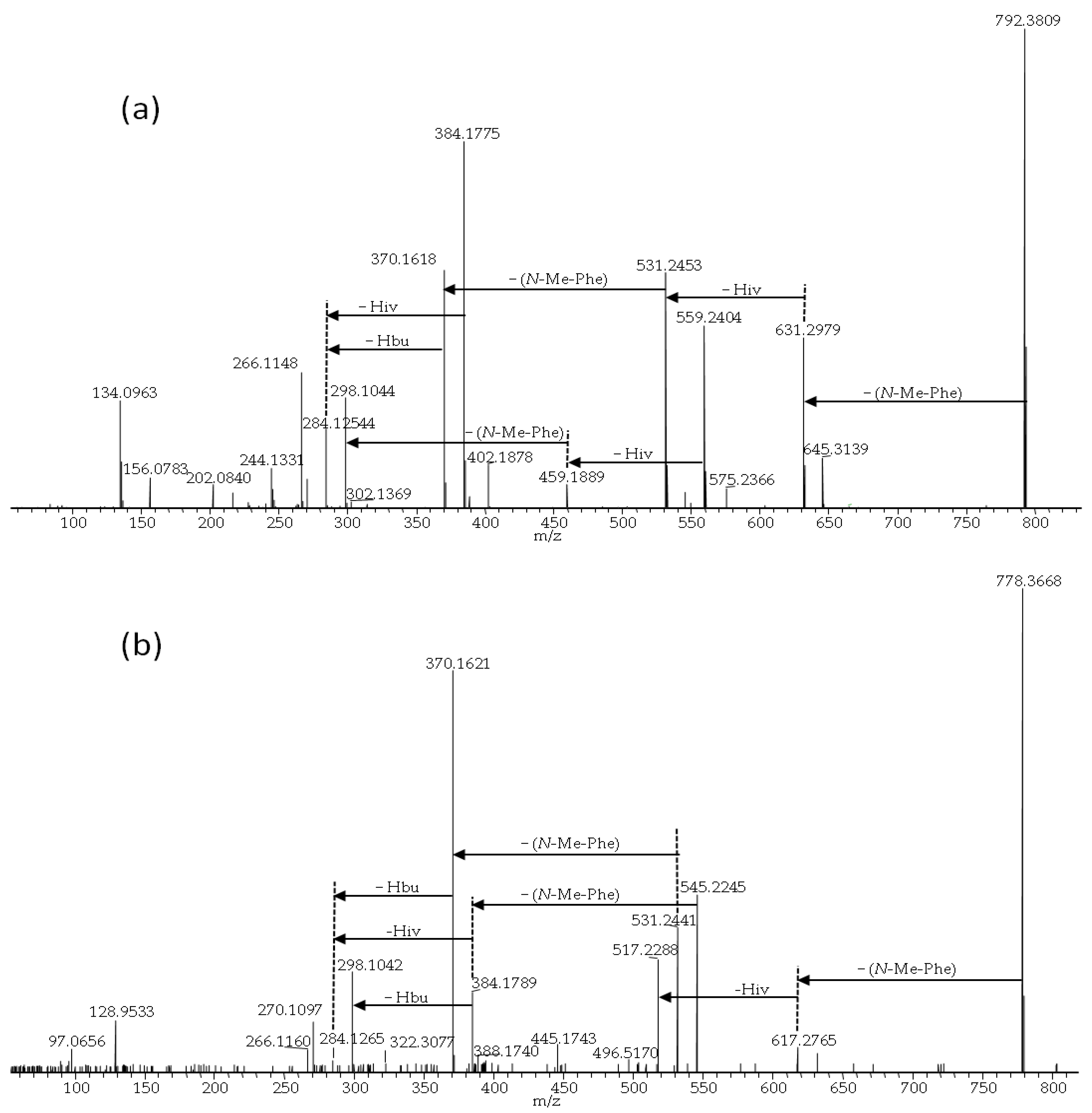

| No. | Strain | Species | Host | Sequence Nucleotide Identity |
|---|---|---|---|---|
| 1 | 1337 | Fusarium avenaceum | wheat | 99.76% identity to the Fusarium avenaceum acc. number KP964905.1 |
| (Triticum L.) | ||||
| 2 | MU2 | Beauveria bassiana | bark beetle | 100% identity to the Beauveria bassiana acc. number KU158425.1 |
| (Trypodendron lineatum) | ||||
| 3 | ENC3 | Beauveria felina | caterpillar | 98.37% identity to the Beauveria felina acc. number MH854578.1 |
| (Lepidoptera sp.) | ||||
| 4 | 15222 | Fusarium acuminatum | pea | 98.19% identity to the Fusarium acuminatum acc. number JF740857.1 |
| (Pisum L.) | ||||
| 5 | P35 | Fusarium concentricum | pineapple | 100% identity to the Fusarium concentricum, acc. number JF740760.1 |
| (Ananas comosus) | ||||
| 6 | RT6.7 | Fusarium proliferatum | rice | 99.78% identity to the Fusarium proliferatum acc. number JF740730.1 |
| (Oryza sativa) | ||||
| 7 | RT5.4 | Fusarium proliferatum | rice | 99.78% identity to the Fusarium proliferatum acc. number JF740730.1 |
| (Oryza sativa) | ||||
| 8 | MU12 | Fusarium verticillioides | banana | 98.66% identity to the Fusarium verticillioides acc. number JF740717.1 |
| (Musa L.) | ||||
| 9 | P36 | Fusarium verticillioides | pineapple | 100% identity to the Fusarium verticillioides acc. number JF740717.1 |
| (Ananas comosus) | ||||
| 10 | 4447 | Isaria farinosa | bark beetle | 100% identity to the Isaria farinosa, acc. number AY624181.1 |
| (Trypodendron lineatum) | ||||
| 11 | ENC5 | Isaria farinosa | bark beetle | 98.92% identity to the Isaria farinosa, acc. number KY646428.1 |
| (Trypodendron lineatum) | ||||
| 12 | ENC9 | Isaria farinosa | beetle | 98.89% identity to the Isaria farinosa, acc. number DQ888729.1 |
| (Coleoptera sp.) | ||||
| 13 | MU5 | Isaria farinosa | pine beetle | 99.53% identity to the Isaria farinosa, acc. number KY646428.1 |
| (Dendroctonus ponderosae) | ||||
| 14 | MU1 | Isaria fumosorosea | beetle | 98.38% identity to the Isaria fumosorosea, acc. number JF792885.1 |
