Characterizing Peri-Implant and Sub-Gingival Microbiota through Culturomics. First Isolation of Some Species in the Oral Cavity. A Pilot Study
Abstract
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Patient Recruitment and Sample Collection
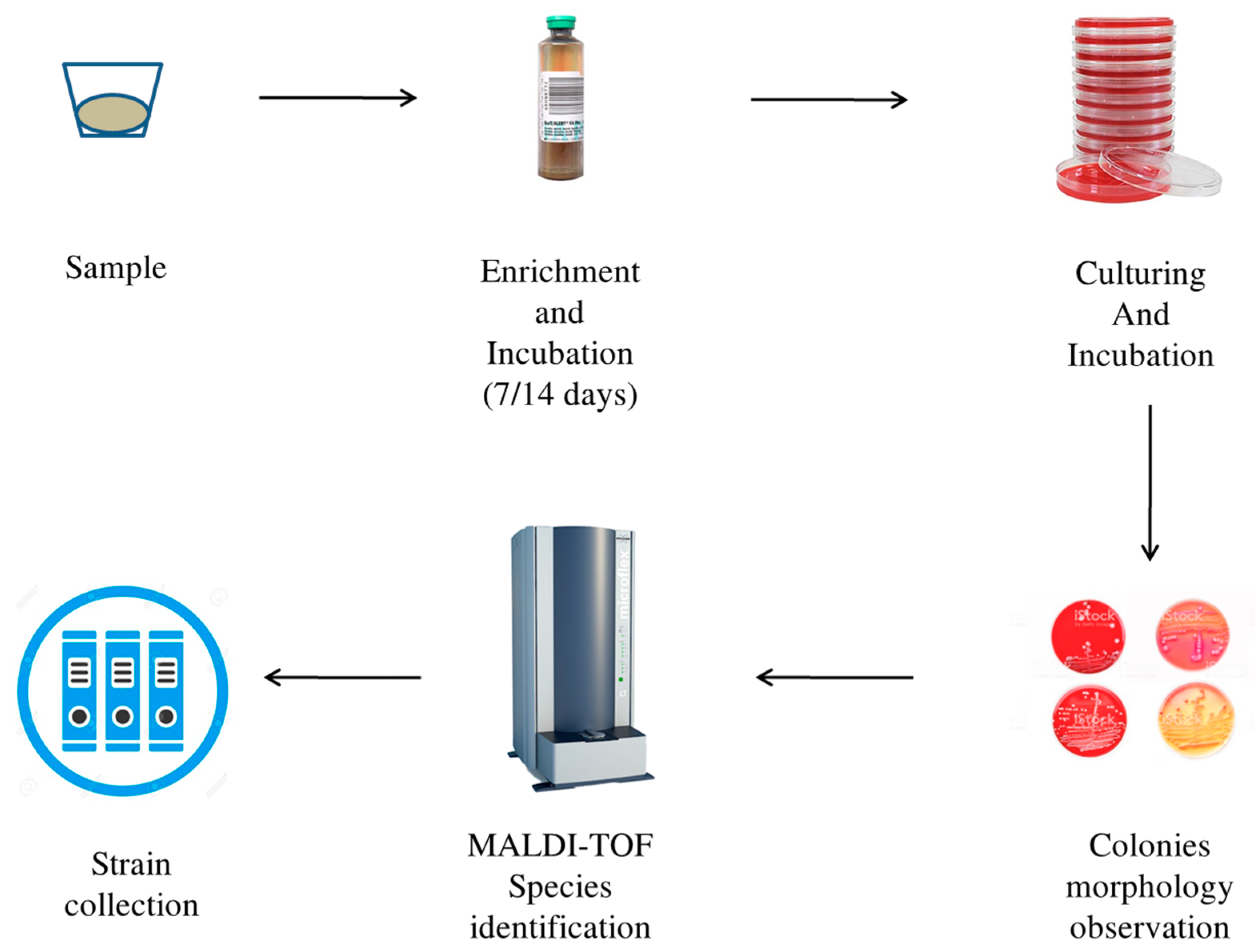
4.2. Culturomics Protocol
4.3. 16S rRNA Gene Sequencing
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Moore, W.E.; Moore, L.V. The bacteria of periodontal diseases. Periodontology 2000 1994, 5, 66–77. [Google Scholar] [CrossRef]
- Teles, R.; Teles, F.; Frias-Lopez, J.; Paster, B.; Haffajee, A. Lessons learned and unlearned in periodontal microbiology. Periodontology 2000 2013, 62, 95–162. [Google Scholar] [CrossRef] [PubMed]
- Socransky, S.S.; Haffajee, A.D.; Smith, G.L.; Dzink, J.L. Difficulties encountered in the search for the etiologic agents of destructive periodontal diseases. J. Clin. Periodontol. 1987, 14, 588–593. [Google Scholar] [CrossRef] [PubMed]
- Leonhardt, A.; Adolfsson, B.; Lekholm, U.; Wikstrom, M.; Dahlen, G. A longitudinal microbiological study on osseointegrated implants in partially edentulous patients. Clin. Oral Implant Res. 1993, 4, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Koka, S.; Razzoog, M.E.; Bloem, T.J.; Syed, S. Microbial colonization of dental implants in partially edentulous subjects. J. Prost. Dent. 1993, 70, 141–144. [Google Scholar] [CrossRef]
- Pontoriero, R.; Tonelli, M.P.; Carnevale, G.; Mombelli, A.; Nyman, S.R.; Lang, N.P. Experimentally induced peri-implant mucositis. A clinical study in humans. Clin. Oral Implant Res. 1994, 5, 254–259. [Google Scholar] [CrossRef]
- Quyrinen, M.; Vogels, R.; Peeters, W.; van Steenberghe, D.; Naert, J.; Haffajee, A.D. Dynamics of initial subgingival colonization of ‘pristine’ peri-implant pockets. Clin. Oral Implant Res. 2006, 17, 25–37. [Google Scholar] [CrossRef]
- Lee, K.H.; Maiden, M.P.; Tanner, A.C.; Weber, H.P. Microbiota of successful osseointegrated dental implants. J. Periodontol. 1999, 70, 131–138. [Google Scholar] [CrossRef]
