Paenibacillus alvei MP1 as a Producer of the Proteinaceous Compound with Activity against Important Human Pathogens, Including Staphylococcus aureus and Listeria monocytogenes †
Abstract
1. Introduction
2. Results
2.1. Strain Growth Condition Associated with an Active Compound Production
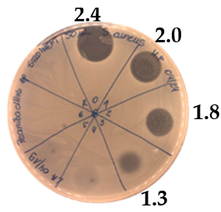
2.2. Inhibitory Spectrum of P. alvei MP1 Cell-Free Precipitates against Indicator Strains
2.3. Effects of Enzymes and Temperature on Antimicrobial Activity of P.alvei MP1 Cell-Free Precipitates
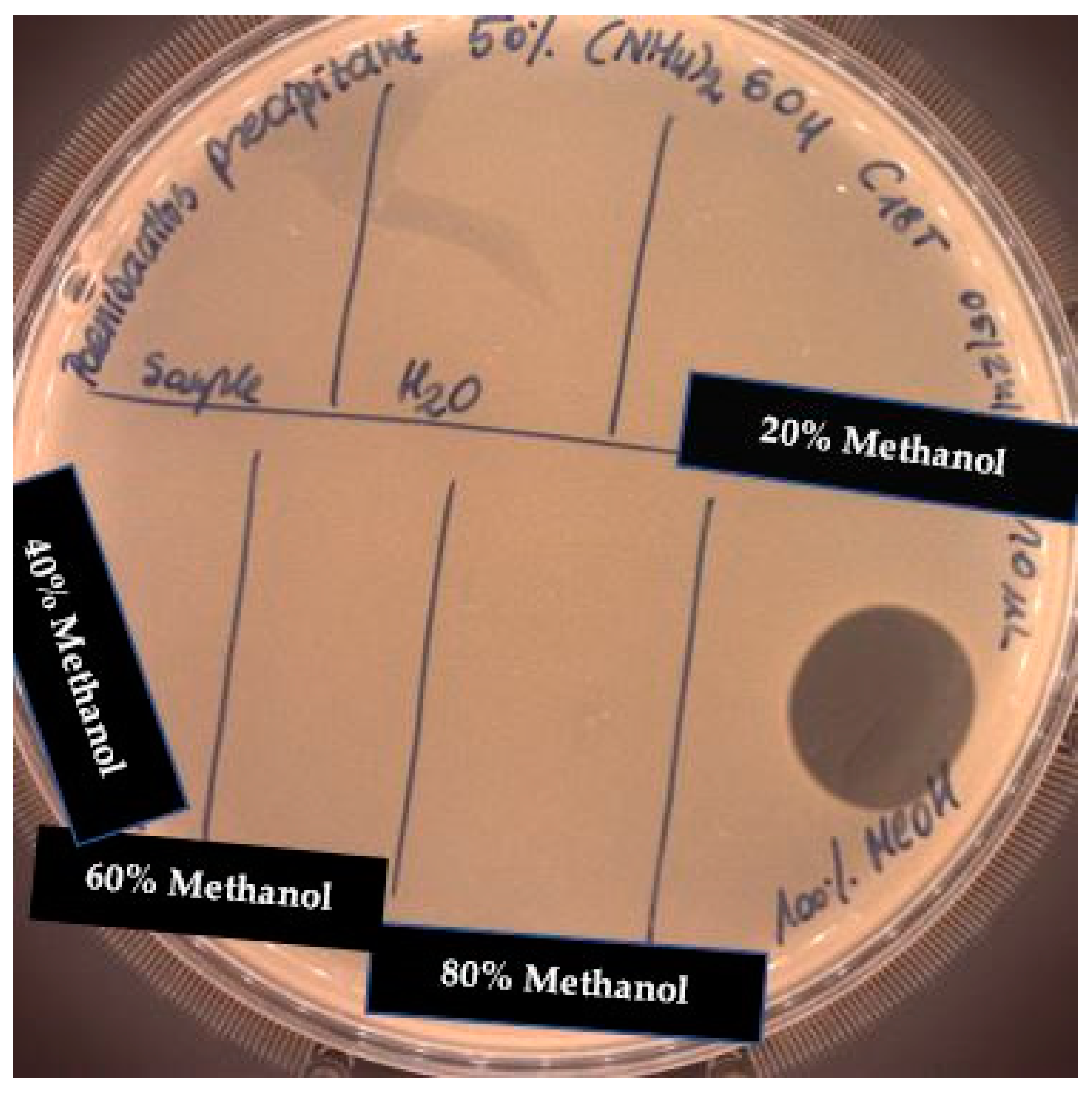
2.4. Solid Phase Extraction (SPE)
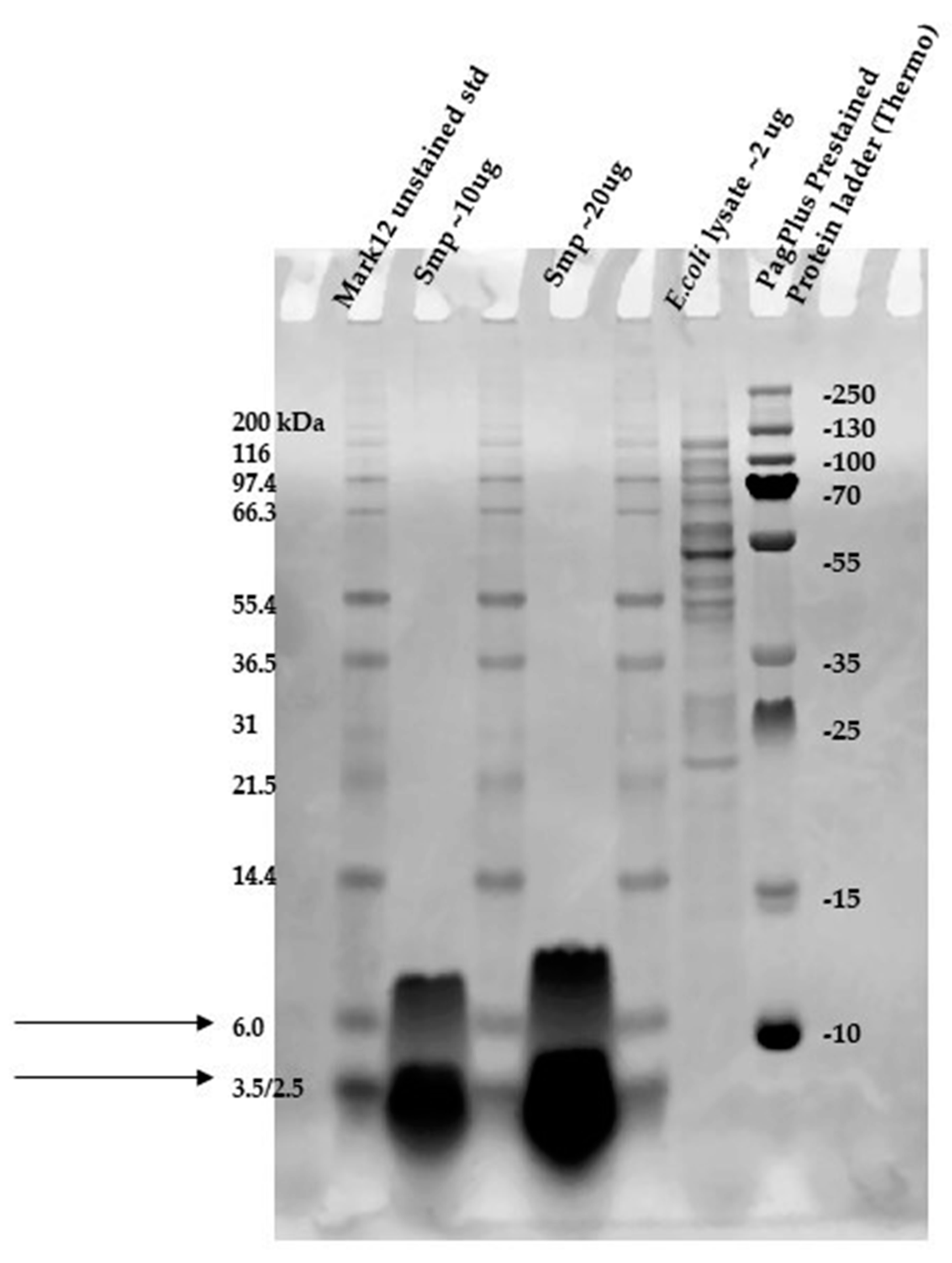
2.5. Direct Detection of Antimicrobial Activity on Tricine SDS-PAGE Gels
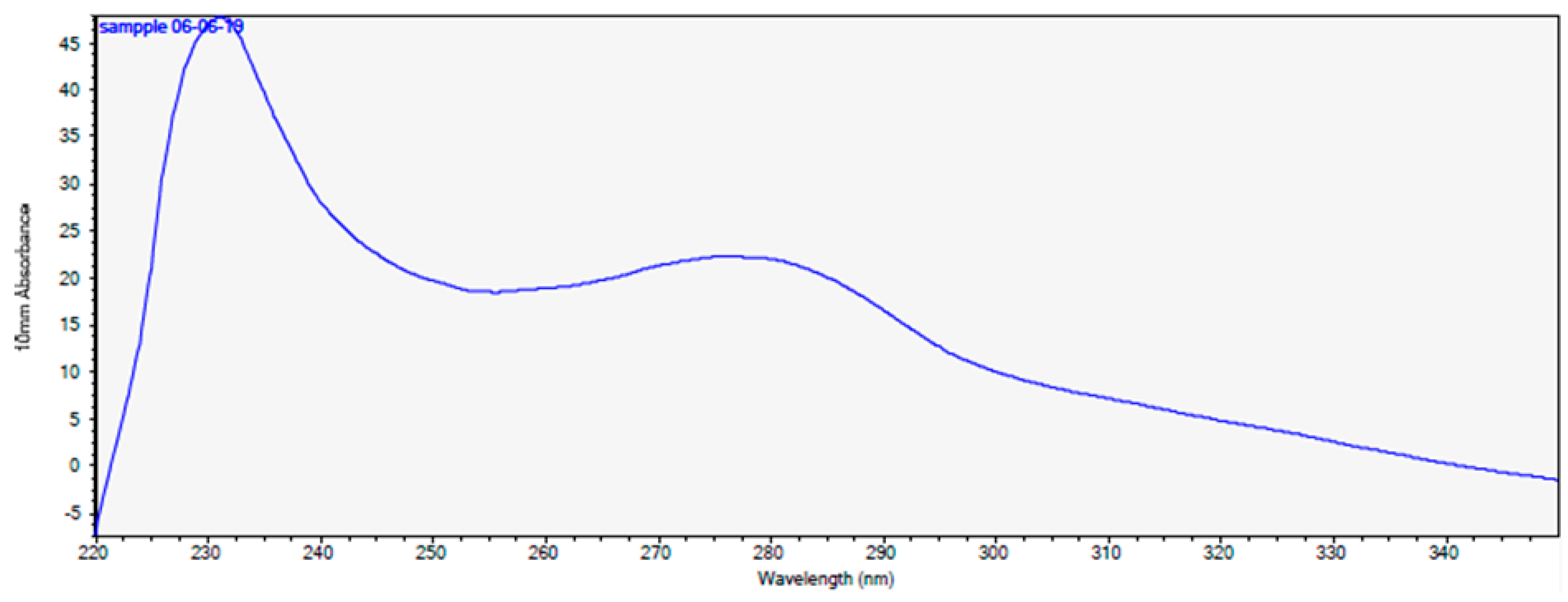
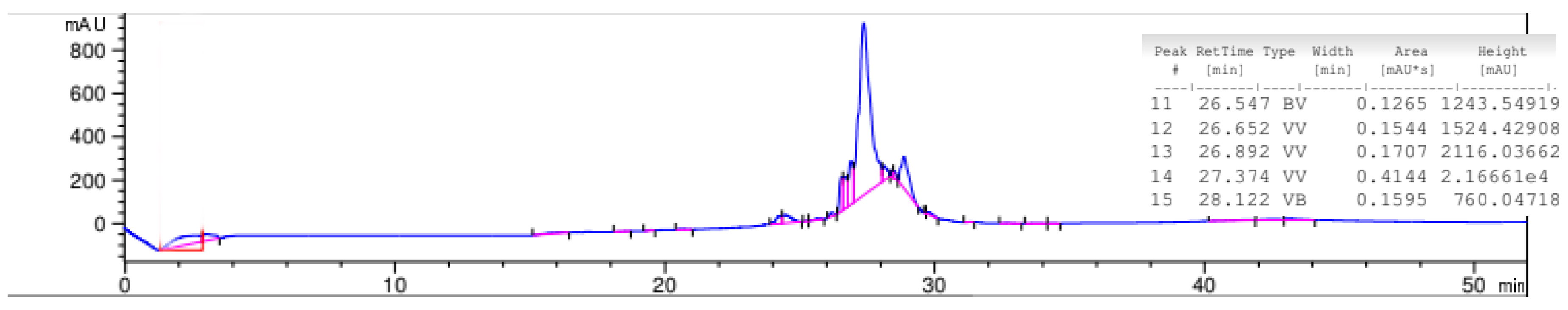
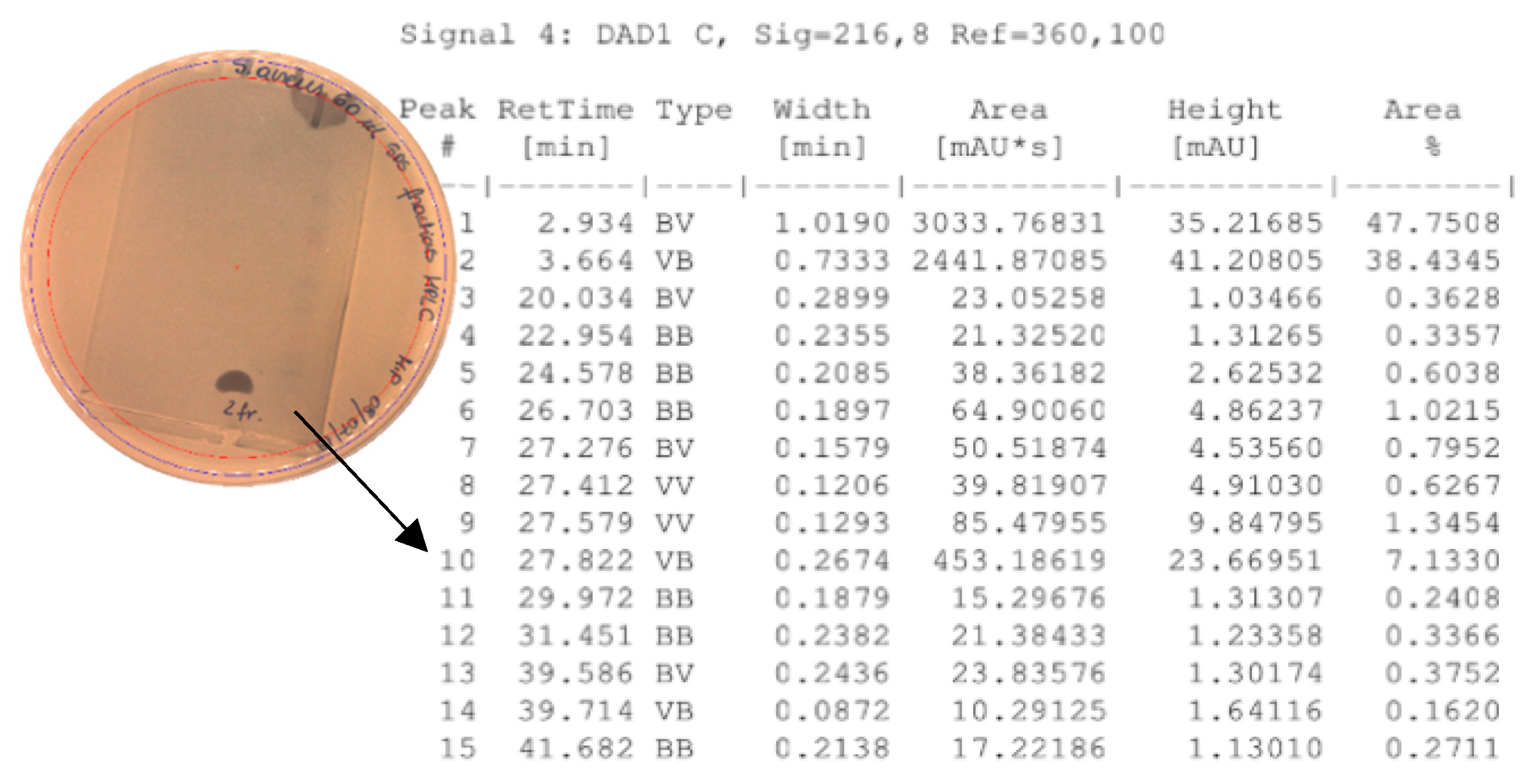
2.6. High-Performance Liquid Chromatography (HPLC)
3. Discussion
4. Materials and Methods
4.1. Strain Growth Conditions Associated with an Active Compound Production
4.2. Partial Purification of the Bioactive Compound with Ammonium Sulfate
4.3. Effects of Proteolytic enzymes and Temperature on Antimicrobial Activity of the Compound
4.4. Solid-Phase Extraction with tC18 Column
4.5. Agar-Overlay Inhibition Assay on SDS-PAGE Gels
4.6. High-Performance Liquid Chromatography (HPLC)
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Shida, O.; Takagi, H.; Kadowaki, K.; Nakamura, L.K.; Komagata, K. Transfer of Bacillus alginolyticus, Bacillus chondroitinus, Bacillus curdlanolyticus, Bacillus glucanolyticus, Bacillus kobensis, and Bacillus thiaminolyticus to the genus Paenibacillus and emended description of the genus Paenibacillus. Int. J. Syst. Bacteriol. 