Abstract
Porcine circovirus type 2 (PCV2) can cause severe disease in infected pigs, resulting in massive economic loss for the swine industry. Transcriptomic and proteomic approaches have been widely employed to identify the underlying molecular mechanisms of the PCV2 infection. Numerous differentially expressed mRNAs, miRNAs, and proteins, together with their associated signaling pathways, have been identified during PCV2 infection, paving the way for analysis of their biological functions. Long noncoding RNAs (lncRNAs) are important regulators of multiple biological processes. However, little is known regarding their role in the PCV2 infection. Hence, in our study, RNA-seq was performed by infecting PK-15 cells with PCV2. Analysis of the differentially expressed genes (DEGs) suggested that the cytoskeleton, apoptosis, cell division, and protein phosphorylation were significantly disturbed. Then, using stringent parameters, six lncRNAs were identified. Additionally, potential targets of the lncRNAs were predicted using both cis- and trans-prediction methods. Interestingly, we found that the HOXB (Homeobox B) gene cluster was probably the target of the lncRNA LOC106505099. Enrichment analysis of the target genes showed that numerous developmental processes were altered during PCV2 infection. Therefore, our study revealed that lncRNAs might affect porcine embryonic development through the regulation of the HOXB genes.
1. Introduction
Porcine circovirus (PCV), which belongs to the family Circoviridae, is a nonenveloped virus with a single-stranded circular DNA [1]. Currently, there are three known types of PCVs. PCV1, which was first identified in the pig kidney cell line, is nonpathogenic to pigs [2,3]. PCV3 is a newly identified pathogen with the first report in North America. The afflicted pigs have symptoms of porcine dermatitis and nephropathy syndrome and reproductive problems [4]. Another PCV is porcine circovirus type 2 (PCV2), which is the primary threat to the global swine industry leading to massive economic loss annually [5]. PCV2 is the cause of pig postweaning multisystemic wasting syndrome (PMWS), which is characterized by progressive loss of weight, respiratory difficulty, jaundice, and reproductive failure [6]. Microscopically, PMWS is associated with severe lymphocyte depletion and multiple organ inflammation [7,8]. Thus, the study of PCV2 is of great importance. PCV2 has a genome size of 1.7 kb with at least three open reading frames (ORFs). ORF1 encodes the proteins Rep and Rep’, both of which are essential for virus replication [9]. ORFs 2 and 3 reside in the opposite strand, encoding the pathogenic structure protein capsid and the peptide involved in the host cell apoptosis, respectively [10,11].
Long noncoding RNAs (lncRNAs) are a group of noncoding RNAs that are longer than 200 nt. Except for their small coding potential, lncRNAs are similar to ordinary mRNAs in terms of exon-intron organization, 5′ cap, and 3′ polyadenine tracts [12,13]. Additionally, lncRNAs can be transcribed from intronic or intergenic regions by the RNA polymerase II in a sense and antisense manner [14]. Previous studies show that lncRNAs have poor sequence conservation, so they are species-specific [15]. However, accumulating evidence suggests that structural information, functions, and synteny are also constituents of the conservation of lncRNAs [16,17]. Since their discovery, lncRNAs have been implicated in numerous biological processes, including cell signaling [18], epigenetic regulation [19], enhancer trapping [20], human diseases [21,22], and virus-host interactions [23,24]. However, until recently, only one study regarding the effect of lncRNAs on PCV2 infection has been published, in which the lncRNA-regulated mRNAs were found to be enriched in the DNA/RNA binding, transcription factor activity, immune response, the MAPK (mitogen-activated protein kinase) signaling pathway, and cytokine–cytokine receptor interaction [25].
In our study, using more stringent screening parameters than were previously used, we identified six differentially expressed lncRNAs upon PCV2 infection. Enrichment analysis of the potential targets of the lncRNAs suggested that developmental processes might be disturbed. Therefore, our study provides novel insight into lncRNAs in PCV2 infection.
2. Materials and Methods
2.1. Cell Culture and PCV2 Infection
PK-15 cells free of PCV2 were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA), and they were cultured in Dulbecco’s modified Eagle medium (HyClone, GE, Beijing, China) supplemented with 10% fetal bovine serum (HyClone, GE, Beijing, China). The cell line was used because it is a widely accepted in vitro infection model, especially in the field of PCV2 virology, from which underlying molecular and cellular mechanisms of PCV2 infection have been learned [26,27,28,29,30,31]. The PCV2 strain 17HNA1 (Accession no.: MT423827) was used in our study. The infection procedures strictly followed a previously published protocol [32] with minor modifications: multiplicity of infection (MOI) was set at 0.1, and the PK-15 cells were harvested at 72 h postinfection. Viral and mock infections were repeated three times. Therefore, the mock infections were named as con-1, con-2, and con-3, while the PCV2 infection repeats were designated as PCV2-1, PCV2-2, and PCV2-3.
