Mycosis Is a Disease State Encountered Rarely in Shore Crabs, Carcinus maenas
Abstract
:1. Introduction
2. Results and Discussion
2.1. General Population Observations
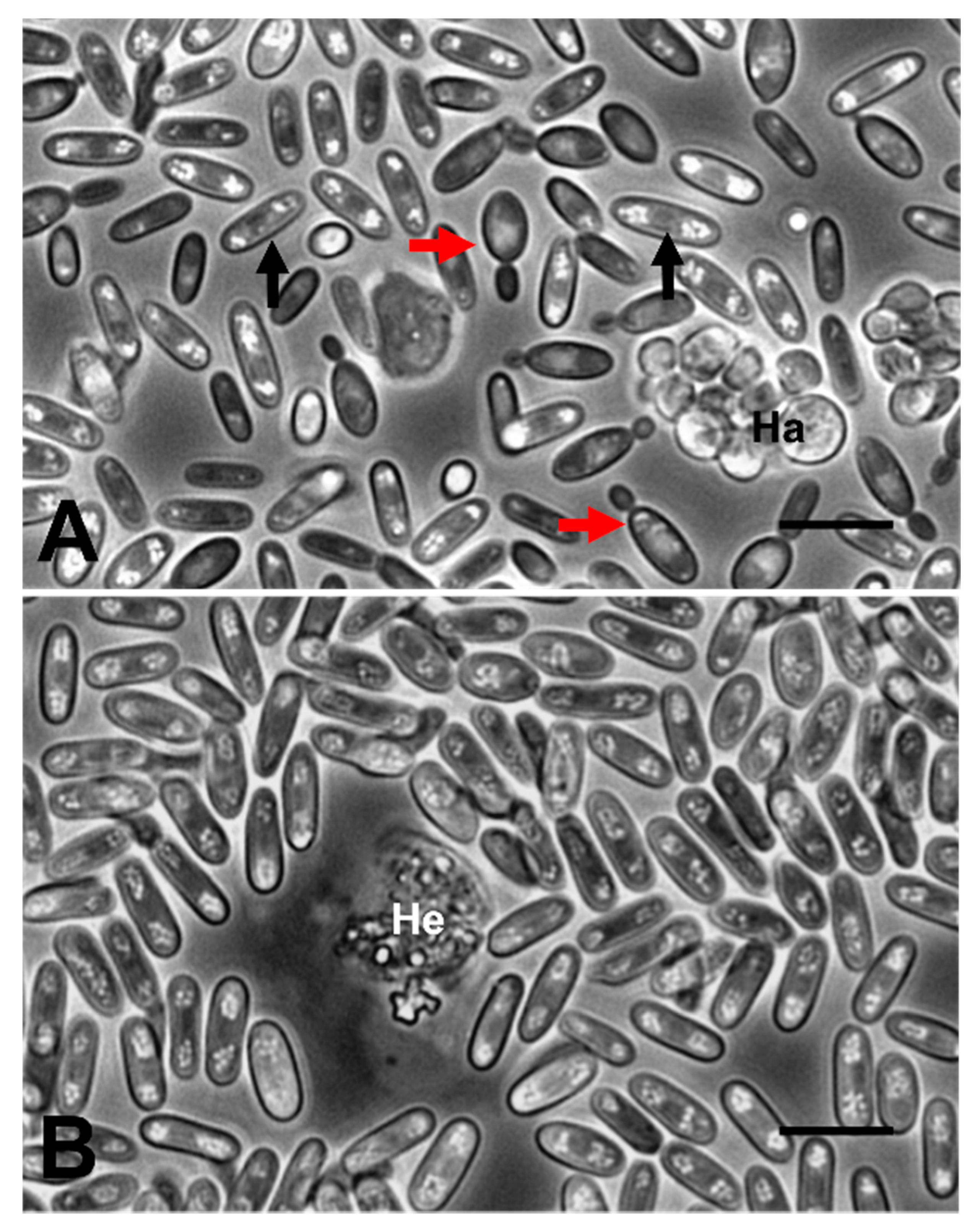
2.2. Haemolymph Preparations
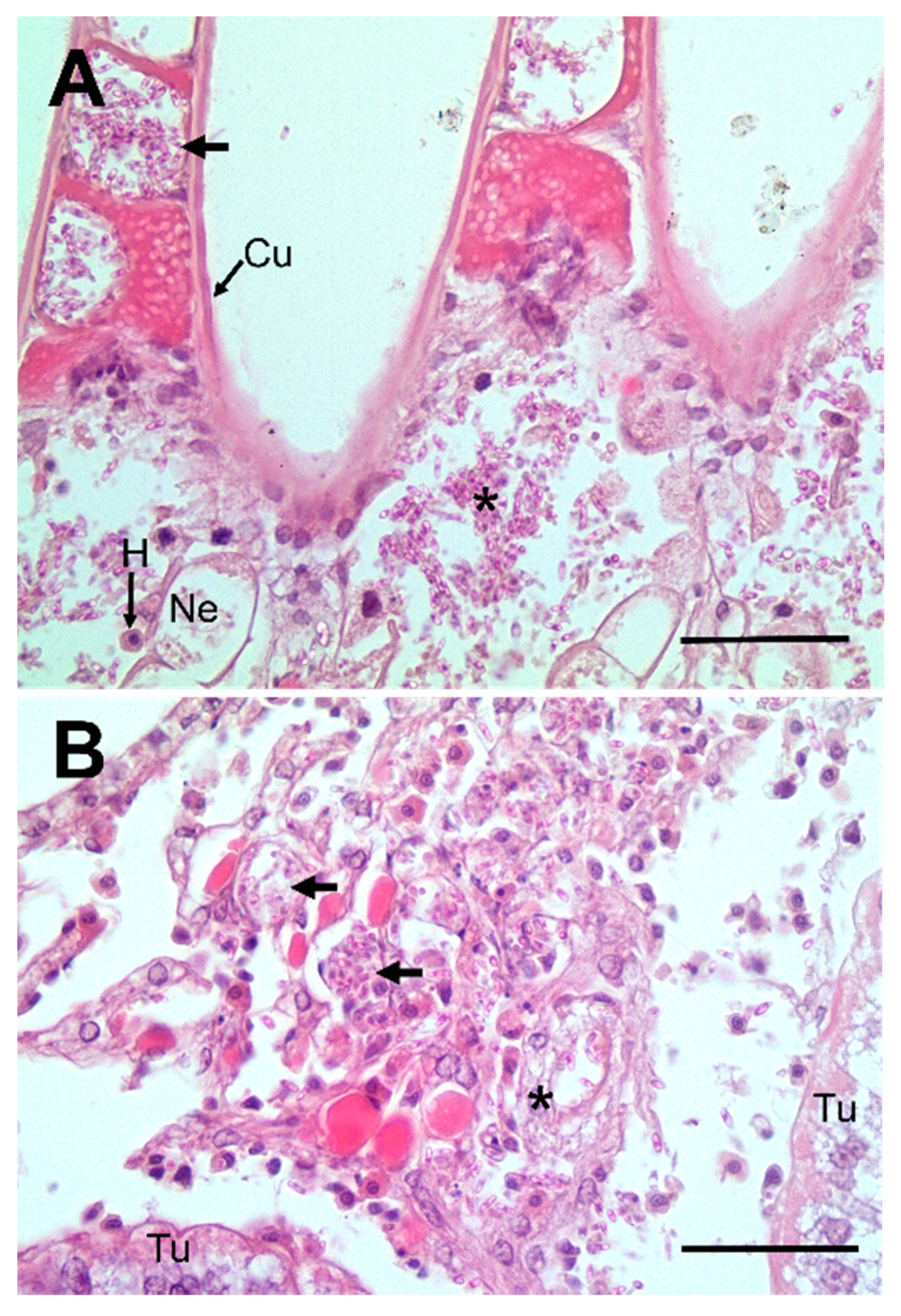
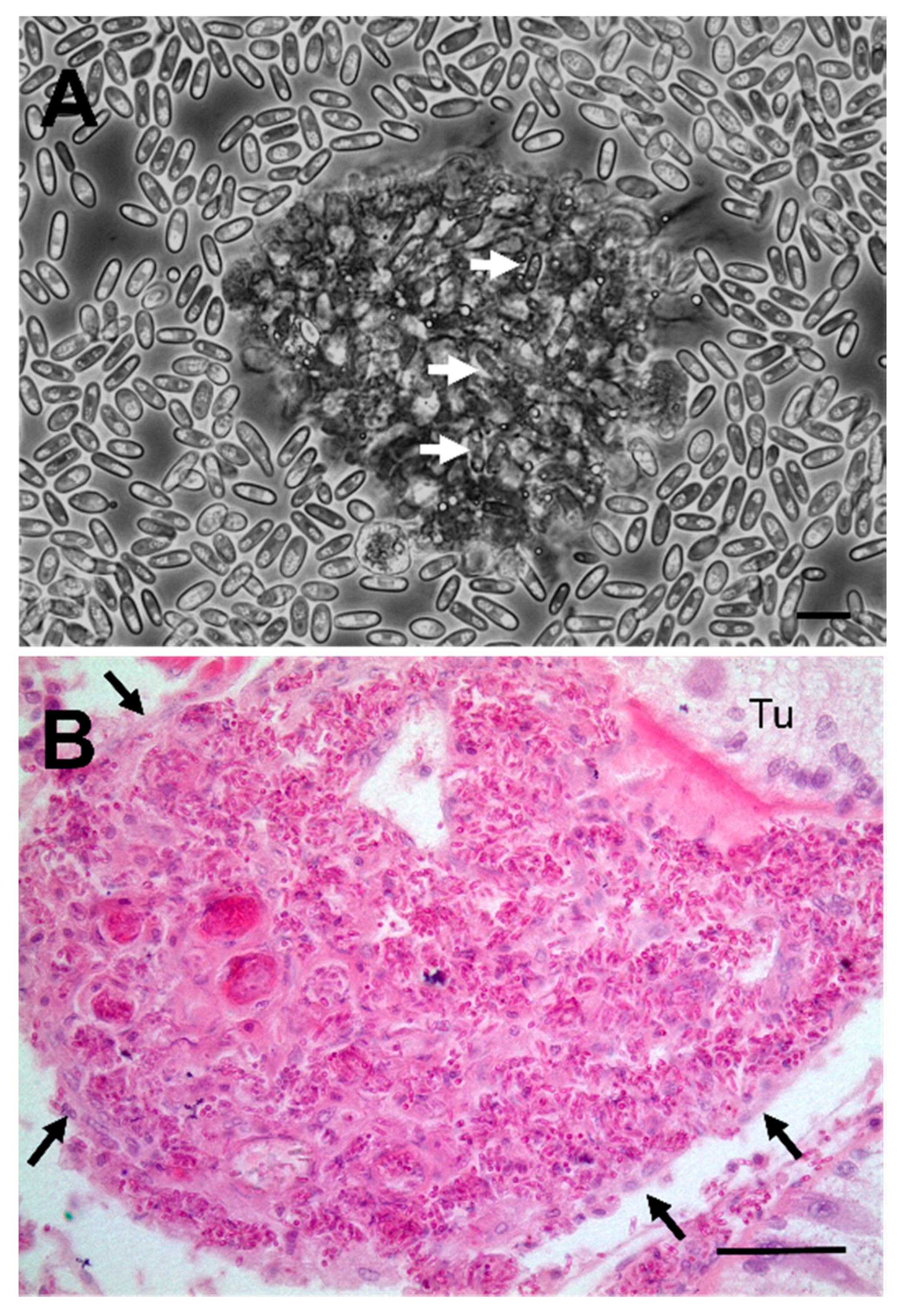
2.3. Histology
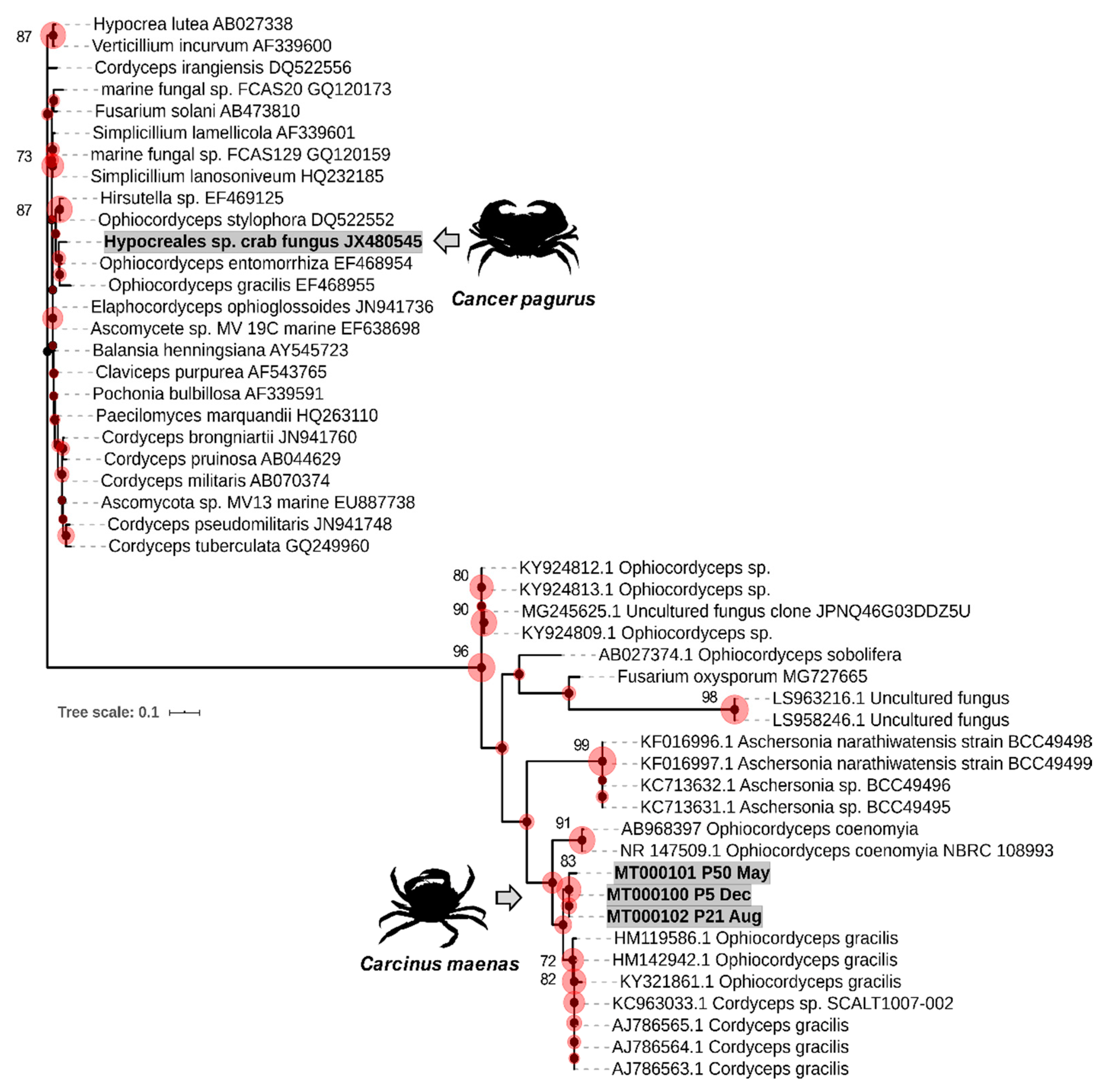
2.4. Phylogenetic Analyses
3. Materials and Methods
3.1. Survey Sites and Sample Collection
3.2. DNA Extraction and Quantification
3.3. PCR and Sequencing Conditions
3.4. Histology
3.5. Phylogenetic Analyses
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Richards, T.A.; Jones, M.D.M.; Leonard, G.; Bass, D. Marine Fungi: Their Ecology and Molecular Diversity. Ann. Rev. Mar. Sci. 2012, 4, 495–522. [Google Scholar] [CrossRef] [PubMed]
- Gladfelter, A.S.; James, T.Y.; Amend, A.S. Marine fungi. Curr. Biol. 2019, 29, R191–R195. [Google Scholar] [CrossRef] [PubMed]
- Jones, E.B.G. Are there more marine fungi to be described? Bot. Mar. 2011, 54, 343–354. [Google Scholar] [CrossRef]
- Amend, A.; Burgaud, G.; Cunliffe, M.; Edgcomb, V.P.; Ettinger, C.L.; Gutiérrez, M.H.; Heitman, J.; Hom, E.F.Y.; Ianiri, G.; Jones, A.C.; et al. Fungi in the marine environment: Open questions and unsolved problems. MBio 2019, 