A New Type of Chronic Wound Infection after Wisdom Tooth Extraction: A Diagnostic Approach with 16S-rRNA Gene Analysis, Next-Generation Sequencing, and Bioinformatics
Abstract
1. Introduction
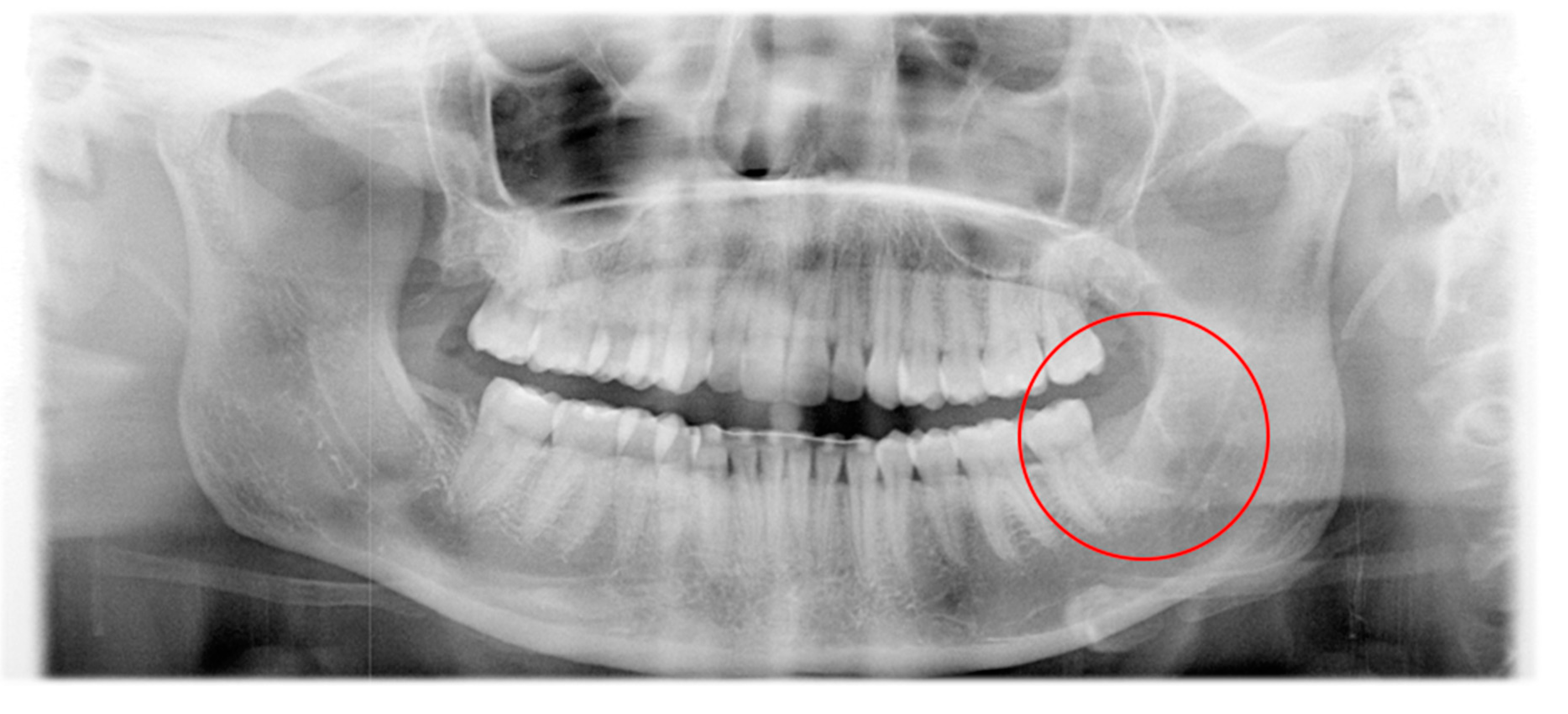