- Hultin, M.; Gustafsson, A.; Klinge, B. Long-term evaluation of osseointegrated dental implants in the treatment of partly edentulous patients. J. Clin. Periodontol. 2000, 27, 128–133. [Google Scholar] [CrossRef]
- Martellacci, L.; Quaranta, G.; Patini, R.; Isola, G.; Gallenzi, P.; Masucci, L. A Literature Review of Metagenomics and Culturomics of the Peri-implant Microbiome: Current Evidence and Future Perspectives. Materials 2019, 12, 3010. [Google Scholar] [CrossRef]
- Dewhirst, F.E.; Chen, T.; Izard, J.; Paster, B.J.; Tanner, A.C.; Yu, W.H.; Lakshmanan, A.; Wade, W.G. The human oral microbiome. J. Bacteriol. 2010, 192, 5002–5017. [Google Scholar] [CrossRef] [PubMed]
- Paster, B.J.; Boches, S.K.; Galvin, J.L.; Ericson, R.E.; Lau, C.N.; Levanos, V.A.; Sahasrabudhe, A.; Dewhirst, F.E. Bacterial diversity in human subgingival plaque. J. Bacteriol. 2001, 183, 3770–3783. [Google Scholar] [CrossRef]
- Lagier, J.C.; Hugon, P.; Khelaifia, S.; Fournier, P.E.; La Scola, B.; Raoult, D. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin. Microbiol. Rev. 2015, 28, 237–264. [Google Scholar] [CrossRef] [PubMed]
- Lagier, J.C.; Dubourg, G.; Million, M.; Cadoret, F.; Bilen, M.; Fenollar, F.; Levasseur, A.; Rolain, J.M.; Fournier, P.E.; Raoult, D. Culturing the human microbiota and culturomics. Nat. Rev. Microbiol. 2018, 16, 540–550. [Google Scholar] [CrossRef] [PubMed]
- Clark, A.E.; Kaleta, E.J.; Arora, A.; Wolk, D.M. Matrix-assisted laser desorption ionization-time of flight mass spectrometry: A fundamental shift in the routine practice of clinical microbiology. Clin. Microbiol. Rev. 2013, 26, 547–603. [Google Scholar] [CrossRef] [PubMed]
- Mukai, T.; Arihara, K.; Ikeda, A.; Nomura, K.; Suzuki, F.; Ohori, H. Lactobacillus kitasatonis sp. nov., from chicken intestine. Int. J. Syst. Evol. Microbiol. 2003, 53, 2055–2059. [Google Scholar] [CrossRef] [PubMed]
- Rossoni, R.D.; Velloso, M.D.S.; de Barros, P.P.; de Alvarenga, J.A.; Santos, J.D.D.; Santos Prado, A.C.C.D.; Ribeiro, F.C.; Anbinder, A.L.; Junqueira, J.C. Inhibitory effect of probiotic Lactobacillus supernatants from the oral cavity on Streptococcus mutans biofilms. Microb. Pathog. 2018, 123, 361–367. [Google Scholar] [CrossRef]