1997, 47, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Sáez-Nieto, J.A.; Medina-Pascual, M.J.; Carrasco, G.; Garrido, N.; Fernandez-Torres, M.A.; Villalón, P.; Valdezate, S. Paenibacillus spp. isolated from human and environmental samples in Spain: Detection of 11 new species. New Microbes New Infect. 2017, 19, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Parte, A.C. LPSN—List of Prokaryotic names with Standing in Nomenclature (bacterio.net), 20 years on. Int. J. Syst. Evol. Microbiol. 2018, 68, 1825–1829. [Google Scholar] [CrossRef] [PubMed]
- Grady, E.N.; MacDonald, J.; Liu, L.; Richman, A.; Yuan, Z.-C. Current knowledge and perspectives of Paenibacillus: A review. Microb. Cell Factories 2016, 15, 203. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Liang, J.; Xiao, R.; Zang, P.; Zhao, Y.; Zhang, L. Effect of four trace elements on Paenibacillus polymyxa Pp-7250 proliferation, activity and colonization in ginseng. AMB Express 2018, 8, 164. [Google Scholar] [CrossRef]
- Vater, J.; Herfort, S.; Doellinger, J.; Weydmann, M.; Borriss, R.; Lasch, P. Genome Mining of the Lipopeptide Biosynthesis of Paenibacillus polymyxa E681 in Combination with Mass Spectrometry: Discovery of the Lipoheptapeptide Paenilipoheptin. Chembiochem 2018, 19, 744–753. [Google Scholar] [CrossRef]
- Chen, W.; Lin, K.; Sheu, S. Paenibacillus lacus sp. nov., isolated from a water reservoir. Int. J. Syst. Evolut. Microbiol. 2019, 67, 1582–1588. [Google Scholar] [CrossRef]
- Satlin, M.J.; Jenkins, S.G. 151 - Polymyxins. In Infectious Diseases, 4th ed.; Cohen, J., Powderly, W.G., Opal, S.M., Eds.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 1285–1288. ISBN 978-0-7020-6285-8. [Google Scholar]
- Timmusk, S.; Copolovici, D.; Copolovici, L.; Teder, T.; Nevo, E.; Behers, L. Paenibacillus polymyxa biofilm polysaccharides antagonise Fusarium graminearum. Sci. Rep. 2019, 9, 662. [Google Scholar] [CrossRef]