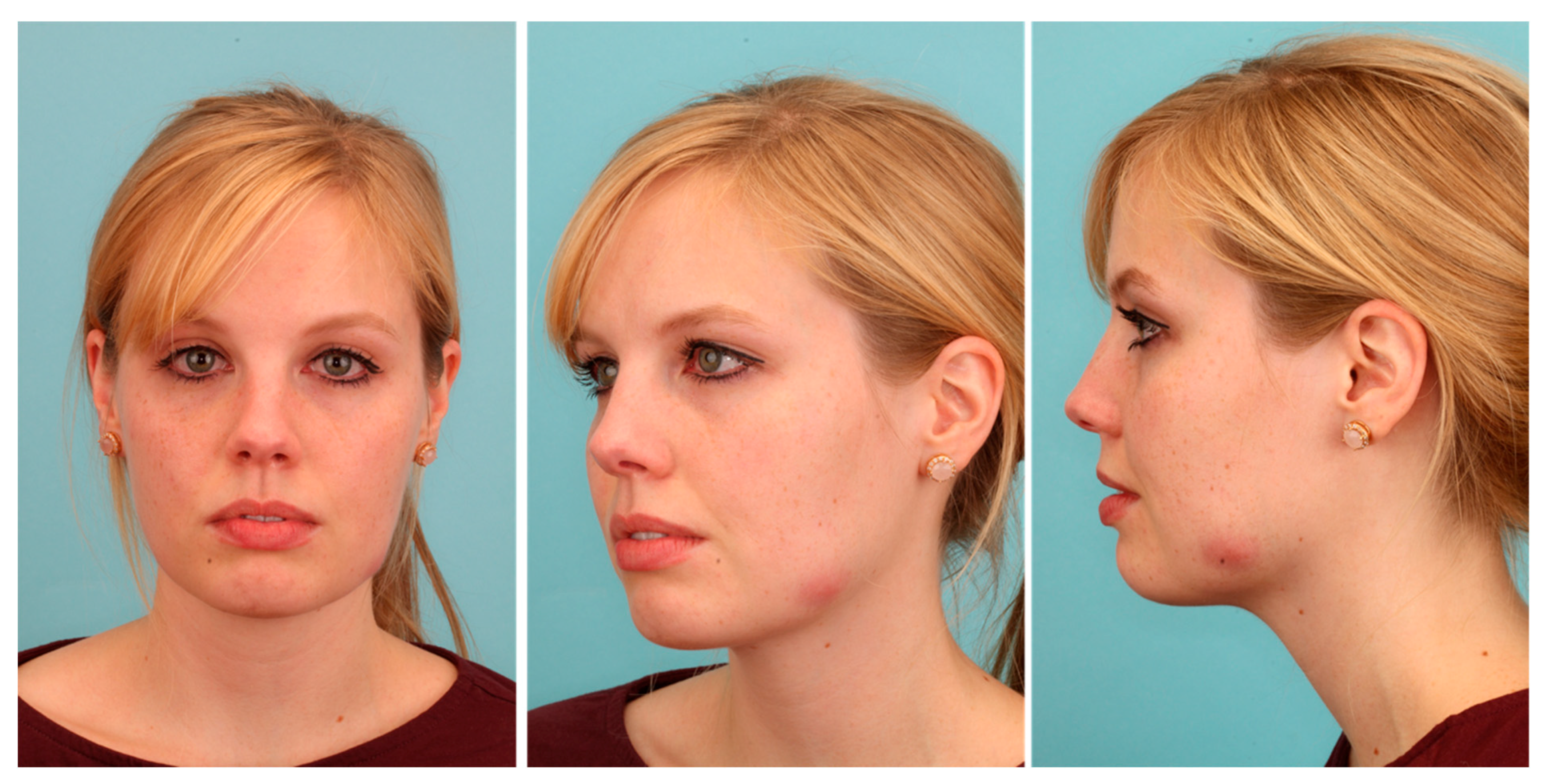
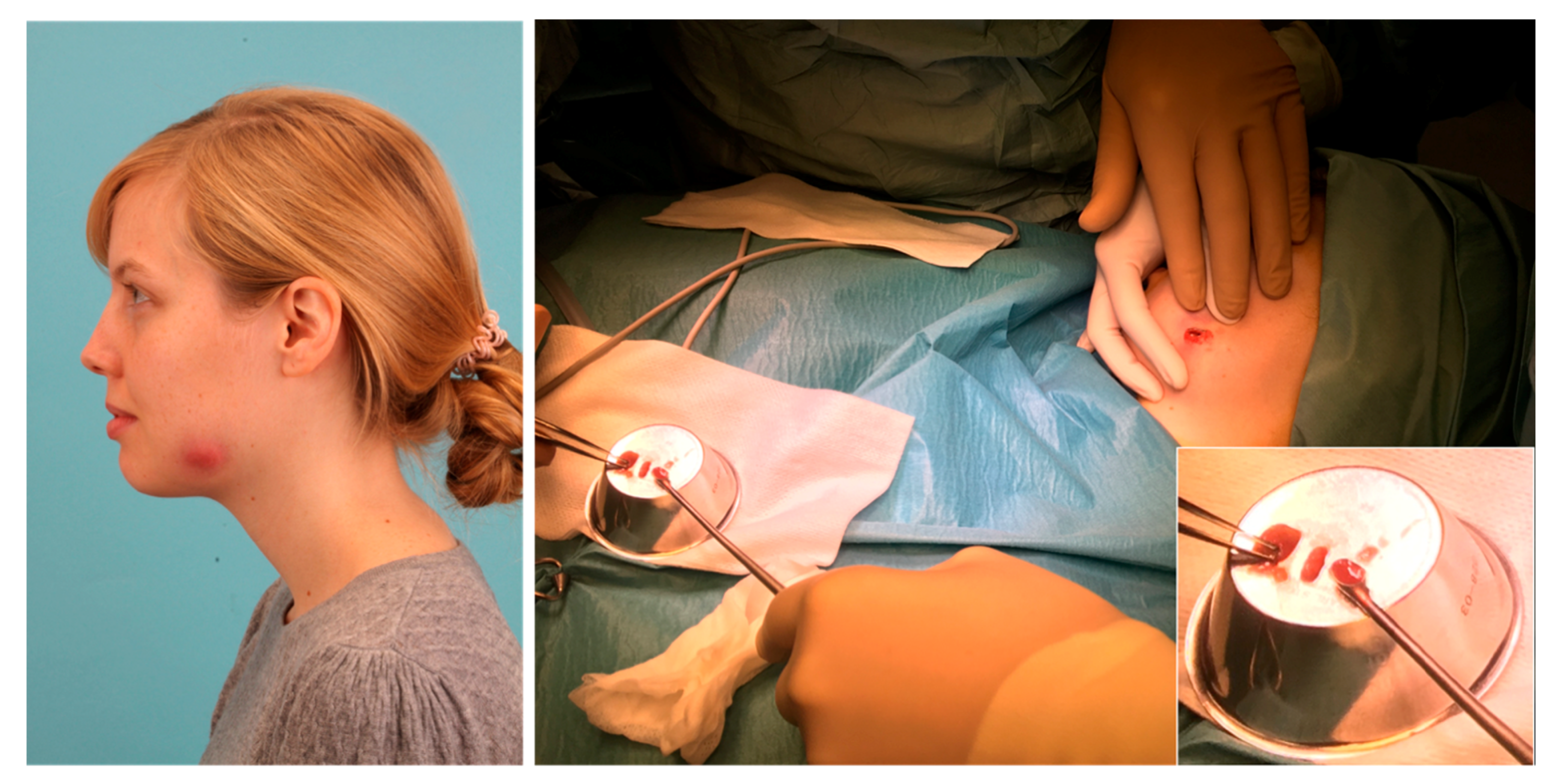
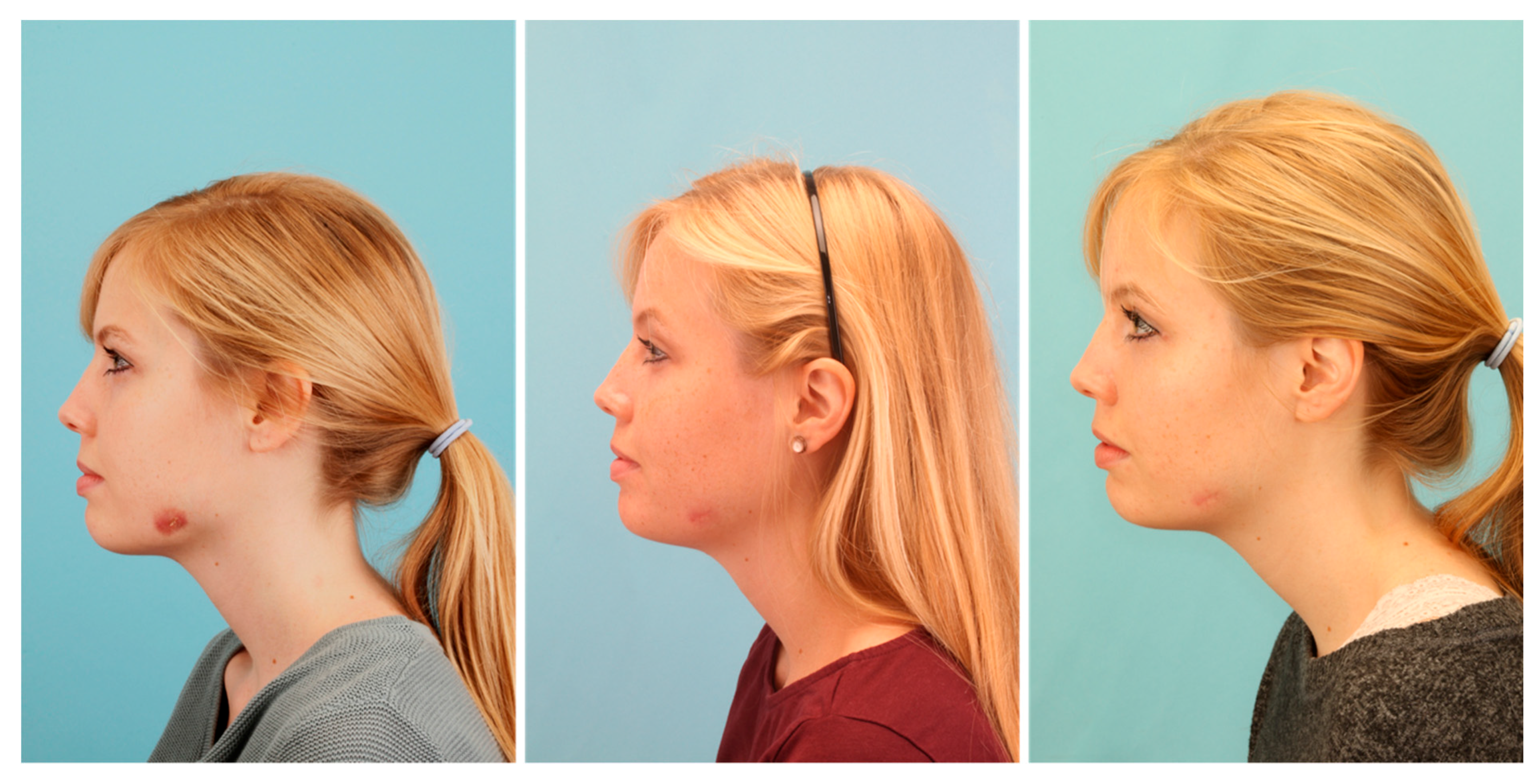
2. Patient and Methods
3. Results
4. Discussion
- The infection of the patient was completely atypical from the clinical point of view. A typical odontogenic infection can usually be managed well with surgery and antibiotic therapy. In the present case, it took more than a year to cure the patient.