2.2. RNA Preparation, Quantitative Real-Time PCR (qRT-PCR), and RNA-Seq
Seventy-two hours postinfection, the PCV2-infected and mock-infected cells were washed with ice-cold PBS (Sigma Aldrich, Shanghai, China) three times before being subjected to total RNA extraction using the Mini Best Universal RNA Extraction kit (Takara, Dalian, China). Next, the extracted RNA was tested using the Agilent 2200 Analyzer for quality control (QC), and those samples with an RNA Integrity Number (RIN) greater than 8 passed the QC process. Then, the RNA was subjected to RNA-seq using the Illumina HiSeq X Ten platform (San Diego, CA, USA) according to the pipeline of Novel Bioinformatics Inc. (Shanghai, China). The raw sequencing data were filtered using the fastp [33] to obtain clean reads, which were subsequently mapped to the porcine genome using the Hisat2 [34]. Then, the raw counts were obtained using the FeatureCounts [35]. Next, differentially expressed transcripts were calculated based on the following parameters: |log2fold change| > 0.2630 (fold change greater than 1.2 or lesser than 5/6) and adjusted p-value (false discovery rate, FDR) < 0.05 using the DESeq2 tool [36]. The identification of lncRNAs used a strategy described in a previous report [25]. The sequenced reads were submitted to the Gene Expression Omnibus with the accession number: GSE149711.
qRT-PCR was then carried out to confirm the expression levels of the selected transcripts, and it was performed according to a previously described protocol [37]. The transcripts tested were ORF2, NGFR, PIGR, SERPINF1, LOC106505099, LOC106509609, TRIM29, and LOC100512687, the latter two of which served as the negative controls. The ACTB was used as an internal control. All primers used are listed in Table S1. qRT-PCR data were analyzed using the 2−ΔΔCt method [38].
2.3. LncRNA Target Prediction
LncRNA cis-regulated target genes were identified by searching the protein-coding genes 50 kb up or downstream of each lncRNA. Then, the expression level of each of the identified lncRNAs was compared with all the sequenced reads. Transcripts with Pearson’s correlation coefficient >0.99 and adjusted p-value < 0.05 were considered potential trans-targets of lncRNAs.
2.4. Enrichment Analysis
The differentially expressed genes (DEGs) identified by RNA-seq and the lncRNA targets were submitted to the Database for Annotation, Visualization and Integrated Discovery (DAVID) [39,40] for the identification of significantly altered Gene Ontology (GO) [41] and Kyoto Encyclopedia of Genes and Genomes (KEGG) [42] terms using the default parameters (EASE score < 0.1).
2.5. Statistics
Student’s t-tests were used to compare the difference in transcripts between PCV2-infected and mock-infected cells. Data are presented as the mean ± standard errors of the means (SEMs). Type I error was set at 0.05.
2.6. Ethical Statement
The experiment was approved by the Animal Welfare Committee of Zhejiang University (No. 11,834 issued on 26 February 2018).
3. Results and Discussion
Total RNA from the PK-15 cells was extracted and subjected to the RNA-seq. After the QC process and removal of ribosomal RNA sequences, an average of 84% of the sequence reads was mapped to the pig genome (Table S2), demonstrating that RNA-seq provided a reliable result. The positions of the reads were mostly located in the coding sequences or exonic regions (Figure S1). A total of 38,841 transcripts were identified in the six sequence samples, of which 111 transcripts were differentially expressed; 87 were upregulated, and 24 were downregulated (Figure 1A and Table S3). Then, a heatmap with clustering analysis using all the DEGs was constructed, in which PCV2-infected cells exhibited significantly different expression patterns compared to the mock-infected cells (Figure 1B). Within this group of transcripts, six differentially expressed lncRNAs were also observed; 4 were upregulated, and 2 were downregulated (Figure 1A and Table S4).
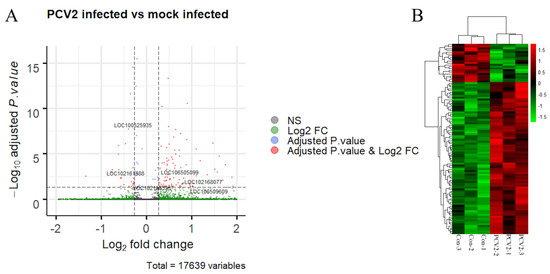
Figure 1.
Analysis of the RNA-seq results. (A) The volcano plot shows the differentially expressed transcripts (red dots) between porcine circovirus type 2 (PCV2) infection versus mock infection. Each dot represents a single transcript, while the blue dots or green dots represent the transcripts that only satisfy either a log2fold change or an adjusted p-value, respectively. The six long noncoding RNAs (lncRNAs) are also labeled. (B) Heatmap with clustering analysis of the differentially expressed genes (DEGs). Each row indicates an individual RNA, with columns representing the samples.