| (Coleoptera sp.) | ||||
| 15 | 4414 | Cordyceps confragosa | parent bug | 100% identity to the Cordyceps confragosa, acc. number MH231312.1 |
| (Elasmucha sp.) | ||||
| 16 | ENC1 | Cordyceps confragosa | bark beetle | 98.08% identity to the Cordyceps confragosa, acc. number MH312006.1 |
| (Trypodendron lineatum) | ||||
| 17 | ENC6 | Cordyceps confragosa | caterpillar | 99.82% identity to the Cordyceps confragosa, acc. number MH312006.1 |
| (Lepidoptera sp.) | ||||
| 18 | MAL 1.4 | Fusarium oxysporum | asparagus | 100% identity to the Fusarium oxysporum acc. number KP964890.1 |
| (Asparagus officinalis L.) | ||||
| 19 | A 4.12 | Fusarium proliferatum | wheat | 100% identity to the Fusarium proliferatum acc. number KU939029.1 |
| (Triticum L.) |
| Strain | BEA [ng/g] | ENN A [ng/g] | ENN A1 [ng/g] | ENN B [ng/g] | ENN B1 [ng/g] |
|---|---|---|---|---|---|
| 1337 (Fav) | 49.3 ± 5.6 | 4300 ± 160 | 63800 ± 2430 | ND | ND |
| MU2 (Bb) | 389 ± 22.4 | ND | ND | ND | ND |
| ENC3 (Bf) | 211 ± 12 | ND | ND | ND | ND |
| 41/9/3 (Fac) | 150 ± 10 | 56600 ± 1960 | 68970 ± 2535 | ND | ND |
| P35 (Fco) | 130400 ± 2300 | 127 ± 20 | ND | 350 ± 37 | ND |
| RT6.7 (Fpr) | 135540 ± 3225 | ND | ND | 1983 ± 37.4 | ND |
| RT5.4 (Fpr) | 55420 ± 411 | ND | ND | 2130 ± 130 | ND |
| MU12 (Fve) | 125 ± 4.3 | ND | ND | 191 ± 12.7 | 51 ± 4.5 |
| P36 (Fve) | 150 ± 8.3 | ND | ND | 23 ± 2.6 | ND |
| 4447 (Ifa) | 10240 ± 246 | ND | ND | ND | ND |
| ENC5 (Ifa) | 215 ± 15 | ND | ND | ND | ND |
| ENC9 (Ifa) | 352 ± 9.6 | ND | ND | ND | ND |
| MU5 (Ifa) | 155 ± 9.3 | ND | ND | ND | ND |
| MU1 (Ifu) | 900 ± 26.2 | ND | ND | 720 ± 23.4 | 71 ± 5 |
| 4414 (Cco) | 125 ± 15.7 | 5730 ± 157 | ND | ND | 9130 ± 128 |
| ENC1 (Cco) | 128±12 | ND | ND | ND | ND |
| ENC6 (Cco) | 115 ± 13 | ND | ND | ND | ND |
| MAL 1.4 (Fox) | 433 ± 11.3 | ND | ND | ND | ND |
| A 4.12 (Fpr) | 730 ± 60 | ND | ND | ND | ND |
| No. | Strain | Species | Metabolic Profile |
|---|---|---|---|
| 1 | 1337 | Fusarium avenaceum | BEA, ENN A, ENN A1 |