- In the case of a normal odontogenic infection, bacteria can usually be cultured from the pus. However, the culture, in this case, was negative, which is probably because the cultural conditions were not optimal or sufficient for the later detected atypical bacterial genera.
- The determined microbiome was completely different from the one we would expect in a normal odontogenic abscess. As described by Figueiredo et al. [9], odontogenic infections show a polymicrobial spectrum with mostly obligate anaerobic bacteria. In the microbiome of the abscess of the present case, we mostly found aerobic environmental bacteria in combination with anaerobic bacteria of the oral cavity. Taking into account that the pus sample was taken from an extra-oral incision, we assume that the infection can be caused by a combination of these bacteria.
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wiese, K.G.; Merten, H.A.; Wiltfang, J.; Luhr, H.G. Clinical studies on the pathophysiology of odontogenic abscesses. Mund Kiefer Gesichtschir. 1999, 3, 242–246. [Google Scholar] [CrossRef] [PubMed]
- Warnke, P.H.; Becker, S.T.; Springer, I.N.; Haerle, F.; Ullmann, U.; Russo, P.A.; Wiltfang, J.; Fickenscher, H.; Schubert, S. Penicillin compared with other advanced broad spectrum antibiotics regarding antibacterial activity against oral pathogens isolated from odontogenic abscesses. J. Craniomaxillofac. Surg. 2008, 36, 462–467. [Google Scholar] [CrossRef] [PubMed]
- Al-Nawas, B.; Maeurer, M. Severe versus local odontogenic bacterial infections: Comparison of microbial isolates. Eur. Surg. Res. 2008, 40, 220–224. [Google Scholar] [CrossRef] [PubMed]
- Al-Nawas, B.; Karbach, J. S3-Leitlinie (Langversion): Odontogene Infektionen. In Leitlinien Zahnmedizin; Arbeitsgemeinschaft der Wissenschaftlichen Medizinischen Fachgesellschaften, 2016; Available online: www.awmf.org (accessed on 10 March 2020).
- Böttger, S.; Lautenbacher, K.; Domann, E.; Howaldt, H.-P.; Attia, S.; Streckbein, P.; Wilbrand, J.-F. Indication for an additional postoperative antibiotic treatment after surgical incision of serious odontogenic abscesses. J. Cranio-Maxillofac. Surg. 2020, 43. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, L.M.; Trindade, S.C.; Sarmento, V.A.; de Oliveira, T.F.; Muniz, W.R.; Valente, R.O. Actinomycotic osteomyelitis of the mandible: An unusual case. Oral Maxillofac. Surg. 2013, 17, 299–302. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, R.; Valmaseda-Castellon, E.; Berini-Aytes, L.; Gay-Escoda, C. Incidence and clinical features of delayed-onset infections after extraction of lower third molars. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 2005, 99, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, R.; Valmaseda-Castellon, E.; Laskin, D.M.; Berini-Aytes, L.; Gay-Escoda, C. Treatment of delayed-onset infections after impacted lower third molar extraction. J. Oral Maxillofac Surg. 2008, 66, 943–947. [Google Scholar] [CrossRef]