Next, qRT-PCR was employed to confirm the expression of some selected genes and lncRNAs. The Cap expression level was significantly higher in the PCV2-infected groups, demonstrating the successful infection of PK-15 cells (Figure 2). Other selected genes and lncRNAs, such as NGFR, PIGR, SERPINF1, LOC106509609, and LOC106505099, showed expression patterns that were similar to the data obtained by RNA-seq (Figure 2 and Table S3). Accordingly, the negative controls (TRIM29 and LOC100512687) had comparable expression levels in the PCV2- and mock-infected cells.
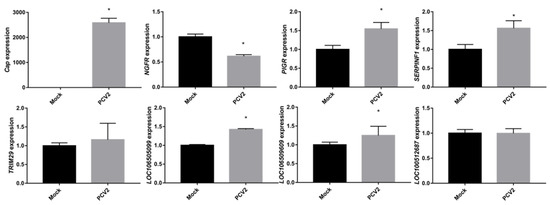
Figure 2.
qRT-PCR validation of selected mRNAs and lncRNAs. Cap (ORF2) levels were higher in PCV2-infected cells than in mock-infected cells, demonstrating successful infection. Other genes or lncRNAs have similar patterns of expression to the RNA-seq results. Data are presented as the mean ± standard errors of the means (SEMs) with * indicating a p-value < 0.05.
Analysis of the underlying molecular mechanisms of PCV2 infection and replication is key to the prevention of the virus. Previously, several high-throughput approaches have been employed to study mRNA, miRNA, and protein changes after PCV2 infection. In these studies, the DEGs analyzed by microarray or RNA-seq were enriched in the immune response, cytokine production, and apoptosis upon viral infection using in vivo or in vitro methods [43,44,45,46,47,48,49]. Later, miRNA was also tested for its role in the disease. Analysis of the targets of miRNAs indicated that disease resistance, MAPK signaling, and transcriptional regulation were involved [47,50]. Direct quantifications of the protein amounts after PCV2 infection revealed that there were altered levels of cytoskeletal proteins, stress response proteins, signal transduction proteins, and proteins involved in the ubiquitin-proteasome processes [32,51]. However, only one study had previously been performed to evaluate the effect of lncRNAs on PCV2 infection [25]. Fang et al. used PCV2 to infect the porcine intestinal epithelial cell line IPEC-J2 to mimic porcine enteritis. In their study, some interesting conclusions were drawn; they found that transcriptional regulation, nucleic acid binding, MAPK signaling, and cytokine-related pathways were disturbed upon PCV2 infection. However, they used the p-value instead of the adjusted p-value to identify the altered lncRNAs and mRNAs, which would substantially increase the type I error, leading to false positive findings. For example, in their lncRNA list, no lncRNA identified has an adjusted p-value of less than 0.05 [25]. Therefore, in the current study, we aimed to study whether some differentially expressed lncRNAs could be found using more stringent screening parameters.
Of the 111 differentially expressed transcripts, 105 were assigned to the protein-coding genes. Thus, these DEGs were analyzed using the DAVID tool for the identification of enriched biological processes (BP), cellular components (CC), and molecular functions (MF). As shown in Table 1, the DEGs were mainly involved in cell division, apoptosis, and the cytoskeleton and protein phosphorylation processes. Cell division and apoptosis have been well studied previously [52,53]. The involvement of cytoskeletal processes was also noticed in proteomic studies, indicating that the virus might affect cell division through the mitotic spindle [32,51]. The protein phosphorylation process is associated with numerous critical biological processes, such as the MAPK signaling pathway, which has been shown to be essential in viral infection [30,31].

Table 1.
Significantly enriched Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) terms by the Database for Annotation, Visualization and Integrated Discovery (DAVID) analysis.
KEGG analysis indicated that Rap1, Ras signaling pathways, and arginine biosynthesis were significantly enriched. Currently, no research has been conducted regarding the roles of arginine biosynthesis and Rap1 signaling in PCV2 infection. However, Ras is the upstream effector of MAPK signaling, which promotes infected cell proliferation [30,31]. In our enrichment analysis, no immune-related terms were found. This disparity may be because we used an MOI of 0.1, resulting in minimally infected PK-15 cells [26].