- Archer, A.C.; Kurrey, N.K.; Halami, P.M. In vitro adhesion and anti-inflammatory properties of native Lactobacillus fermentum and Lactobacillus delbrueckii spp. J. Appl. Microbiol. 2018, 125, 243–256. [Google Scholar] [CrossRef]
- Wasfi, R.; Abd El-Rahman, O.A.; Zafer, M.M.; Ashour, H.M. Probiotic Lactobacillus sp. inhibit growth, biofilm formation and gene expression of caries-inducing Streptococcus mutans. J. Cell. Mol. Med. 2018, 22, 1972–1983. [Google Scholar] [CrossRef]
- Storms, V.; Devriese, L.A.; Coopman, R.; Schumann, P.; Vyncke, F.; Gillis, M. Arthrobacter gandavensis sp. nov., for strains of veterinary origin. Int. J. Syst. Evol. Microbiol. 2003, 53, 1881–1884. [Google Scholar] [CrossRef][Green Version]
- Assadian, O.; Assadian, A.; Senekowitsch, C.; Makristathis, A.; Hagmüller, G. Gas gangrene due to Clostridium perfringens in two injecting drug users in Vienna, Austria. Wien. Klin. Wochenschr. 2004, 116, 264–267. [Google Scholar] [CrossRef] [PubMed]
- Aronoff, D.M.; Kazanjian, P.H. Historical and contemporary features of infections due to Clostridium novyi. Anaerobe 2018, 50, 80–84. [Google Scholar] [CrossRef] [PubMed]
- Bonifaz, A.; Ramírez-Ricarte, I.; Rodríguez-Leviz, A.; Hernández, M.A.; Mena, C.; Valencia, A. Trichomycosis (trichobacteriosis) capitis in an infant: Microbiological, dermoscopic and ultrastructural features. Rev. Chil. Pediatr. 2017, 88, 258–262. [Google Scholar] [PubMed]
- Funke, G.; Hutson, R.A.; Hilleringmann, M.; Heizmann, W.R.; Collins, M.D. Corynebacterium lipophiloflavum sp. nov. isolated from a patient with bacterial vaginosis. FEMS Microbiol. Lett. 1997, 150, 219–224. [Google Scholar] [CrossRef]
- Lee, J.Y.; Kim, S.H.; Jeong, H.S.; Oh, S.H.; Kim, H.R.; Kim, Y.H.; Lee, J.N.; Kook, J.K.; Kho, W.G.; Bae, I.K.; et al. Two cases of peritonitis caused by Kocuria marina in patients undergoing continuous ambulatory peritoneal dialysis. J. Clin. Microbiol. 2009, 47, 3376–3378. [Google Scholar] [CrossRef]
- Szczerba, I.; Krzemiński, Z. Occurrence of bacteria in the mouth from genera of Micrococcus, Kocuria, Nesterenkonia, Kytococcus and Dermacoccus. Med. Dosw. Mikrobiol. 2002, 54, 29–34. [Google Scholar] [PubMed]