10, e01189-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgerton, B.F.; Henttonen, P.; Jussila, J.; Mannonen, A.; Paasonen, P.; Taugbøl, T.; Edsman, L.; Souty-Grosset, C. Understanding the causes of disease in European freshwater crayfish. Conserv. Biol. 2004, 18, 1466–1474. [Google Scholar] [CrossRef]
- Phillips, A.J.; Anderson, V.L.; Robertson, E.J.; Secombes, C.J.; van West, P. New insights into animal pathogenic oomycetes. Trends Microbiol. 2008, 16, 13–19. [Google Scholar] [CrossRef]
- Raffaele, S.; Kamoun, S. Genome evolution in filamentous plant pathogens: Why bigger can be better. Nat. Rev. Microbiol. 2012, 10, 417–430. [Google Scholar] [CrossRef]
- Fisher, W.S.; Nilson, E.H.; Shleser, R.A. Effect of the fungus Haliphthoros milfordensis on the juvenile stages of the American lobster Homarus americanus. J. Invertebr. Pathol. 1975, 26, 41–45. [Google Scholar] [CrossRef]
- Fisher, W.; Nilson, E.; Steenbergen, J.; Lightner, D. Microbial diseases of cultured lobsters: A review. Aquaculture 1978, 14, 115–140. [Google Scholar] [CrossRef]
- Nilson, E.H.; Fisher, W.S.; Shleser, R.A. Filamentous infestations observed on eggs and larvae of cultured crustaceans. Proc. Annu. Meet. World Maric. Soc. 1975. [Google Scholar] [CrossRef]
- Davies, C.E.; Wootton, E.C. Current and emerging diseases of the European lobster (Homarus gammarus): A review. Bull. Mar. Sci. 2018, 94, 959–978. [Google Scholar] [CrossRef]