With our experimental conditions in combination with the stringent screening parameters, only six lncRNAs were found to be differentially expressed (adjusted p-value < 0.05, Table S4). Thus, these lncRNAs may be sensitive to PCV2 infection. Next, to functionally annotate these lncRNAs, the lncRNA targets were predicted. Currently, there are three main prediction approaches. The first one is cis-regulated target prediction, which is based on the fact that lncRNA can regulate its nearby gene transcription [54]. Thus, the genes adjacent to the lncRNA within an appropriate range are all potentially regulated. The second approach predicts the targets based on the coexpressing pattern. The underlying mechanism is that the interacting lncRNA and mRNA might have a similar expression pattern. The third method is directly computing the interaction potential of lncRNA and its targets using parameters such as free energy change, secondary structural information [55,56,57]. The former two methods can produce fast prediction results; however, the prediction is indirectly based on correlation, which needs to be validated experimentally. The computational approach can calculate the direct interaction of lncRNA and its targets. However, it also needs intensive computational facilities. After weighing the pros and cons, cis-regulated target prediction and coexpression methods were employed in our current study, and the results were then shown in Table 2. Analysis of the neighboring genes showed that 18 genes were potential targets; however, no cis-target was found for LOC102158335. One interesting finding is that lncRNA LOC106505099 was located within the HOXB (Homeobox B) gene family, which is critical for the development of embryos [58,59,60]. Coexpression analysis using stringent parameters predicted that only three genes, DNAAF3, PPP3CB, and CKMT1A, were correlated with LOC102168077, LOC100525935, and LOC102161888, respectively. Thus, the three genes might be regulated by their respective lncRNAs in a trans manner.

Table 2.
The predicted targets of lncRNAs.
Next, to predict the potential biological roles of these lncRNAs in the infected PK-15 cells, GO analysis was performed using the potential target genes of the lncRNAs. The enrichment analysis showed that these target genes were mainly located in the nucleus (CC) and were involved in two MF processes: DNA binding and transcription factor regulation (Table 3). GO_BP analysis revealed the involvement of DNA transcription and several developmental processes, e.g., embryonic skeletal development, facial nerve structural organization, and rhombomere 4 development, suggesting the role of PCV2 in organ development. Previous studies on pig embryos showed that PCV2-exposed embryos had a higher mortality rate, especially when the zona pellucida was compromised, resulting in a low pregnancy rate of sows [61,62,63]. These results indicated that PCV2 might affect the reproductive systems through lncRNA-mediated modulation of HOXB genes. Virus infection mediated dysregulation of HOX genes has been reported before. In one study, the hepatitis B virus can induce HOXA10 gene expression in human cells [64]. Another report showed that human papillomavirus gene expression could be modulated by HOXD9 in cervical cancer [65]. The Epstein-Barr virus activates NKL homeobox genes in B-cell lymphoma [66]. Thus, it is reasonable to link PCV2 infection with HOXB gene regulation. Subsequently, we searched the human genome for the presence of possible lncRNA orthologs. Based on syntenic information, LOC106505099 and LOC102161888 have potential human orthologs HOXB-AS1 and SMIM15-AS1, respectively. LOC106505099 and HOXB-AS1 are both located between HOXB2 and HOXB3 and are in the antisense direction, while LOC102161888 and SMIM15-AS1 are adjacent to the SMIM15 gene in a tail-to-tail manner. However, only HOXB-AS1 and LOC106505099 showed some extent of sequence conservation according to a Basic Local Alignment Search Tool (BLAST) search of the pig transcriptome using the HOXB-AS1 sequence (ENST00000435312.5). Functionally, some lncRNAs residing in the HOX gene cluster have been associated with developmental regulation through the modulation of neighboring HOX genes [67,68]. Moreover, two functional studies regarding HOXB-AS1 in two cancer cell lines indicated that the lncRNA might promote proliferation and migration of cancer cells by either acting as a miRNA sponge or by participating in the Wnt/beta-catenin signaling pathways [69,70]. Another research project on mouse ortholog Hoxb3os showed that the lncRNA was downregulated in cystic kidneys, and regulated EIF2, mTOR, and EIF4 signaling pathways [71]. However, the direct link between HOXB-AS1/LOC106505099 and development has yet to be established, and further functional validation is required to unveil the biological roles of HOXB-AS1 or LOC106505099 in porcine embryo development and PCV2 infection.

Table 3.
The enriched GO terms using the lncRNA targeted mRNAs.
4. Conclusions
Using the RNA-seq technique, we identified differentially expressed lncRNAs upon PCV2 infection with stringent screening parameters. Through analysis of the targets of these lncRNAs, we found that LOC106505099 might be involved in the stages of embryonic development through the regulation of HOXB genes. These findings suggest a novel association between PCV2 infection and embryonic development, which would shed new light on the pathological mechanisms of PCV2 infection.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-0817/9/6/479/s1, Figure S1: Reads distribution of sequencing results. Table S1: List of primer sequences. Table S2: The mapping statistics of the sequenced reads. Table S3: List of differentially expressed genes of PCV2 infected vs. mock infected. Table S4: List of differentially expressed lncRNAs.