- Cochrane, S.A.; Vederas, J.C. Lipopeptides from Bacillus and Paenibacillus spp.: A Gold Mine of Antibiotic Candidates. Med. Res. Rev. 2016, 36, 4–31. [Google Scholar] [CrossRef]
- Prévost, G. Chapter 562—Exfoliative Toxins—Staphylococcal Epidermolysins. In Handbook of Proteolytic Enzymes, 3rd ed.; Rawlings, N.D., Salvesen, G.B.T., Eds.; Academic Press: Cambridge, MA, USA, 2013; pp. 2539–2543. ISBN 978-0-12-382219-2. [Google Scholar]
- Liu, D. Chapter 55—Enterotoxin-Producing Staphylococcus aureus. In Molecular Medical Microbiology, 2nd ed.; Tang, Y.-W., Sussman, M., Liu, D., Poxton, I., Schwartzman, J.B.T., Eds.; Academic Press: Boston, MA, USA, 2015; pp. 979–995. ISBN 978-0-12-397169-2. [Google Scholar]
- Gnanamani, A. Staphylococcus aureus: Overview of Bacteriology, Clinical Diseases, Epidemiology, Antibiotic Resistance and Therapeutic Approach. In Frontiers in Staphylococcus aureus; Enany, S., Crotty Alexander, L., Eds.; IntechOpen Limited: London, UK, 2017. [Google Scholar]
- Véghová, A.; Minarovičová, J.; Koreňová, J.; Drahovská, H.; Kaclíková, E. Prevalence and tracing of persistent Listeria monocytogenes strains in meat processing facility production chain. J. Food Saf. 2017, 37, e12315. [Google Scholar] [CrossRef]
- Lomonaco, S.; Nucera, D.; Filipello, V. The evolution and epidemiology of Listeria monocytogenes in Europe and the United States. Infect. Genet. Evol. 2015, 35, 172–183. [Google Scholar] [CrossRef] [PubMed]
- Campos, C.; Castro, M.; Gliemmo, M.F.; Schelegueda, L. Use of natural antimicrobials for the control of Listeria monocytogenes in foods. In Science against Microbial Pathogens: Communicating Current Research and Technological Advances; Formatex: Badajoz, Spain, 2011; pp. 1112–1123. [Google Scholar]