- Figueiredo, R.; Valmaseda-Castellon, E.; Formoso-Senande, M.F.; Berini-Aytes, L.; Gay-Escoda, C. Delayed-onset infections after impacted lower third molar extraction: Involved bacteria and sensitivity profiles to commonly used antibiotics. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2012, 114, 43–48. [Google Scholar] [CrossRef]
- Brunello, G.; De Biagi, M.; Crepaldi, G.; Rodrigues, F.I.; Sivolella, S. An Observational Cohort Study on Delayed-Onset Infections after Mandibular Third-Molar Extractions. Int. J. Dent. 2017, 2017, 1435348. [Google Scholar] [CrossRef]
- Sukegawa, S.; Yokota, K.; Kanno, T.; Manabe, Y.; Sukegawa-Takahashi, Y.; Masui, M.; Furuki, Y. What are the risk factors for postoperative infections of third molar extraction surgery: A retrospective clinical study? Med. Oral Patol Oral Cir. Bucal. 2019, 24, e123–e129. [Google Scholar] [CrossRef]
- Valour, F.; Sénéchal, A.; Dupieux, C.; Karsenty, J.; Lustig, S.; Breton, P.; Gleizal, A.; Boussel, L.; Laurent, F.; Braun, E.; et al. Actinomycosis: Etiology, clinical features, diagnosis, treatment, and management. Infect. Drug Resist. 2014, 7, 183–197. [Google Scholar] [CrossRef]
- Wong, V.K.; Turmezei, T.D.; Weston, V.C. Actinomycosis. BMJ 2011, 343, d6099. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Navarro, B.; Arranz-Obispo, C.; Albuquerque, R.; Jane-Salas, E.; Lopez-Lopez, J. Osteomyelitis of the jaw (with pathological fracture) following extraction of an impacted wisdom tooth. A case report. J. Stomatol Oral Maxillofac Surg. 2017, 118, 306–309. [Google Scholar] [CrossRef] [PubMed]
- Bayazit, Y.A.; Bayazit, N.; Namiduru, M. Mycobacterial cervical lymphadenitis. ORL J. Otorhinolaryngol Relat Spec. 2004, 66, 275–280. [Google Scholar] [CrossRef] [PubMed]
- Thavagnanam, S.; McLoughlin, L.M.; Hill, C.; Jackson, P.T. Atypical Mycobacterial infections in children: The case for early diagnosis. Ulster Med. J. 2006, 75, 192–194. [Google Scholar]
- Keller, P.M.; Hombach, M.; Bloemberg, G.V. 16S-rRNA-Gen-basierte Identifikation bakterieller Infektionen. Biospektrum: Das Magazin für Biowissenschaften 2010, 16, 755–758. [Google Scholar]
- Woo, P.C.; Lau, S.K.; Teng, J.L.; Tse, H.; Yuen, K.Y. Then and now: Use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin. Microbiol. Infect. 2008, 14, 908–934. [Google Scholar] [CrossRef]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ. 2016, 4, e2584. [Google Scholar] [CrossRef]