Author Contributions
J.H. and H.W. conceived the study and designed the experiment. J.H. performed the computational analysis. C.L. and F.C. conducted the virus-related experiments. J.P., A.L., and H.Z. carried out the molecular and cellular experiments. J.H. and H.W. wrote the manuscripts. All authors have read and agreed to the published version of the manuscript.
Funding
This research and the APC was funded by the Natural Science Foundation of Zhejiang Province (No. LY20C170001) and the National Natural Science Foundation (No. 81930040).
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
BLAST, basic local alignment search tool; BP, biological process; CC, cellular component; DAVID, Database for Annotation, Visualization and Integrated Discovery tool; DEG, differentially expressed gene; FDR, false discovery rate; GO, gene ontology; HOXB, homeobox B; KEGG, Kyoto Encyclopedia of Genes and Genomes; lncRNA, long noncoding RNA; MAPK, mitogen-activated protein kinase; MF, molecular function; MOI, multiplicity of infection; ORF, open reading frame; PCV2, porcine circovirus type 2; PMWS, postweaning multisystemic wasting syndrome; QC, quality control; qRT-PCR, quantitative real-time polymerase chain reaction; RIN, RNA integrity number; SEM, standard errors of the mean.
References
- Breitbart, M.; Delwart, E.; Rosario, K.; Segalés, J.; Varsani, A. ICTV virus taxonomy profile: Circoviridae. J. Gen. Virol. 2017, 98, 199–1998. [Google Scholar] [CrossRef] [PubMed]
- Tischer, I.; Mields, W.; Wolff, D.; Vagt, M.; Griem, W. Studies on epidemiology and pathogenicity of porcine circovirus. Arch. Virol. 1986, 91, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Tischer, I.; Rasch, R.; Tochtermann, G. Characterization of papovavirus and picornavirus like particles in permanent pig kidney cell lines. Zbl.bakt.r. A 1974, 226, 153–167. [Google Scholar]
- Palinski, R.; Piñeyro, P.; Shang, P.; Yuan, F.; Guo, R.; Fang, Y.; Byers, E.; Hause, B.M. A Novel Porcine Circovirus Distantly Related to Known Circoviruses Is Associated with Porcine Dermatitis and Nephropathy Syndrome and Reproductive Failure. J. Virol. 2016, 91, e01879-e16. [Google Scholar] [CrossRef] [PubMed]
- Ellis, J. Porcine Circovirus: A Historical Perspective. Vet. Pathol. 2014, 51, 315–327. [Google Scholar] [CrossRef] [PubMed]
- Harding, J.C.S.; Clark, E.G. Recognizing and diagnosing postweaning multisystemic wasting syndrome (PMWS). J. Swine Heal. Prod. 1997, 5, 5. [Google Scholar]
- Chianini, F.; Majó, N.; Segalés, J.; Domínguez, J.; Domingo, M. Immunohistochemical characterisation of PCV2 associate lesions in lymphoid and non-lymphoid tissues of pigs with natural postweaning multisystemic wasting syndrome (PMWS). Vet. Immunol. Immunopathol. 2003, 94, 63–75. [Google Scholar] [CrossRef]
- Sorden, S.D. Update on porcine circovirus and postweaning multisystemic wasting syndrome (PMWS). J. Swine Heal. Prod. 2000, 8, 133–136. [Google Scholar]
- Mankertz, A.; Mankertz, J.; Wolf, K.; Buhk, H.J. Identification of a protein essential for replication of porcine circovirus. J. Gen. Virol. 1998, 79, 381–384. [Google Scholar] [CrossRef] [PubMed]
- Nawagitgul, P.; Morozov, I.; Bolin, S.R.; Harms, P.A.; Sorden, S.D.; Paul, P.S. Open reading frame 2 of porcine circovirus type 2 encodes a major capsid protein. J. Gen. Virol. 2000, 81, 2281–2287. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, I.; Kwang, J. Characterization of a Previously Unidentified Viral Protein in Porcine Circovirus Type 2-Infected Cells and Its Role in Virus-Induced Apoptosis. J. Virol. 2005, 79, 8262–8274. [Google Scholar] [CrossRef]
- Cech, T.R.; Steitz, J.A. The noncoding RNA revolution-Trashing old rules to forge new ones. Cell 2014, 157, 77–94. [Google Scholar] [CrossRef] [PubMed]