- Insulkar, P.; Kerkar, S.; Lele, S.S. Purification and structural-functional characterization of an exopolysaccharide from Bacillus licheniformis PASS26 with in-vitro antitumor and wound healing activities. Int. J. Biol. Macromol. 2018, 120, 1441–1450. [Google Scholar] [CrossRef]
- Rostami, N.; Shields, R.C.; Yassin, S.A.; Hawkins, A.R.; Bowen, L.; Luo, T.L.; Rickard, A.H.; Holliday, R.; Preshaw, P.M.; Jakubovics, N.S. A Critical Role for Extracellular DNA in Dental Plaque Formation. J. Dent. Res. 2017, 96, 208–216. [Google Scholar] [CrossRef]
- Hadad, D.; Geresh, S.; Sivan, A. Biodegradation of polyethylene by the thermophilic bacterium Brevibacillus borstelensis. J. Appl. Microbiol. 2005, 98, 93–100. [Google Scholar] [CrossRef]
- Heyrman, J.; Vanparys, B.; Logan, N.A.; Balcaen, A.; Rodríguez-Díaz, M.; Felske, A.; De Vos, P. Bacillus novalis sp. nov., Bacillus vireti sp. nov., Bacillus soli sp. nov., Bacillus bataviensis sp. nov. and Bacillus drentensis sp. nov., from the Drentse A grasslands. Int. J. Syst. Evol. Microbiol. 2004, 54, 47–57. [Google Scholar] [CrossRef]
- Luna, V.A.; King, D.S.; Peak, K.K.; Reeves, F.; Heberlein-Larson, L.; Veguilla, W.; Heller, L.; Duncan, K.E.; Cannons, A.C.; Amuso, P.; et al. Bacillus anthracis virulent plasmid pX02 genes found in large plasmids of two other Bacillus species. J. Clin. Microbiol. 2006, 44, 2367–2377. [Google Scholar] [CrossRef] [PubMed]
- Patini, R.; Cattani, P.; Marchetti, S.; Isola, G.; Quaranta, G.; Gallenzi, P. Evaluation of Predation Capability of Periodontopathogens Bacteria by Bdellovibrio Bacteriovorus HD100. An in Vitro Study. Materials 2019, 12, 2008. [Google Scholar] [CrossRef] [PubMed]
- Patini, R.; Mangino, G.; Martellacci, L.; Quaranta, G.; Masucci, L.; Gallenzi, P. The effect of different antibiotic regimens on bacterial resistance: A systematic review. Antibiotics 2020, 9, 22. [Google Scholar] [CrossRef] [PubMed]
- Isola, G.; Matarese, G.; Ramaglia, L.; Pedullà, E.; Rapisarda, E.; Iorio-Siciliano, V. Association between periodontitis and glycosylated haemoglobin before diabetes onset: A cross-sectional study. Clin. Oral Investig. 2019. [Google Scholar] [CrossRef] [PubMed]