- Holt, C.; Foster, R.; Daniels, C.L.; van der Giezen, M.; Feist, S.W.; Stentiford, G.D.; Bass, D. Halioticida noduliformans infection in eggs of lobster (Homarus gammarus) reveals its generalist parasitic strategy in marine invertebrates. J. Invertebr. Pathol. 2018, 154, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Leignel, V.; Stillman, J.H.; Baringou, S.; Thabet, R.; Metais, I. Overview on the European green crab Carcinus spp. (Portunidae, Decapoda), one of the most famous marine invaders and ecotoxicological models. Environ. Sci. Pollut. Res. 2014, 21, 9129–9144. [Google Scholar] [CrossRef]
- Davies, C.E.; Batista, F.M.; Malkin, S.H.; Thomas, J.E.; Bryan, C.C.; Crocombe, P.; Coates, C.J.; Rowley, A.F. Spatial and temporal disease dynamics of the parasite Hematodinium sp. in shore crabs, Carcinus maenas. Parasit. Vectors 2019, 12, 472. [Google Scholar] [CrossRef]
- Bookelaar, B.E.; O’Reilly, A.J.; Lynch, S.A.; Culloty, S.C. Role of the intertidal predatory shore crab Carcinus maenas in transmission dynamics of ostreid herpesvirus-1 microvariant. Dis. Aquat. Organ. 2018, 130, 221–233. [Google Scholar] [CrossRef] [Green Version]