- Goldberg, M.H.; Nemarich, A.N.; Marco, W.P., 2nd. Complications after mandibular third molar surgery: A statistical analysis of 500 consecutive procedures in private practice. J. Am. Dent. Assoc. 1985, 111, 277–279. [Google Scholar] [CrossRef] [PubMed]
- Christiaens, I.; Reychler, H. [Complications after third molar extractions: Retrospective analysis of 1,213 teeth]. Rev. Stomatol. Chir. Maxillofac. 2002, 103, 269–274. [Google Scholar]
- Goldberg, M.H.; Galbraith, D.A. Late onset of mandibular and lingual dysesthesia secondary to postextraction infection. Oral Surg. Oral Med. Oral Pathol. 1984, 58, 269–271. [Google Scholar] [CrossRef]
- Piecuch, J.F.; Arzadon, J.; Lieblich, S.E. Prophylactic antibiotics for third molar surgery: A supportive opinion. J. Oral Maxillofac Surg. 1995, 53, 53–60. [Google Scholar] [CrossRef]
- Boon, L.C. Late presentation of actinomycosis after third molar surgery. Med. J. Malaysia 1987, 42, 207–208. [Google Scholar] [PubMed]
- Bonnefond, S.; Catroux, M.; Melenotte, C.; Karkowski, L.; Rolland, L.; Trouillier, S.; Raffray, L. Clinical features of actinomycosis: A retrospective, multicenter study of 28 cases of miscellaneous presentations. Medicine (Baltimore) 2016, 95. [Google Scholar] [CrossRef]
- Bennhoff, D.F. Actinomycosis: Diagnostic and therapeutic considerations and a review of 32 cases. Laryngoscope 1984, 94, 1198–1217. [Google Scholar] [CrossRef]
- Eckert, A.W.; Hohne, C.; Schubert, J. Pathogen spectrum and resistance status of exclusively anaerobic odontogenic infections. Mund Kiefer Gesichtschir. 2000, 4, 153–158. [Google Scholar] [CrossRef]
- Riggio, M.P.; Aga, H.; Murray, C.A.; Jackson, M.S.; Lennon, A.; Hammersley, N.; Bagg, J. Identification of bacteria associated with spreading odontogenic infections by 16S rRNA gene sequencing. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 2007, 103, 610–617. [Google Scholar] [CrossRef]
- Khan, S.; Sistla, S.; Dhodapkar, R.; Parija, S.C. Fatal Delftia acidovorans infection in an immunocompetent patient with empyema. Asian Pac. J. Trop Biomed. 2012, 2, 923–924. [Google Scholar] [CrossRef]
- Bilgin, H.; Sarmis, A.; Tigen, E.; Soyletir, G.; Mulazimoglu, L. Delftia acidovorans: A rare pathogen in immunocompetent and immunocompromised patients. Can. J. Infect. Dis Med. Microbiol. 2015, 26, 277–279. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, I.; Yagi, T.; Hatakeyama, K.; Ohkura, T.; Ohkusu, K.; Takahashi, Y.; Kojima, S.; Hasegawa, Y. Recurrent vascular catheter-related bacteremia caused by Delftia acidovorans with different antimicrobial susceptibility profiles. J. Infect. Chemother. 2011, 17, 111–113. [Google Scholar] [CrossRef] [PubMed]