- Ponting, C.P.; Oliver, P.L.; Reik, W. Evolution and Functions of Long Noncoding RNAs. Cell 2009, 136, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Mallory, A.C.; Shkumatava, A. LncRNAs in vertebrates: Advances and challenges. Biochimie 2015, 117, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Hezroni, H.; Koppstein, D.; Schwartz, M.G.; Avrutin, A.; Bartel, D.P.; Ulitsky, I. Principles of Long Noncoding RNA Evolution Derived from Direct Comparison of Transcriptomes in 17 Species. Cell Rep. 2015, 11, 1110–1122. [Google Scholar] [CrossRef] [PubMed]
- Diederichs, S. The four dimensions of noncoding RNA conservation. Trends Genet. 2014, 30, 121–123. [Google Scholar] [CrossRef] [PubMed]
- Johnsson, P.; Lipovich, L.; Grandér, D.; Morris, K.V. Evolutionary conservation of long non-coding RNAs; Sequence, structure, function. Biochim. Biophys. Acta Gen. Subj. 2014, 1840, 1063–1071. [Google Scholar] [CrossRef]
- Peng, W.X.; Koirala, P.; Mo, Y.Y. LncRNA-mediated regulation of cell signaling in cancer. Oncogene 2017, 36, 5661–5667. [Google Scholar] [CrossRef]
- Tsai, M.C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef]
- Cho, S.W.; Xu, J.; Sun, R.; Mumbach, M.R.; Carter, A.C.; Chen, Y.G.; Yost, K.E.; Kim, J.; He, J.; Nevins, S.A.; et al. Promoter of lncRNA Gene PVT1 Is a Tumor-Suppressor DNA Boundary Element. Cell 2018, 173, 1398–1412. [Google Scholar] [CrossRef]
- Qureshi, I.A.; Mattick, J.S.; Mehler, M.F. Long non-coding RNAs in nervous system function and disease. Brain Res. 2010, 1338, 20–35. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.C.; Spitale, R.C.; Chang, H.Y. Long intergenic noncoding RNAs: New links in cancer progression. Cancer Res. 2011, 71, 3–7. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, J.; Zhu, X.; Chen, Y.; Wei, H.; Chen, Q.; Chi, X.; Qi, B.; Zhang, L.; Zhao, Y.; Gao, G.F.; et al. NRAV, a long noncoding RNA, modulates antiviral responses through suppression of interferon-stimulated gene transcription. Cell Host Microbe 2014, 16, 616–626. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Xu, J.; Wang, Y.; Cao, X. An interferon-independent lncRNA promotes viral replication by modulating cellular metabolism. Science 2017, 358, 1051–1055. [Google Scholar] [CrossRef]
- Fang, M.; Yang, Y.; Wang, N.; Wang, A.; He, Y.; Wang, J.; Jiang, Y.; Deng, Z. Genome-wide analysis of long non-coding RNA expression profile in porcine circovirus 2-infected intestinal porcine epithelial cell line by RNA sequencing. PeerJ. 2019, 7, e6577. [Google Scholar] [CrossRef]
- Li, C.; Sun, Y.; Li, J.; Jiang, C.; Zeng, W.; Zhang, H.; Fan, S.; He, Q. PCV2 Regulates cellular inflammatory responses through dysregulating cellular mirna-mrna networks. Viruses 2019, 11, 1055. [Google Scholar] [CrossRef]
- Zhou, C.X.; Elsheikha, H.M.; Zhou, D.H.; Liu, Q.; Zhu, X.Q.; Suo, X. Dual identification and analysis of differentially expressed transcripts of porcine PK-15 cells and Toxoplasma gondii during in vitro infection. Front. Microbiol. 2016, 7, 721. [Google Scholar] [CrossRef]
- Cai, J.; Xie, X.; Hu, Y.; Zhan, Y.; Yu, W.; Wang, A.; Wang, N. Generation of PCV2 in PK15 cells transfected with recombinant baculovirus containing A 1.1 copy of the PCV2 genome. Acta Vet. Hung. 2017, 65, 278–290. [Google Scholar] [CrossRef]
- Zhang, T.; Chen, H.; Qi, L.; Zhang, J.; Wu, R.; Zhang, Y.; Sun, Y. Transcript profiling identifies early response genes against FMDV infection in PK-15 cells. Viruses 2018, 10, 364. [Google Scholar] [CrossRef]
- Wei, L.; Zhu, Z.; Wang, J.; Liu, J. JNK and p38 Mitogen-Activated Protein Kinase Pathways Contribute to Porcine Circovirus Type 2 Infection. J. Virol. 2009, 83, 6039–6047. [Google Scholar] [CrossRef][Green Version]