- Isola, G.; Alibrandi, A.; Currò, M.; Matarese, M.; Ricca, S.; Matarese, G.; Ientile, R.; Kocher, T. Evaluation of salivary and serum ADMA levels in patients with periodontal and cardiovascular disease as subclinical marker of cardiovascular risk. J. Periodontol. 2020. [Google Scholar] [CrossRef]
- Isola, G.; Polizzi, A.; Alibrandi, A.; Indelicato, F.; Ferlito, S. Analysis of Endothelin-1 concentrations in individuals with periodontitis. Sci. Rep. 2020, 10, 1652. [Google Scholar] [CrossRef]
- Isola, G.; Alibrandi, A.; Rapisarda, E.; Matarese, G.; Williams, R.C.; Leonardi, R. Association of Vitamin d in patients with periodontitis: A cross-sectional study. J. Periodontal Res. 2020. [Google Scholar] [CrossRef]
- Isola, G.; Lo Giudice, A.; Polizzi, A.; Alibrandi, A.; Patini, R.; Ferlito, S. Periodontitis and tooth loss have negative systemic impact on circulating progenitors cell levels: A clinical study. Genes 2019, 10, 1022. [Google Scholar] [CrossRef]
- Mascitti, M.; Togni, L.; Troiano, G.; Caponio, V.; Gissi, D.; Montebugnoli, L.; Procaccini, M.; Lo Muzio, L.; Santarelli, A. Beyond Head and Neck Cancer: The Relationship between Oral Microbiota and Tumour Development in Distant Organs. Front. Cell. Infect. Microbiol. 2019, 9, 232. [Google Scholar] [CrossRef]
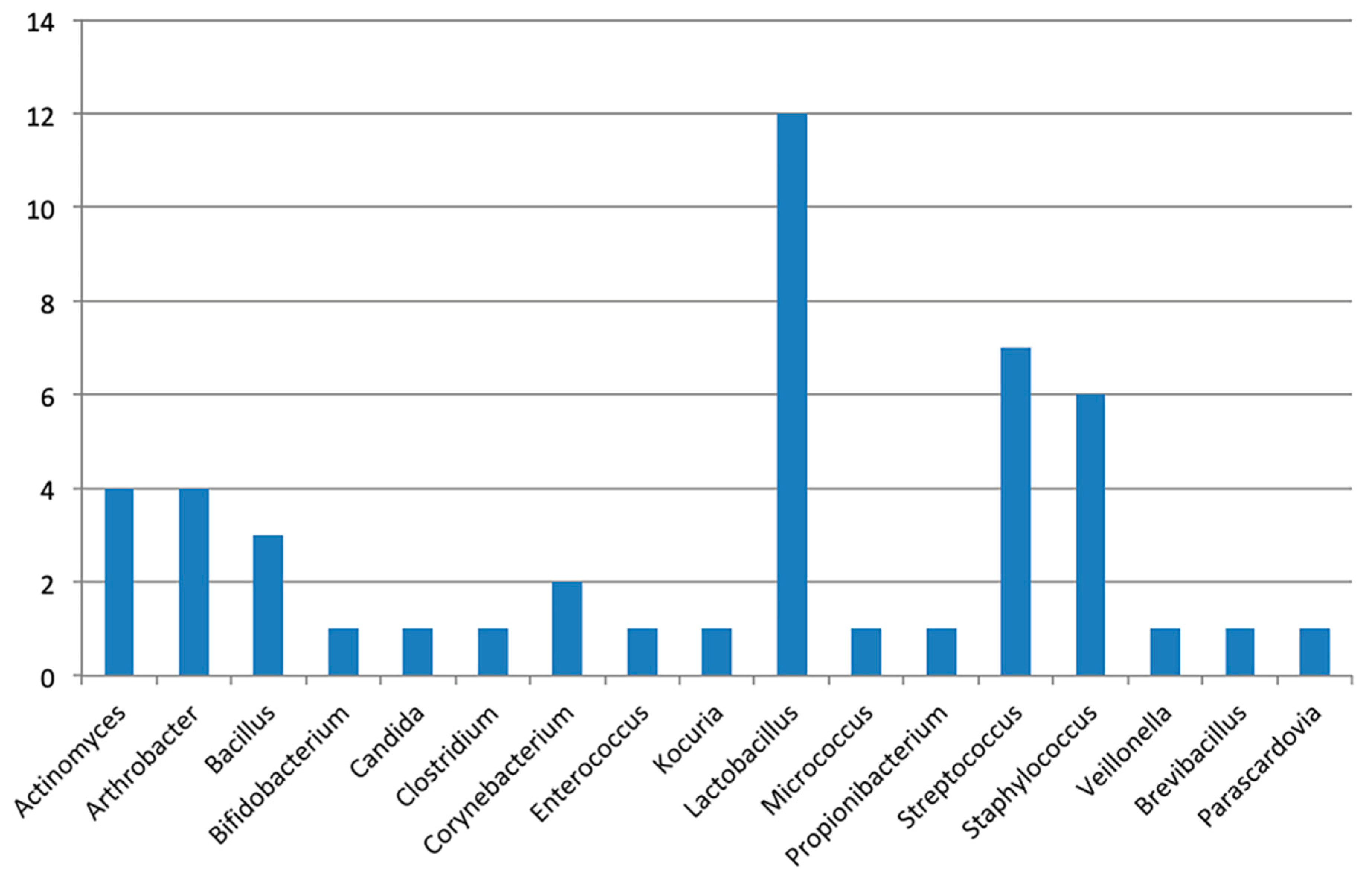
| Patient (Sex) | Age | Smoke | Antibiotic Therapy | Systemic Disease | Active Oral Disease | Implant Site | Implant PD/BoP | Tooth Site | Tooth PD/BoP | Prosthesis |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 (F) | 36 | No | None | None | None | 3.7 | 2/Absent | 3.6 | 3/None | Crown |
| 2 (F) | 62 | No | None | HP | None | 4.6 | 2/Absent | NA | NA | Toronto Bridge |
| 3 (F) | 73 | No | None | None | None | 4.5 | 1/Absent | 4.4 | 1/None | Crown |
| 4 (F) | 68 | No | None | HP | None | 3.6 | 2/Absent | 3.5 | 2/None | Crown |
| Patient | Samples Site | Culture Time | Species Identified by Culturomics |
|---|---|---|---|
| P 1 | implant | 7 days | A. odontolyticus, A. gandavensis *, B. dentium, L. casei, L. fermentum, L. gasseri, L. rhamnosus, S. capitis, S. gordonii, S. haemolyticus |