- Chotikanatis, K.; Backer, M.; Rosas-Garcia, G.; Hammerschlag, M.R. Recurrent intravascular-catheter-related bacteremia caused by Delftia acidovorans in a hemodialysis patient. J. Clin. Microbiol. 2011, 49, 3418–3421. [Google Scholar] [CrossRef] [PubMed]
- Mahmood, S.; Taylor, K.E.; Overman, T.L.; McCormick, M.I. Acute infective endocarditis caused by Delftia acidovorans, a rare pathogen complicating intravenous drug use. J. Clin. Microbiol. 2012, 50, 3799–3800. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.M.; Kim, M.K.; Lee, J.L.; Wee, W.R.; Lee, J.H. Experience of Comamonas acidovorans keratitis with delayed onset and treatment response in immunocompromised cornea. Korean J. Ophthalmol. 2008, 22, 49–52. [Google Scholar] [CrossRef]
- Camargo, C.H.; Ferreira, A.M.; Javaroni, E.; Reis, B.A.; Bueno, M.F.; Francisco, G.R.; Gallo, J.F.; Garcia Dde, O. Microbiological characterization of Delftia acidovorans clinical isolates from patients in an intensive care unit in Brazil. Diagn. Microbiol. Infect. Dis. 2014, 80, 330–333. [Google Scholar] [CrossRef]
- Muchesa, P.; Leifels, M.; Jurzik, L.; Hoorzook, K.B.; Barnard, T.G.; Bartie, C. Coexistence of free-living amoebae and bacteria in selected South African hospital water distribution systems. Parasitol. Res. 2017, 116, 155–165. [Google Scholar] [CrossRef]
- Szymanska, J.; Sitkowska, J.; Dutkiewicz, J. Microbial contamination of dental unit waterlines. Ann. Agric. Environ. Med. 2008, 15, 173–179. [Google Scholar]
- Yakimov, M.M.; Golyshin, P.N.; Lang, S.; Moore, E.R.; Abraham, W.R.; Lunsdorf, H.; Timmis, K.N. Alcanivorax borkumensis gen. nov., sp. nov., a new, hydrocarbon-degrading and surfactant-producing marine bacterium. Int. J. Syst. Bacteriol. 1998, 48, 339–348. [Google Scholar] [CrossRef]
- Naether, D.J.; Slawtschew, S.; Stasik, S.; Engel, M.; Olzog, M.; Wick, L.Y.; Timmis, K.N.; Heipieper, H.J. Adaptation of the hydrocarbonoclastic bacterium Alcanivorax borkumensis SK2 to alkanes and toxic organic compounds: A physiological and transcriptomic approach. Appl. Environ. Microbiol. 2013, 79, 4282–4293. [Google Scholar] [CrossRef]
- Kasai, Y.; Kishira, H.; Syutsubo, K.; Harayama, S. Molecular detection of marine bacterial populations on beaches contaminated by the Nakhodka tanker oil-spill accident. Environ. Microbiol. 2001, 3, 246–255. [Google Scholar] [CrossRef] [PubMed]
- Godfrin, M.P.; Sihlabela, M.; Bose, A.; Tripathi, A. Behavior of Marine Bacteria in Clean Environment and Oil Spill Conditions. Langmuir 2018, 34, 9047–9053. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.F.; Johnston, A.M.; Johnson, B.; Huntington, M.K.; Mackenzie, C.D. Microbial contamination of dental unit waterlines: Prevalence, intensity and microbiological characteristics. J. Am. Dent. Assoc. 1993, 124, 59–65. [Google Scholar] [CrossRef] [PubMed]