- Allocati, N.; Masulli, M.; Alexeyev, F.M.; Di Ilio, C. Escherichia coli in Europe: An Overview. Int. J. Environ. Res. Public Health 2013, 10, 6235–6254. [Google Scholar] [CrossRef]
- Kim, H.W.; Rhee, M.S. Novel Antibiotic Testing Approaches Reveal Reduced Antibiotic Efficacy Against Shiga Toxin-Producing Escherichia coli O157:H7 Under Simulated Microgravity. Front. Microbiol. 2018, 9, 3214. [Google Scholar] [CrossRef]
- Pajor, M.; Worobo, R.W.; Milewski, S.; Szweda, P. The Antimicrobial Potential of Bacteria Isolated from Honey Samples Produced in the Apiaries Located in Pomeranian Voivodeship in Northern Poland. Int. J. Environ. Res. Public Health 2018, 15, 2002. [Google Scholar] [CrossRef] [PubMed]
- Cohen, I.; Ron, I.G.; Ben-Jacob, E. From branching to nebula patterning during colonial development of the Paenibacillus alvei bacteria. Phys. A Stat. Mech. Appl. 2000, 286, 321–336. [Google Scholar] [CrossRef]
- Seal, B.S.; Drider, D.; Oakley, B.B.; Brüssow, H.; Bikard, D.; Rich, J.O.; Miller, S.; Devillard, E.; Kwan, J.; Bertin, G.; et al. Microbial-derived products as potential new antimicrobials. Vet. Res. 2018, 49, 66. [Google Scholar] [CrossRef]
- Tortorella, E.; Tedesco, P.; Palma Esposito, F.; January, G.G.; Fani, R.; Jaspars, M.; de Pascale, D. Antibiotics from Deep-Sea Microorganisms: Current Discoveries and Perspectives. Mar. Drugs 2018, 16, 355. [Google Scholar] [CrossRef]
- Boakye, Y.D. Antimicrobial Agents: Antibacterial Agents, Anti-biofilm Agents, Antibacterial Natural Compounds, and Antibacterial Chemicals. In Antimicrobials, Antibiotic Resistance, Antibiofilm Strategies and Activity Methods; Osafo, N., Ed.; IntechOpen: Rijeka, Croatia, 2019; ISBN 978-1-78985-790-0. [Google Scholar]