- Bojko, J.; Stebbing, P.D.; Dunn, A.M.; Bateman, K.S.; Clark, F.; Kerr, R.C.; Stewart-Clark, S.; Johannesen, Á.; Stentiford, G.D. Green crab Carcinus maenas symbiont profiles along a North Atlantic invasion route. Dis. Aquat. Organ. 2018, 128, 147–168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bojko, J.; Subramaniam, K.; Waltzek, T.B.; Stentiford, G.D.; Behringer, D.C. Genomic and developmental characterisation of a novel bunyavirus infecting the crustacean Carcinus maenas. Sci. Rep. 2019, 9, 12957. [Google Scholar] [CrossRef]
- Rowley, A.F.; Davies, C.E.; Malkin, S.H.; Bryan, C.C.; Thomas, J.E.; Batista, F.M.; Coates, C.J. Prevalence and histopathology of the parasitic barnacle, Sacculina carcini in shore crabs, Carcinus maenas. J. Invertebr. Pathol. 2020, 171, 107338. [Google Scholar] [CrossRef]
- Subramaniam, K.; Behringer, D.C.; Bojko, J.; Yutin, N.; Clark, A.S.; Bateman, K.S.; van Aerle, R.; Bass, D.; Kerr, R.C.; Koonin, E.V.; et al. A new family of DNA viruses causing disease in crustaceans from diverse aquatic biomes. MBio 2020, 11. [Google Scholar] [CrossRef] [Green Version]
- Davies, C.E.; Bass, D.; Ward, G.M.; Batista, F.M.; Malkin, S.H.; Thomas, J.E.; Bateman, K.; Feist, S.W.; Coates, C.J.; Rowley, A.F. Diagnosis and prevalence of two new species of haplosporidians infecting shore crabs Carcinus maenas: Haplosporidium carcini n. sp., and H. cranc n. sp. Parasitology 2020, in press. [Google Scholar]