- Wei, L.; Liu, J. Porcine circovirus type 2 replication is impaired by inhibition of the extracellular signal-regulated kinase (ERK) signaling pathway. Virology 2009, 386, 203–209. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhou, J.; Wu, Y.; Zheng, X.; Ma, G.; Wang, Z.; Jin, Y.; He, J.; Yan, Y. Differential proteome analysis of host cells infected with porcine circovirus type 2. J. Proteome Res. 2009, 8, 5111–5119. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; GU, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- He, J.; Ye, J.; Li, Q.; Feng, Y.; Bai, X.; Chen, X.; Wu, C.; Yu, Z.; Zhao, Y.; Hu, X.; et al. Construction of a transgenic pig model overexpressing polycystic kidney disease 2 (PKD2) gene. Transgenic Res. 2013, 22, 861–867. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, H.; Wang, P.; Wang, L.; Sun, Y.; Liu, G.; Zhang, P.; Kang, L.; Jiang, S.; Jiang, Y. RNA-seq analysis reveals genes underlying different disease responses to porcine circovirus type 2 in pigs. PLoS ONE 2016, 11, e0155502. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Liu, S.; Wang, Y.; Deng, F.; Yan, W.; Yang, K.; Chen, H.; He, Q.; Charreyre, C.; Audoneet, J.C. Transcription analysis of the porcine alveolar macrophage response to porcine circovirus type 2. BMC Genom. 2013, 14, 353. [Google Scholar] [CrossRef]
- Mavrommatis, B.; Offord, V.; Patterson, R.; Watson, M.; Kanellos, T.; Steinbach, F.; Grierson, S.; Werling, D. Global gene expression profiling of myeloid immune cell subsets in response to in vitro challenge with porcine circovirus 2b. PLoS ONE 2014, 9, e911081. [Google Scholar] [CrossRef]
- Miller, L.C.; Fleming, D.S.; Lager, K.M. Comparison of the transcriptome response within the swine tracheobronchial lymphnode following infection with PRRSV, PCV-2 or IAV-S. Pathogens 2020, 9, 99. [Google Scholar] [CrossRef]
- Hong, J.S.; Kim, N.H.; Choi, C.Y.; Lee, J.S.; Na, D.; Chun, T.; Lee, Y.S. Changes in cellular microRNA expression induced by porcine circovirus type 2-encoded proteins. Vet. Res. 2015, 46, 39. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tomás, A.; Fernandes, L.T.; Sánchez, A.; Segalés, J. Time course differential gene expression in response to porcine circovirus type 2 subclinical infection. Vet. Res. 2010, 41, 12. [Google Scholar] [CrossRef]
- Fernandes, L.T.; Tomás, A.; Bensaid, A.; Sibila, M.; Sánchez, A.; Segalés, J. Microarray analysis of mediastinal lymph node of pigs naturally affected by postweaning multisystemic wasting syndrome. Virus Res. 2012, 165, 134–142. [Google Scholar] [CrossRef]
- Zhang, P.; Wang, L.; Li, Y.; Jiang, P.; Wang, Y.; Wang, P.; Kang, L.; Wang, Y.; Sun, Y.; Jiang, Y. Identification and characterization of microRNA in the lung tissue of pigs with different susceptibilities to PCV2 infection. Vet. Res. 2018, 49, 18. [Google Scholar] [CrossRef]
- Cheng, S.; Zhang, M.; Li, W.; Wang, Y.; Liu, Y.; He, Q. Proteomic analysis of porcine alveolar macrophages infected with porcine circovirus type 2. J. Proteomics 2012, 75, 3258–3269. [Google Scholar] [CrossRef] [PubMed]
- Tsai, Y.C.; Jeng, C.R.; Hsiao, S.H.; Chang, H.W.; Liu, J.J.; Chang, C.C.; Lin, C.M.; Chia, M.Y.; Pang, V.F. Porcine circovirus type 2 (PCV2) induces cell proliferation, fusion, and chemokine expression in swine monocytic cells in vitro. Vet. Res. 2010, 41, 60. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Resendes, A.R.; Majó, N.; van den Ingh, T.S.G.A.M.; Mateu, E.; Domingo, M.; Calsamiglia, M.; Segalés, J. Apoptosis in postweaning multisystemic wasting syndrome (PMWS) hepatitis in pigs naturally infected with porcine circovirus type 2 (PCV2). Vet. J. 2011, 189, 72–76. [Google Scholar] [CrossRef] [PubMed]
- Yan, P.; Luo, S.; Lu, J.Y.; Shen, X. Cis- and trans-acting lncRNAs in pluripotency and reprogramming. Curr. Opin. Genet. Dev. 2017, 46, 170–178. [Google Scholar] [CrossRef]