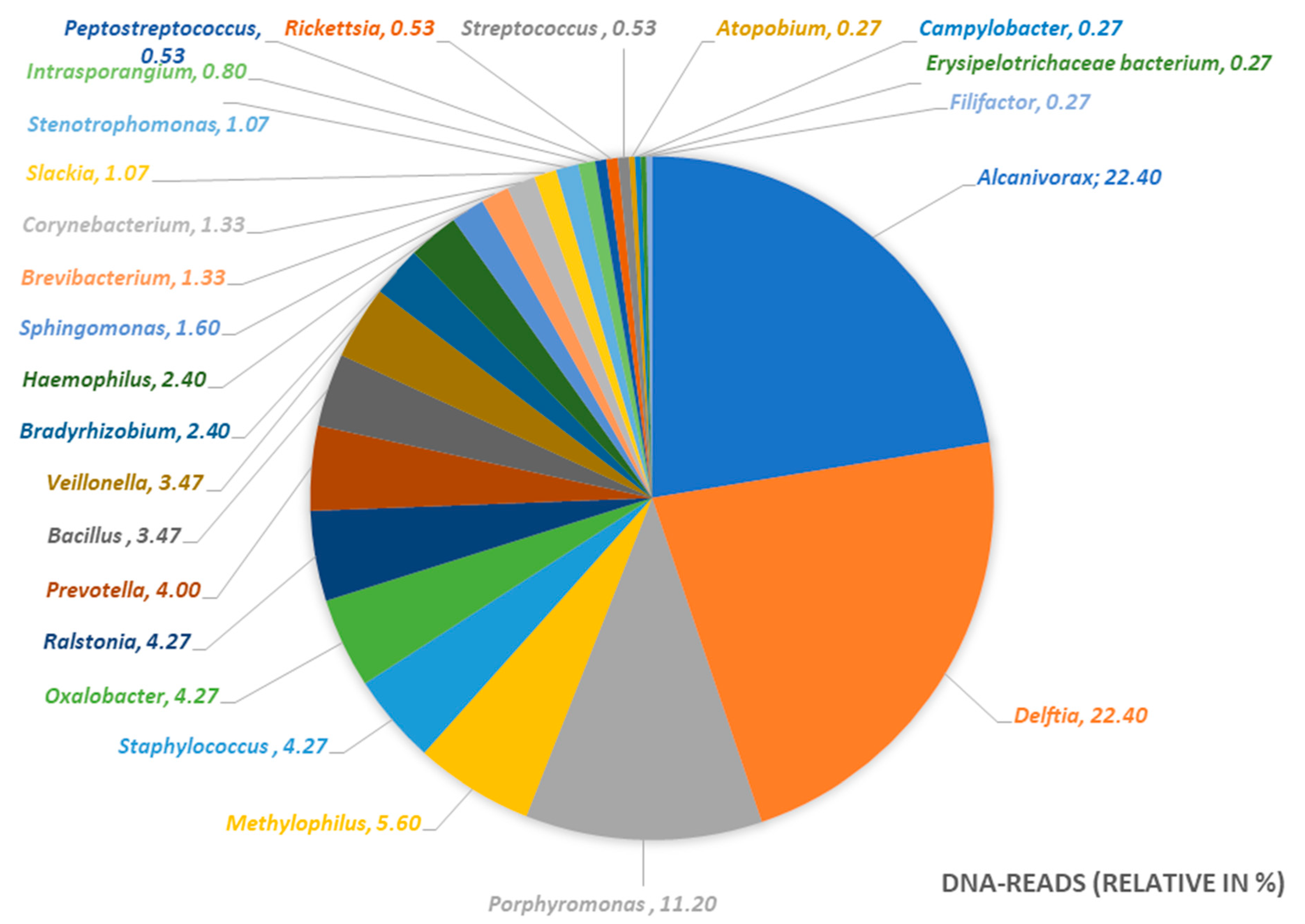
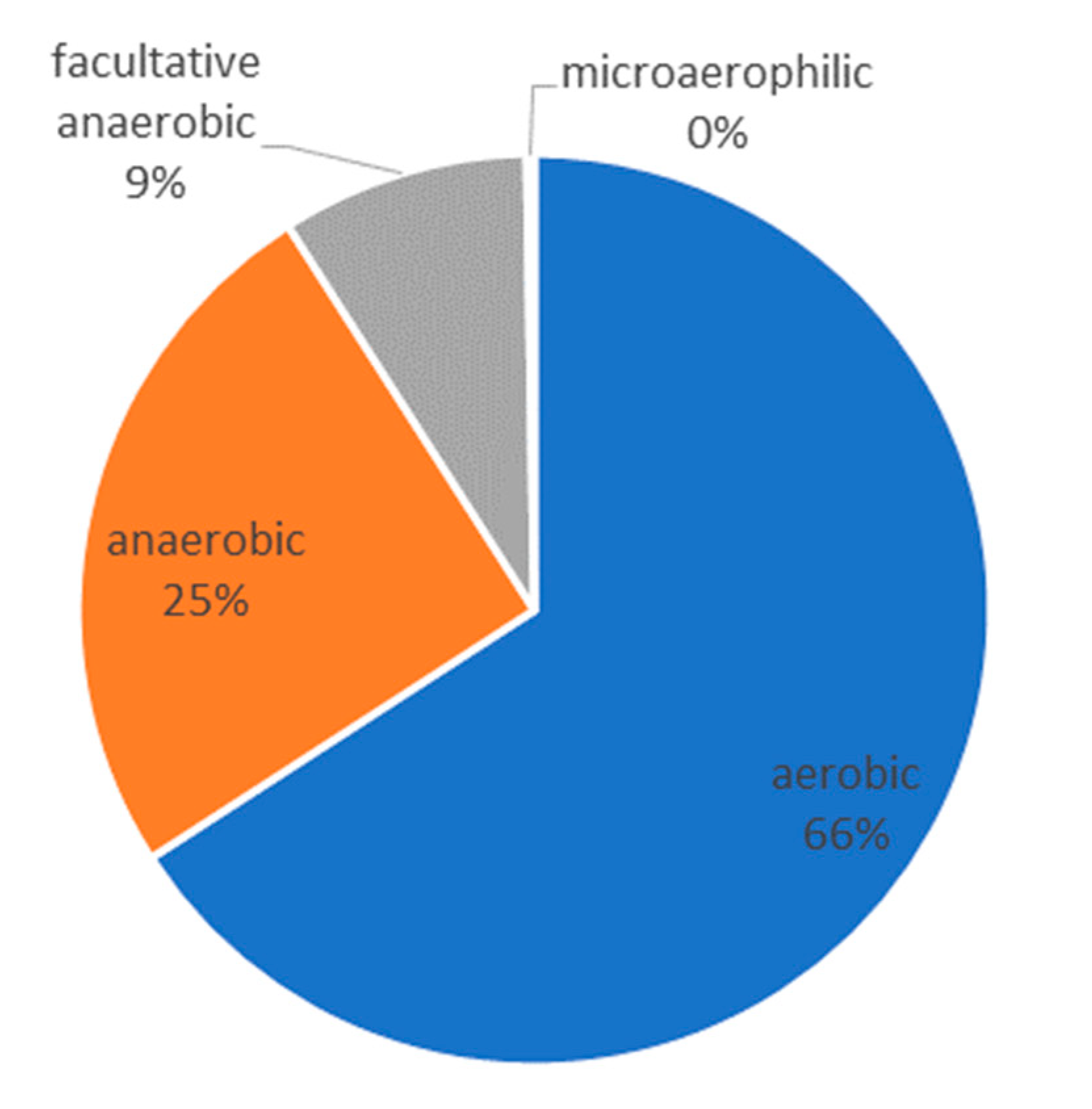
| Bacterial Genus | DNA-Reads (Relative in %) | Metabolism |
|---|---|---|
| Alcanivorax | 22.40 | aerobic |
| Delftia | 22.40 | aerobic |
| Porphyromonas | 11.20 | anaerobic |
| Methylophilus | 5.60 | aerobic |
| Staphylococcus | 4.27 | facultative anaerobic |
| Oxalobacter | 4.27 | anaerobic |
| Ralstonia | 4.27 | aerobic |
| Prevotella | 4.00 | anaerobic |
| Bacillus | 3.47 | aerobic |
| Veillonella | 3.47 | anaerobic |
| Bradyrhizobium | 2.40 | aerobic |
| Haemophilus | 2.40 | facultative anaerobic |
| Sphingomonas | 1.60 | aerobic |
| Brevibacterium | 1.33 | aerobic |
| Corynebacterium | 1.33 | facultative anaerobic |
| Slackia | 1.07 | anaerobic |
| Stenotrophomonas | 1.07 | aerobic |
| Intrasporangium | 0.80 | aerobic |
| Peptostreptococcus | 0.53 | anaerobic |
| Rickettsia | 0.53 | aerobic |
| Streptococcus | 0.53 | facultative anaerobic |
| Atopobium | 0.27 | facultative anaerobic |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Böttger, S.; Zechel-Gran, S.; Streckbein, P.; Knitschke, M.; Hain, T.; Weigel, M.; Wilbrand, J.-F.; Domann, E.; Howaldt, H.-P.; Attia, S. A New Type of Chronic Wound Infection after Wisdom Tooth Extraction: A Diagnostic Approach with 16S-rRNA Gene Analysis, Next-Generation Sequencing, and Bioinformatics. Pathogens 2020, 9, 798. https://doi.org/10.3390/pathogens9100798
Böttger S, Zechel-Gran S, Streckbein P, Knitschke M, Hain T, Weigel M, Wilbrand J-F, Domann E, Howaldt H-P, Attia S. A New Type of Chronic Wound Infection after Wisdom Tooth Extraction: A Diagnostic Approach with 16S-rRNA Gene Analysis, Next-Generation Sequencing, and Bioinformatics. Pathogens. 2020; 9(10):798. https://doi.org/10.3390/pathogens9100798
Chicago/Turabian StyleBöttger, Sebastian, Silke Zechel-Gran, Philipp Streckbein, Michael Knitschke, Torsten Hain, Markus Weigel, Jan-Falco Wilbrand, Eugen Domann, Hans-Peter Howaldt, and Sameh Attia. 2020. "A New Type of Chronic Wound Infection after Wisdom Tooth Extraction: A Diagnostic Approach with 16S-rRNA Gene Analysis, Next-Generation Sequencing, and Bioinformatics" Pathogens 9, no. 10: 798. https://doi.org/10.3390/pathogens9100798
APA StyleBöttger, S., Zechel-Gran, S., Streckbein, P., Knitschke, M., Hain, T., Weigel, M., Wilbrand, J.-F., Domann, E., Howaldt, H.-P., & Attia, S. (2020). A New Type of Chronic Wound Infection after Wisdom Tooth Extraction: A Diagnostic Approach with 16S-rRNA Gene Analysis, Next-Generation Sequencing, and Bioinformatics. Pathogens, 9(10), 798. https://doi.org/10.3390/pathogens9100798