- Edwards, M.; Coates, C.J.; Rowley, A.F. Host Range of the Mikrocytid parasite Paramikrocytos canceri in decapod crustaceans. Pathogens 2019, 8, 252. [Google Scholar] [CrossRef] [Green Version]
- Stentiford, G.D.; Evans, M.; Bateman, K.; Feist, S.W. Co-infection by a yeast-like organism in Hematodinium-infected European edible crabs Cancer pagurus and velvet swimming crabs Necora puber from the English Channel. Dis. Aquat. Organ. 2003, 54, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.L.; Hamilton, K.M.; Hirschle, L.; Wootton, E.C.; Vogan, C.L.; Pope, E.C.; Eastwood, D.C.; Rowley, A.F. Characterization and molecular epidemiology of a fungal infection of edible crabs (Cancer pagurus) and interaction of the fungus with the dinoflagellate parasite Hematodinium. Appl. Environ. Microbiol. 2013, 79, 783–793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, A.L.; Hirschle, L.; Vogan, C.L.; Rowley, A.F. Parasitization of juvenile edible crabs (Cancer pagurus) by the dinoflagellate, Hematodinium sp.: Pathobiology, seasonality and its potential effects on commercial fisheries. Parasitology 2015, 142, 428–438. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.L.; Rowley, A.F. Effects of experimental infection of juvenile edible crabs Cancer pagurus with the parasitic dinoflagellate Hematodinium sp. J. Shellfish Res. 2015, 34, 511–519. [Google Scholar] [CrossRef]
