Genetic Diversity and Recombination in the Plant Pathogen Sclerotinia sclerotiorum Detected in Sri Lanka
Abstract
1. Introduction
2. Results
2.1. Mycelial Compatibility Group (MCG) Diversity
2.2. Multilocus Haplotype (MLH) Diversity
2.3. Detection of Recombination
2.4. Distribution of Mating Type Genes
2.5. Detection of Multiple Infections per Cabbage Head
3. Discussion
4. Materials and Methods
4.1. Sample Collection, Establishment of Pure Cultures, and MCG Assays
4.2. DNA Isolation, Microsatellite Genotyping, and Data Analysis
4.3. Detection of Recombination
4.4. Distribution of Mating Type Genes
4.5. Detection of Multiple Infections per Cabbage Head and Detached Leaf Assay with Multiple Genotypes
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Boland, G.; Hall, R. Index of Plant Hosts of Sclerotinia sclerotiorum. Can. J. Plant Pathol. 1994, 16, 93–108. [Google Scholar] [CrossRef]
- Hudyncia, J.; Shew, H.D.; Cody, B.R.; Cubeta, M.A. Evaluation of Wounds as a Factor to Infection of Cabbage by Ascospores of Sclerotinia sclerotiorum. Plant Dis. 2000, 84, 316–320. [Google Scholar] [CrossRef]
- Atallah, Z.K.; Larget, B.; Chen, X.; Johnson, D.A. High Genetic Diversity, Phenotypic Uniformity, and Evidence of Outcrossing in Sclerotinia Sclerotiorum in the Columbia Basin of Washington State. Phytopathology 2004, 94, 737–742. [Google Scholar] [CrossRef]
- Attanayake, R.N.; Tennekoon, V.; Johnson, D.A.; Porter, L.D.; Del Río-Mendoza, L.; Jiang, D.; Chen, W. Inferring Outcrossing in the Homothallic Fungus Sclerotinia sclerotiorum Using Linkage Disequilibrium Decay. Heredity 2014, 113, 353–363. [Google Scholar] [CrossRef]
- Hemmati, R.; Javan-Nikkhah, M.; Linde, C. Population Genetic Structure of Sclerotinia sclerotiorum on Canola in Iran. Eur. J. Plant Pathol. 2009, 125, 617–628. [Google Scholar] [CrossRef]
- Mahalingam, T.; Rajapakse, C.S.K.; Somachandra, K.P.; Attanayake, R.N. Carbon Source Dependent-Anaerobic Soil Disinfestation (ASD) Mitigates the Sclerotial Germination of Sclerotinia sclerotiorum. Trop. Plant Pathol. 2020, 45, 13–24. [Google Scholar] [CrossRef]
- Li, Z.; Wang, Y.; Chen, Y.; Zhang, J.; Fernando, W.D. Genetic and Differentiation of Sclerotinia sclerotiorum Populations in Sunflower. Phytoparasitica 2008, 37, 77–85. [Google Scholar] [CrossRef]
- Kohn, L.M. Mycelial Incompatibility and Molecular Markers Identify Genetic Variability in Field Populations of Sclerotinia sclerotiorum. Phytopathology 1991, 81, 480. [Google Scholar] [CrossRef]
- Cubeta, M.A.; Cody, B.R.; Kohli, Y.; Kohn, L.M. Clonality in Sclerotinia sclerotiorum on Infected Cabbage in Eastern North Carolina. Phytopathology 1997, 87, 1000–1004. [Google Scholar] [CrossRef]
- Anderson, J.B.; Kohn, L.M. Clonality in Soil Borne Plant-pathogenic Fungi. Ann. Rev. Phytopathol. 1995, 33, 369–391. [Google Scholar] [CrossRef]
- Malvárez, G.; Carbone, I.; Grünwald, N.J.; Subbarao, K.V.; Schafer, M.; Kohn, L.M. New Populations of Sclerotinia sclerotiorum from Lettuce in California and Peas and Lentils in Washington. Phytopathology 2007, 97, 470–483. [Google Scholar] [CrossRef] [PubMed]