- Pham, J.V.; Yilma, M.A.; Feliz, A.; Majid, M.T.; Maffetone, N.; Walker, J.R.; Kim, E.; Cho, H.J.; Reynolds, J.M.; Song, M.C.; et al. A Review of the Microbial Production of Bioactive Natural Products and Biologics. Front. Microbiol. 2019, 10, 1404. [Google Scholar] [CrossRef]
- Singh, H.; Kaur, M.; Jangra, M.; Mishra, S.; Nandanwar, H.; Pinnaka, A.K. Antimicrobial properties of the novel bacterial isolate Paenibacilllus sp. SMB1 from a halo-alkaline lake in India. Sci. Rep. 2019, 9, 11561. [Google Scholar] [CrossRef]
- Bennett, V.; Koch, M.; Aldarhami, A.; Upton, M. Purification and characterisation of antimicrobial agents isolated from a member of the Paenibacillus genus. Access Microbiol. 2019, 1. [Google Scholar] [CrossRef]
- Knolhoff, A.M.; Zheng, J.; McFarland, M.A.; Luo, Y.; Callahan, J.H.; Brown, E.W.; Croley, T.R. Identification and Structural Characterization of Naturally-Occurring Broad-Spectrum Cyclic Antibiotics Isolated from Paenibacillus. J. Am. Soc. Mass Spectrom. 2015, 26, 1768–1779. [Google Scholar] [CrossRef] [PubMed]
- Alkotaini, B.; Anuar, N.; Kadhum, A.; Sani, A.A.A. Detection of secreted antimicrobial peptides isolated from cell-free culture supernatant of Paenibacillus alvei AN5. J. Ind. Microbiol. Biotechnol. 2013, 40, 571–579. [Google Scholar] [CrossRef] [PubMed]
- Jagadeesan, Y.; Athinarayanan, S.; Ayub, S.B.M.; Balaiah, A. Assessment of Synthesis Machinery of Two Antimicrobial Peptides from Paenibacillus alvei NP75. Probiotics Antimicrob. Proteins 2020, 12, 39–47. [Google Scholar] [CrossRef]
- Huang, E.; Yousef, A.E. Biosynthesis of paenibacillin, a lantibiotic with N-terminal acetylation, by Paenibacillus polymyxa. Microbiol. Res. 2015, 181, 15–21. [Google Scholar] [CrossRef] [PubMed]
- “The LIPID MAPS® Lipidomics Gateway”. Available online: http://www.lipidmaps.org/ (accessed on 20 April 2020).