- Hu, R.; Sun, X. LncRNATargets: A platform for lncRNA target prediction based on nucleic acid thermodynamics. J. Bioinform. Comput. Biol. 2016, 14, 1650016. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ma, W.; Zeng, P.; Wang, J.; Geng, B.; Yang, J.; Cui, Q. LncTar: A tool for predicting the RNA targets of long noncoding RNAs. Brief Bioinform. 2014, 16, 806–812. [Google Scholar] [CrossRef]
- Tafer, H.; Hofacker, I.L. RNAplex: A fast tool for RNA-RNA interaction search. Bioinformatics 2008, 24, 2657–2663. [Google Scholar] [CrossRef]
- Deschamps, J.; van Nes, J. Developmental regulation of the Hox genes during axial morphogenesis in the mouse. Development 2005, 132, 2931–2942. [Google Scholar] [CrossRef]
- Iimura, T.; Pourquié, O. Collinear activation of Hoxb genes during gastrulation is linked to mesoderm cell ingression. Nature 2006, 442, 568–571. [Google Scholar] [CrossRef]
- Tschopp, P.; Tarchini, B.; Spitz, F.; Zakany, J.; Duboule, D. Uncoupling time and space in the collinear regulation of Hox genes. PLoS Genet. 2009, 5, e1000398. [Google Scholar] [CrossRef]
- Mateusen, B.; Sanchez, R.E.; Van Soom, A.; Meerts, P.; Maes, D.G.D.; Nauwynck, H.J. Susceptibility of pig embryos to porcine circovirus type 2 infection. Theriogenology 2004, 61, 91–101. [Google Scholar] [CrossRef]
- Zhao, H.; Ji, Q.; Zhao, G.; Song, Z.; Du, B.; Nie, Y.; Chen, Y.; Cong, P. Damage of zona pellucida reduces the developmental potential and quality of porcine circovirus type 2-infected oocytes after parthenogenetic activation. Theriogenology 2014, 82, 790–799. [Google Scholar] [CrossRef] [PubMed]
- Mateusen, B.; Maes, D.G.D.; Van Soom, A.; Lefebvre, D.; Nauwynck, H.J. Effect of a porcine circovirus type 2 infection on embryos during early pregnancy. Theriogenology 2007, 68, 896–901. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Zhang, Q.; Zhang, X.; You, L.; Wang, W.; Liu, W.; Han, Y.; Ma, C.; XU, W.; Chen, J.; et al. HoxA10 Facilitates SHP-1-Catalyzed Dephosphorylation of p38 MAPK/STAT3 To Repress Hepatitis B Virus Replication by a Feedback Regulatory Mechanism. J. Virol. 2019, 93, e01607–e01625. [Google Scholar] [CrossRef]
- Hirao, N.; Iwata, T.; Tanaka, K.; Nishio, H.; Nakamura, M.; Morisada, T.; Morii, K.; Maruyama, N.; Katoh, Y.; Yaguchi, T.; et al. Transcription factor homeobox D9 is involved in the malignant phenotype of cervical cancer through direct binding to the human papillomavirus oncogene promoter. Gynecol. Oncol. 2019, 155, 340–348. [Google Scholar] [CrossRef]
- Nagel, S.; Uphoff, C.C.; Dirks, W.G.; Pommerenke, C.; Meyer, C.; Drexler, H.G. Epstein-Barr virus (EBV) activates NKL homeobox gene HLX in DLBCL. PLoS ONE 2019, 14, e0216898. [Google Scholar] [CrossRef]
- Li, L.; Liu, B.; Wapinski, O.L.; Tsai, M.C.; Ou, K.; Zhang, J.; Carlson, J.C.; Lin, M.; Fang, F.; Gupta, R.A.; et al. Targeted disruption of Hotair leads to homeotic transformation and gene derepression. Cell Rep. 2013, 5, 3–12. [Google Scholar] [CrossRef]
- Wang, K.C.; Yang, Y.W.; Liu, B.; Sanyal, A.; Corces-Zimmerman, R.; Chen, Y.; Lajoie, B.R.; Protacio, A.; Flynn, R.A.; Gupta, R.A.; et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene experssion. Nature 2011, 472, 120–124. [Google Scholar] [CrossRef]
- Chen, R.; Zhang, X.; Wang, C. LncRNA HOXB-AS1 promotes cell growth in multiple myeloma via FUT4 mRNA stability by ELAVL1. J. Cell. Biochem. 2019. [Google Scholar] [CrossRef]
- Chen, X.; Li, L.Q.; Qiu, X.; Wu, H. Long non-coding RNA HOXB-AS1 promotes proliferation, migration and invasion of glioblastoma cells via HOXB-AS1/miR-885-3p/HOXB2 axis. Neoplasma 2019, 66, 386–396. [Google Scholar] [CrossRef]
- Aboudehen, K.; Farahani, S.; Kanchwala, M.; Chan, S.C.; Avdulov, S.; Mickelson, A.; Lee, D.; Gearhart, M.D.; Patel, V.; Xing, C.; et al. Long noncoding RNA Hoxb3os is dysregulated in autosomal dominant polycystic kidney disease and regulates mTOR signaling. J. Biol. Chem. 2018, 293, 9388–9398. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).