- Zhong, X.; Peng, Q.; Qi, L.; Lei, W.; Liu, X. rDNA-targeted PCR primers and FISH probe in the detection of Ophiocordyceps sinensis hyphae and conidia. J. Microbiol. Methods 2010, 83, 188–193. [Google Scholar] [CrossRef]
- Sung, G.; Hywel-jones, N.L.; Sung, J.; Luangsa-ard, J.J.; Shrestha, B.; Joseph, W. Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Stud. Mycol. 2007, 57, 5–59. [Google Scholar] [CrossRef] [Green Version]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes—Application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J.; White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols; Academic Press (Elsevier): Amsterdam, The Netherlands, 1990; pp. 315–322. ISBN 008088671X. [Google Scholar]
- Op De Beeck, M.; Lievens, B.; Busschaert, P.; Declerck, S.; Vangronsveld, J.; Colpaert, J.V. Comparison and Validation of Some ITS Primer Pairs Useful for Fungal Metabarcoding Studies. PLoS ONE 2014, 9, e97629. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| P5_Dec | P50_Dec | P50_May | P21_Aug | |
|---|---|---|---|---|
| Date | December 2017 | December 2017 | May 2018 | August 2018 |
| Carapace size | 42.9 mm | 47.5 mm | 33 mm | 39 mm |
| Sex | Female | Male | Female | Female |
| Fouling * | Yes | No | No | No |
| Carapace colour | Yellow | Yellow | Yellow | Yellow |
| Limb loss | 0 | 3 | 0 | 0 |
| Haemolymph colour | Milky | Cloudy | Milky | Milky |