- Giuliani, A.; Giovanna, P.; Silvia, N. Antimicrobial peptides: An overview of a promising class of therapeutics. Open Life Sci. 2007, 2, 1–33. [Google Scholar] [CrossRef]
- Wu, X.-C.; Shen, X.-B.; Ding, R.; Qian, C.-D.; Fang, H.-H.; Li, O. Isolation and partial characterization of antibiotics produced by Paenibacillus elgii B69. FEMS Microbiol. Lett. 2010, 310, 32–38. [Google Scholar] [CrossRef][Green Version]
- Qian, C.-D.; Wu, X.-C.; Teng, Y.; Zhao, W.-P.; Li, O.; Fang, S.-G.; Huang, Z.-H.; Gao, H.-C. Battacin (Octapeptin B5), a new cyclic lipopeptide antibiotic from Paenibacillus tianmuensis active against multidrug-resistant Gram-negative bacteria. Antimicrob. Agents Chemother. 2012, 56, 1458–1465. [Google Scholar] [CrossRef]
- Henkel, M.; Hausmann, R. Chapter 2—Diversity and Classification of Microbial Surfactants. In Biobased Surfactants, 2nd ed.; Hayes, D.G., Solaiman, D.K.Y., Ashby, R.D.B.T., Eds.; AOCS Press: Urbana, IL, USA, 2019; pp. 41–63. ISBN 978-0-12-812705-6. [Google Scholar]
- Huang, E.; Guo, Y.; Yousef, A.E. Biosynthesis of the new broad-spectrum lipopeptide antibiotic paenibacterin in Paenibacillus thiaminolyticus OSY-SE. Res. Microbiol. 2014, 165, 243–251. [Google Scholar] [CrossRef]
- Huang, Z.; Hu, Y.; Shou, L.; Song, M. Isolation and partial characterization of cyclic lipopeptide antibiotics produced by Paenibacillus ehimensis B7. BMC Microbiol. 2013, 13, 87. [Google Scholar] [CrossRef]
- Luo, Y.; Wang, C.; Allard, S.; Strain, E.; Allard, M.W.; Brown, E.W.; Zheng, J. Draft Genome Sequences of Paenibacillus alvei A6-6i and TS-15. Genome Announc. 2013, 1, e00673-13. [Google Scholar] [CrossRef]
- Zhu, S.; Hegemann, J.D.; Fage, C.D.; Zimmermann, M.; Xie, X.; Linne, U.; Marahiel, M.A. Insights into the Unique Phosphorylation of the Lasso Peptide Paeninodin. J. Biol. Chem. 2016, 291, 13662–13678. [Google Scholar] [CrossRef] [PubMed]
- Zulkhairi Amin, F.A.; Sabri, S.; Ismail, M.; Chan, K.W.; Ismail, N.; Mohd Esa, N.; Mohd Lila, M.A.; Zawawi, N. Probiotic Properties of Bacillus Strains Isolated from Stingless Bee (Heterotrigona itama) Honey Collected across Malaysia. Int. J. Environ. Res. Public Health 2019, 17, 278. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Churey, J.J.; Worobo, R.W. Antimicrobial activity of bacterial isolates from different floral sources of honey. Int. J. Food Microbiol. 2008, 126, 240–244. [Google Scholar] [CrossRef] [PubMed]
- Pizzo, E.; Pane, K.; Bosso, A.; Landi, N.; Ragucci, S.; Russo, R.; Gaglione, R.; Torres, M.D.T.; de la Fuente-Nunez, C.; Arciello, A.; et al. Novel bioactive peptides from PD-L1/2, a type 1 ribosome inactivating protein from Phytolacca dioica L. Evaluation of their antimicrobial properties and anti-biofilm activities. Biochim. Biophys. Acta Biomembr. 2018, 1860, 1425–1435. [Google Scholar] [CrossRef]