| 14 days | L. fermentum, L. gasseri, L. rhamnosus, M. luteus, P. acnes, S. epidermidis, S. mutans | ||
| tooth | 7 days | A. odontolyticus, A. oris, C. albicans, C. novyi **, E. faecalis, L. fermentum, L. gasseri, L. salivarius, S. epidermidis, S. capitis, S. cohnii, | |
| 14 days | A. odontolyticus, A. oris, C. albicans, L. fermentum, L. kitasatonis, L. salivarius, S.simulans | ||
| P 2 | implant | 7 days | A. odontolyticus, B. dentium, C. flavescens **, K. marina **, L. reuteri, S. epidermidis, S. gordonii |
| 14 days | A. odontolyticus, A. oris, A. oxydans **, A. protophormiae **, B. licheniformis **, B. dentium, B. borstelensis *, C. lipophiloflavum **, L. plantarum, L. reuteri, M. luteus, P. denticolens, P. acnes, S. capitis, S. epidermidis, S. haemolyticus | ||
| P 3 | implant | 7 days | B. dentium, S. epidermidis, S. hominis, S. intermedius, S. mutans, S. salivarius |
| 14 days | A. naeslundii, B. dentium, L. gasseri, S. epidermidis, S. hominis, S. mutans | ||
| tooth | 7 days | A. oris, S. mitis | |
| 14 days | P. acnes, S. epidermidis, S. agalactiae | ||
| P 4 | implant | 7 days | A. odontolyticus, B. dentium, C. albicans, E. faecalis, L. delbrueckii, L. fermentum, L. paracasei, L. rhamnosus, M. luteus, S. epidermidis, V. parvula |
| 14 days | A. dentalis, A. odontolyticus, A. tumbae*, B. bataviensis *, B. dentium, C. albicans, E. faecalis, L. casei, L. fermentum, L. gasseri, L. paracasei, L. rhamnosus | ||
| tooth | 7 days | A. odontolyticus, B. luciferensis *, C. albicans, E. faecalis, L. gasseri, L. rhamnosus, S. capitis | |
| 14 days | E. faecalis, L. gasseri, L. rhamnosus, L. buchneri, L. parabuchneri |
| Pre-incubation in aerobic blood culture bottle for 7 and 14 days at 30 °C. Culturing on TSA, CNA, PVX in aerobic condition at 30 °C. |
| Pre-incubation in aerobic blood culture bottle for 7 and 14 days at 30 °C. Culturing on TSA, CNA, PVX in microaerophilic condition at 30 °C. |
| Pre-incubation in anaerobic blood culture bottle for 7 and 14 days at 30 °C. Culturing on CNA and SCH in anaerobic condition at 30 °C. |