| Co-infection | Hematodinium sp. | Encysted trematodes; Haplosporidium carcini | N/A | Hematodinium sp. |
| PCR Amplicons | ||||
| -Accession no. | MT000100 | N/A | MT000101 | MT000102 |
| -Sequence length | 242 bp | 244 bp | 244 bp | |
| Top BLASTn | ~89% | N/A | ~88% | ~88% |
| HM119586.1 | HM119586.1 | HM119586.1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Davies, C.E.; Malkin, S.H.; Thomas, J.E.; Batista, F.M.; Rowley, A.F.; Coates, C.J. Mycosis Is a Disease State Encountered Rarely in Shore Crabs, Carcinus maenas. Pathogens 2020, 9, 462. https://doi.org/10.3390/pathogens9060462
Davies CE, Malkin SH, Thomas JE, Batista FM, Rowley AF, Coates CJ. Mycosis Is a Disease State Encountered Rarely in Shore Crabs, Carcinus maenas. Pathogens. 2020; 9(6):462. https://doi.org/10.3390/pathogens9060462
Chicago/Turabian StyleDavies, Charlotte E., Sophie H. Malkin, Jessica E. Thomas, Frederico M. Batista, Andrew F. Rowley, and Christopher J. Coates. 2020. "Mycosis Is a Disease State Encountered Rarely in Shore Crabs, Carcinus maenas" Pathogens 9, no. 6: 462. https://doi.org/10.3390/pathogens9060462
APA StyleDavies, C. E., Malkin, S. H., Thomas, J. E., Batista, F. M., Rowley, A. F., & Coates, C. J. (2020). Mycosis Is a Disease State Encountered Rarely in Shore Crabs, Carcinus maenas. Pathogens, 9(6), 462. https://doi.org/10.3390/pathogens9060462