| 2 | MU2 | Beauveria bassiana | BEA, BEA B, BEA C, BEA F/A, ALLOBEA A, ALLOBEA B, ALLOBEA C |
| 3 | ENC3 | Beauveria felina | BEA, |
| 4 | 15222 | Fusarium acuminatum | BEA, BEA C, BEA D, BEA G1, ALLOBEA C, ENN A, ENN A1 |
| 5 | P35 | Fusarium concentricum | BEA, BEA B, BEA C, BEA D, BEA E, BEA F/A, BEA J, BEA G1, BEA G2, BEAE A, BEAE B, BEAE G1/G2/G3, BEAE *, ALLOBEA A, ALLOBEA B, ALLOBEA C, BEAE L, BEA K, BEA L, ENN A, ENN B |
| 6 | RT6.7 | Fusarium proliferatum | BEA, BEA B, BEA C, BEA D, BEA E, BEA F/A, BEA J, BEA G1, BEA G2, BEAE A, BEAE B, ALLOBEA A, ALLOBEA B, ALLOBEA C, BEAE L, BEA K, ENN B |
| 7 | RT5.4 | Fusarium proliferatum | BEA, BEA D, BEA J, BEA G1, BEAE L, ENN B |
| 8 | MU12 | Fusarium verticillioides | BEA, BEA D, BEA G1, BEAE A, BEA K, ENN B, ENN B1 |
| 9 | P36 | Fusarium verticillioides | BEA, BEA C, ALLOBEA C, ENN B |
| 10 | 4447 | Isaria farinosa | BEA, BEA B, BEA C, BEA D, BEA E, BEA F/A, BEA J, BEA G1, BEA G2, BEAE A, ALLOBEA A, ALLOBEA B, ALLOBEA C, BEAE L, BEA K, BEA L |
| 11 | ENC5 | Isaria farinosa | BEA |
| 12 | ENC9 | Isaria farinosa | BEA |
| 13 | MU5 | Isaria farinosa | BEA |
| 14 | MU1 | Isaria fumosorosea | BEA, ENN B, ENN B1 |
| 15 | 4414 | Cordyceps confragosa | BEA, ENN A, ENN B1 |
| 16 | ENC1 | Cordyceps confragosa | BEA |
| 17 | ENC6 | Cordyceps confragosa | BEA |
| 18 | MAL 1.4 | Fusarium oxysporum | BEA, BEA B, BEA C, BEA D, BEA E, BEA F/A, BEA J, BEA G1, BEA G2, BEAE A, BEAE B, ALLOBEA A, ALLOBEA B, ALLOBEA C, BEAE L |
| 19 | A 4.12 | Fusarium proliferatum | BEA, BEA B, BEA C, BEA D, BEA E, BEA F/A, BEA J, BEA G1, BEA G2, BEAE A, BEAE B, ALLOBEA A, ALLOBEA B, ALLOBEA C, BEAE L |
| Compound | Measured (m/z) [M + H]+ | Measured (m/z) [M + NH4]+ | Measured (m/z) [M + Na]+ | Retention Time (min) | Elemental Composition | Mass Error (ppm) [M + H]+ | Mass Error (ppm) [M + NH4]+ | Mass Error (ppm) [M + Na]+ |
|---|---|---|---|---|---|---|---|---|
| 1 | 784.4144 | 801.4426 | 806.3947 | 7.4 | C45H57N309 | −3.7 | −1.6 | −5 |
| 2/11 | 812.4446 | 829.4738 | 834.4267 | 12.4 | C47H61N3O9 | −4.9 | −1.7 | −4.8 |