- Nawrot, R.; Barylski, J.; Nowicki, G.; Broniarczyk, J.; Buchwald, W.; Goździcka-Józefiak, A. Plant antimicrobial peptides. Folia Microbiol. 2014, 59, 181–196. [Google Scholar] [CrossRef]
- Gorlenko, L.C.; Kiselev, Y.H.; Budanova, V.E.; Zamyatnin, A.A.; Ikryannikova, N.L. Plant Secondary Metabolites in the Battle of Drugs and Drug-Resistant Bacteria: New Heroes or Worse Clones of Antibiotics? Antibiotics 2020, 9, 170. [Google Scholar] [CrossRef]
- Zaynab, M.; Fatima, M.; Abbas, S.; Sharif, Y.; Umair, M.; Zafar, M.H.; Bahadar, K. Role of secondary metabolites in plant defense against pathogens. Microb. Pathog. 2018, 124, 198–202. [Google Scholar] [CrossRef]
- Fira, D.; Dimkic, I.; Beric, T.; Lozo, J.; Stankovic, S. Biological control of plant pathogens by Bacillus species. J. Biotechnol. 2018, 285, 44–55. [Google Scholar] [CrossRef]
- Murtey, M. Das Sample Preparations for Scanning Electron Microscopy—Life Sciences. In Modern Electron Microscopy in Physical and Life Sciences; Kral, R., Ed.; IntechOpen: Rijeka, Croatia, 2016. [Google Scholar]
- Schägger, H. Tricine—SDS-PAGE. Nat. Protoc. 2006, 1, 16–23. [Google Scholar] [CrossRef]
- Grecka, K.; Kuś, P.M.; Okińczyc, P.; Worobo, R.W.; Walkusz, J.; Szweda, P. The Anti-Staphylococcal Potential of Ethanolic Polish Propolis Extracts. Molecules 2019, 24, 1732. [Google Scholar] [CrossRef]
- Grecka, K.; Kuś, P.M.; Worobo, R.W.; Szweda, P. Study of the anti-staphylococcal potential of honeys produced in Northern Poland. Molecules 2018, 23, 260. [Google Scholar] [CrossRef] [PubMed]
- Kuś, P.M.; Szweda, P.; Jerković, I.; Tuberoso, C.I.G. Activity of Polish unifloral honeys against pathogenic bacteria and its correlation with colour, phenolic content, antioxidant capacity and other parameters. Lett. Appl. Microbiol. 2016, 62, 269–276. [Google Scholar] [CrossRef] [PubMed]

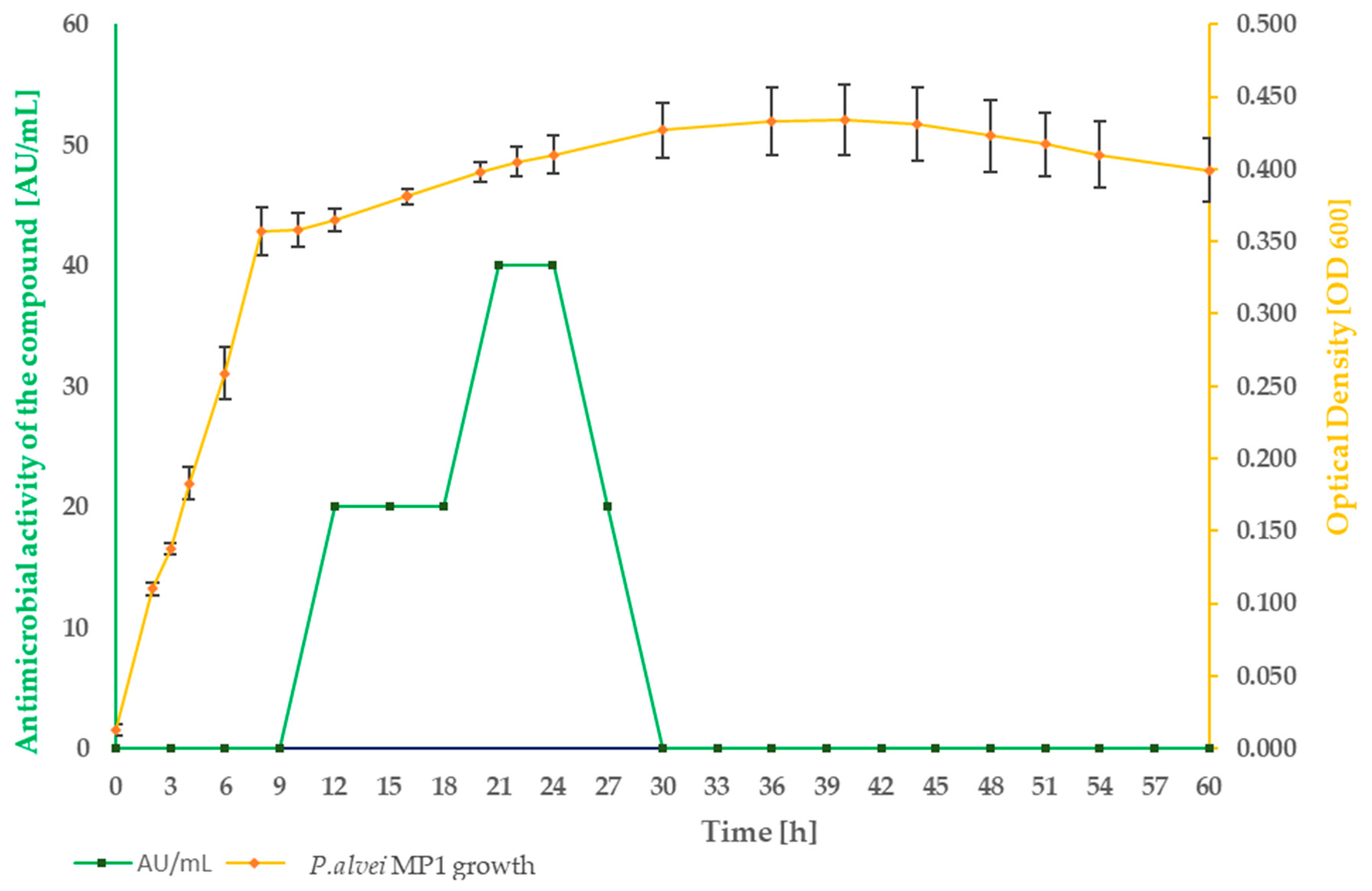
| Hours of Experiment [h] | Two-fold Serial Dilutions of Cell-Free Supernatant Spotted onto TSA Soft Agar, with S. aureus L1- 0030 as Indicator Strain | Antibacterial Activity [AU/mL] | Inhibition Zones Diameter Ø [cm] |
|---|---|---|---|
| 9 |  | 0 | - |
| 12 | 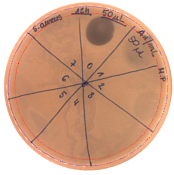 | 20 | 1.9 |
| 15 |  | 20 | 1.7 |
| 18 |  | 20 | 1.9 |
| 21 | 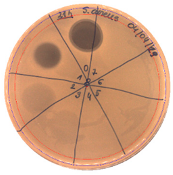 | 40 | 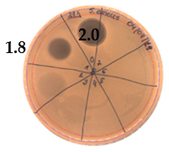 |
| 24 | 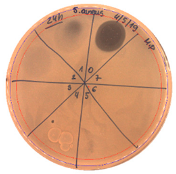 | 20 | 2.0 |
| 27 |  | - | - |
| 30 |  | - | - |
| Two-fold Serial Dilutions – 50 µL Spots | Inhibition Zone Diameter Ø [cm] |
|---|---|
| 20% (NH4)2SO4 | |
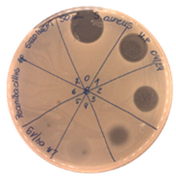 |  |
| Precipitate Activity—160 AU/mL | |
| 40% (NH4)2SO4 | |