| Pre-incubation in aerobic blood culture bottle for 7 and 14 days at 37 °C. Culturing on TSA, CNA, PVX in aerobic condition at 37 °C. |
| Pre-incubation in aerobic blood culture bottle for 7 and 14 days at 37 °C. Culturing on TSA, CNA, PVX in microaerophilic condition at 37 °C. |
| Pre-incubation in anaerobic blood culture bottle for 7 and 14 days at 37 °C. Culturing on CNA and SCH in anaerobic condition at 37 °C. |
| Pre-incubation in aerobic blood culture bottle for 7 and 14 days at 42 °C. Culturing on TSA, CNA, PVX in aerobic condition at 42 °C. |
| Pre-incubation in aerobic blood culture bottle for 7 and 14 days at 42 °C. Culturing on TSA, CNA, PVX in microaerophilic condition at 42 °C. |
| Pre-incubation in anaerobic blood culture bottle for 7 and 14 days at 42 °C. Culturing on CNA and SCH in anaerobic condition at 42 °C. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martellacci, L.; Quaranta, G.; Fancello, G.; D’Addona, A.; Sanguinetti, M.; Patini, R.; Masucci, L. Characterizing Peri-Implant and Sub-Gingival Microbiota through Culturomics. First Isolation of Some Species in the Oral Cavity. A Pilot Study. Pathogens 2020, 9, 365. https://doi.org/10.3390/pathogens9050365
Martellacci L, Quaranta G, Fancello G, D’Addona A, Sanguinetti M, Patini R, Masucci L. Characterizing Peri-Implant and Sub-Gingival Microbiota through Culturomics. First Isolation of Some Species in the Oral Cavity. A Pilot Study. Pathogens. 2020; 9(5):365. https://doi.org/10.3390/pathogens9050365
Chicago/Turabian StyleMartellacci, Leonardo, Gianluca Quaranta, Giovanni Fancello, Antonio D’Addona, Maurizio Sanguinetti, Romeo Patini, and Luca Masucci. 2020. "Characterizing Peri-Implant and Sub-Gingival Microbiota through Culturomics. First Isolation of Some Species in the Oral Cavity. A Pilot Study" Pathogens 9, no. 5: 365. https://doi.org/10.3390/pathogens9050365
APA StyleMartellacci, L., Quaranta, G., Fancello, G., D’Addona, A., Sanguinetti, M., Patini, R., & Masucci, L. (2020). Characterizing Peri-Implant and Sub-Gingival Microbiota through Culturomics. First Isolation of Some Species in the Oral Cavity. A Pilot Study. Pathogens, 9(5), 365. https://doi.org/10.3390/pathogens9050365