| 3 | 736.4155 | 753.4431 | 758.3968 | 6.4 | C41H57N3O9 | −2.5 | −0.9 | −3.3 |
| 4 | 770.3997 | 787.4274 | 792.3809 | 7 | C44H55N3O9 | −2.6 | −1 | −3.5 |
| 5 | 756.384 | 773.4124 | 778.3668 | 6.7 | C43H53N3O9 | −1.9 | −0.2 | −1.5 |
| 6 | 688.4184 | 705.4425 | 710.3965 | 6.3 | C37H57N3O9 | 1.6 | −1.1 | −3.9 |
| 7/8/9 | 716.4469 | 733.4745 | 738.4263 | 6.7 | C39H61N3O9 | −2.4 | −0.9 | −5.8 |
| 10 | 702.4344 | 719.4581 | 724.413 | 5.7 | C38H59N3O9 | 2.1 | −2 | −2.6 |
| Compound | Parent ion (m/z) [M+NH4]+ | Primary Daughter Ion (m/z) | Secondary Daughter Ion (m/z) | Collision Energy (eV) | LOD a (ng/g) | LOQ b (ng/g) |
|---|---|---|---|---|---|---|
| ENN A | 699.4 | 228.2 * | 210.1 | 36 | 2 | 6 |
| ENN A1 | 685.4 | 214.2 * | 210.1 | 38 | 3 | 9 |
| ENN B | 657.3 | 214.1 * | 196.0 | 40 | 2 | 6 |
| ENN B1 | 671.2 | 228.0 * | 214.1 | 57 | 2 | 6 |
| BEA | 801.2 | 784.0 | 244.1 * | 28 | 1 | 3 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Urbaniak, M.; Waśkiewicz, A.; Trzebny, A.; Koczyk, G.; Stępień, Ł. Cyclodepsipeptide Biosynthesis in Hypocreales Fungi and Sequence Divergence of The Non-Ribosomal Peptide Synthase Genes. Pathogens 2020, 9, 552. https://doi.org/10.3390/pathogens9070552
Urbaniak M, Waśkiewicz A, Trzebny A, Koczyk G, Stępień Ł. Cyclodepsipeptide Biosynthesis in Hypocreales Fungi and Sequence Divergence of The Non-Ribosomal Peptide Synthase Genes. Pathogens. 2020; 9(7):552. https://doi.org/10.3390/pathogens9070552
Chicago/Turabian StyleUrbaniak, Monika, Agnieszka Waśkiewicz, Artur Trzebny, Grzegorz Koczyk, and Łukasz Stępień. 2020. "Cyclodepsipeptide Biosynthesis in Hypocreales Fungi and Sequence Divergence of The Non-Ribosomal Peptide Synthase Genes" Pathogens 9, no. 7: 552. https://doi.org/10.3390/pathogens9070552
APA StyleUrbaniak, M., Waśkiewicz, A., Trzebny, A., Koczyk, G., & Stępień, Ł. (2020). Cyclodepsipeptide Biosynthesis in Hypocreales Fungi and Sequence Divergence of The Non-Ribosomal Peptide Synthase Genes. Pathogens, 9(7), 552. https://doi.org/10.3390/pathogens9070552