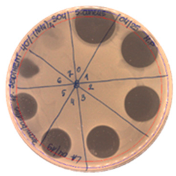 | 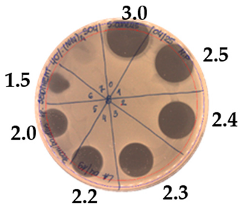 |
| Precipitate Activity—1280 AU/mL | |
| 50% (NH4)2SO4 | |
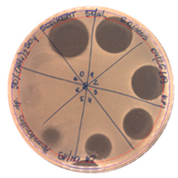 |  |
| Precipitate Activity—640 AU/mL | |
| 60% (NH4)2SO4 | |
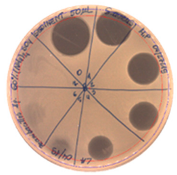 | 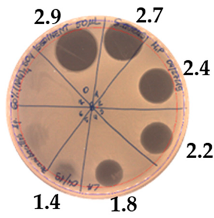 |
| Precipitate Activity—640 AU/mL | |
| 70% (NH4)2SO4 | |
 | 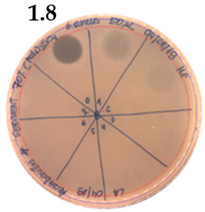 |
| Precipitate Activity—20 AU/mL | |
| Indicator Strain | |
|---|---|
| Two-fold serial dilutions – 50 µL spots | |
| L. monocytogenes FSL – X1-0001 | E. coli O157: H7 |
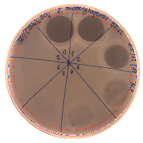 | 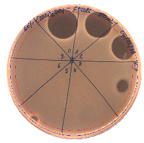 |
| (160 AU/mL) | (160 AU/mL) |
| Inhibition zone diameter Ø [cm] | |
| L. monocytogenes FSL – X1-0001 | E. coli O157: H7 |
 |  |
| Ammonium Sulfate Saturation [%] | Indicator Strain - S. aureus L1- 0030 |
|---|---|
| Two-fold Serial Dilutions – 50 µL Spots | |
| 40 |  |
| 50 |  |
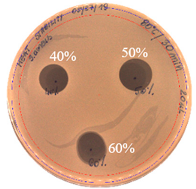
| 60 | 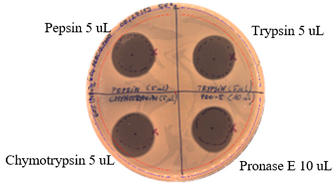 |
| Proteinase K Test |
|---|
 |
| 60 °C for 30 min. | 80 °C for 30 min | 100 °C for 30 min |
|---|---|---|
 |  | 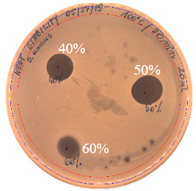 |
| Incubation Time [h] | Indicator Strain | ||
|---|---|---|---|
| L. monocytogenes FSL – X1 - 0001 | S. aureus L1 - 0030 | E. coli O157: H7 | |
| 7 | 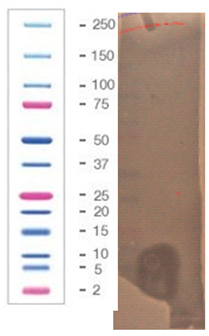 | 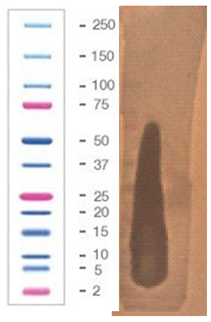 | 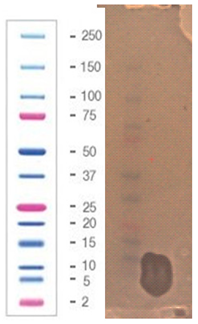 |
| 24 |  | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pajor, M.; Xiong, Z.R.; Worobo, R.W.; Szweda, P. Paenibacillus alvei MP1 as a Producer of the Proteinaceous Compound with Activity against Important Human Pathogens, Including Staphylococcus aureus and Listeria monocytogenes. Pathogens 2020, 9, 319. https://doi.org/10.3390/pathogens9050319
Pajor M, Xiong ZR, Worobo RW, Szweda P. Paenibacillus alvei MP1 as a Producer of the Proteinaceous Compound with Activity against Important Human Pathogens, Including Staphylococcus aureus and Listeria monocytogenes. Pathogens. 2020; 9(5):319. https://doi.org/10.3390/pathogens9050319
Chicago/Turabian StylePajor, Magdalena, Zirui Ray Xiong, Randy W. Worobo, and Piotr Szweda. 2020. "Paenibacillus alvei MP1 as a Producer of the Proteinaceous Compound with Activity against Important Human Pathogens, Including Staphylococcus aureus and Listeria monocytogenes" Pathogens 9, no. 5: 319. https://doi.org/10.3390/pathogens9050319
APA StylePajor, M., Xiong, Z. R., Worobo, R. W., & Szweda, P. (2020). Paenibacillus alvei MP1 as a Producer of the Proteinaceous Compound with Activity against Important Human Pathogens, Including Staphylococcus aureus and Listeria monocytogenes. Pathogens, 9(5), 319. https://doi.org/10.3390/pathogens9050319