- Clarkson, J.P.; Coventry, E.; Kitchen, J.; Carter, H.E.; Whipps, J.M. Population Structure of Sclerotinia sclerotiorum in Crop and Wild Hosts in the UK. Plant Pathol. 2012, 62, 309–324. [Google Scholar] [CrossRef]
- Gomes, E.V.; Nascimento, L.B.D.; De Freitas, M.A.; Nasser, L.C.B.; Petrofeza, S. Microsatellite Markers Reveal Genetic Variation within Sclerotinia sclerotiorum Populations in Irrigated Dry Bean Crops in Brazil. J. Phytopathol. 2011, 159, 94–99. [Google Scholar] [CrossRef]
- Carpenter, M.; Frampton, C.; Stewart, A. Genetic Variation in New Zealand Populations of the Plant Pathogen Sclerotinia sclerotiorum. N. Z. J. Crop. Hortic. Sci. 1999, 27, 13–21. [Google Scholar] [CrossRef][Green Version]
- Lehner, M.S.; Júnior, T.J.P.; Júnior, B.T.H.; Teixeira, H.; Vieira, R.F.; Carneiro, J.E.S.; Mizubuti, E.S.G. Low Genetic Variability in Sclerotinia sclerotiorum Populations from Common Bean Fields in Minas Gerais State, Brazil, at Regional, Local and Micro-scales. Plant Pathol. 2015, 64, 921–931. [Google Scholar] [CrossRef]
- Lehner, M.S.; Júnior, T.J.D.P.; Del Ponte, E.M.; Mizubuti, E.S.G.; Pethybridge, S.J. Independently Founded Populations of Sclerotinia sclerotiorum from a Tropical and a Temperate Region have Similar Genetic Structure. PLoS ONE 2017, 12, e0173915. [Google Scholar] [CrossRef]
- Júnior, C.L.; Gomes, E.; Junior, M.L.; Nasser, L.; Petrofeza, S. Genetic Diversity and Mycelial Compatibility Groups of the Plant-pathogenic Fungus Sclerotinia sclerotiorum in Brazil. Genet. Mol. Res. 2011, 10, 868–877. [Google Scholar] [CrossRef]
- Mahalingam, T.; Guruge, B.M.A.; Somachandra, K.P.; Rajapakse, C.S.; Attanayake, R.N. First Report of White Mold Caused by Sclerotinia sclerotiorum on Cabbage in Sri Lanka. Plant Dis. 2017, 101, 249. [Google Scholar] [CrossRef]
- Mahalingam, T.; Guruge, B.M.A.; Somachandra, K.; Jayasekara, E.A.E.S.S.; Rajapakse, C.S.K.; Attanayake, R.N. Phenotypic Variation of Cabbage White Mold Pathogen, Sclerotinia sclerotiorum in the Upcountry Commercial Cabbage Fields in Sri Lanka. J. Natl. Sci. Found. Sri Lanka 2018, 46, 159. [Google Scholar] [CrossRef]
- Read, A.F.; Zachos, J.; Pagani, M.; Sloan, L.; Thomas, E.; Billups, K. The Ecology of Genetically Diverse Infections. Science 2001, 292, 1099–1102. [Google Scholar] [CrossRef]
- Abdullah, A.S.; Moffat, C.S.; Lopez-Ruiz, F.J.; Gibberd, M.R.; Hamblin, J.; Zerihun, A. Host–Multi-Pathogen Warfare: Pathogen Interactions in Co-infected Plants. Front. Plant Sci. 2017, 8, 1806. [Google Scholar] [CrossRef] [PubMed]
- Tollenaere, C.; Susi, H.; Laine, A.-L. Evolutionary and Epidemiological Implications of Multiple Infection in Plants. Trends. Plant Sci. 2016, 21, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Alizon, S.; De Roode, J.C.; Michalakis, Y. Multiple Infections and the Evolution of Virulence. Ecol. Lett. 2013, 16, 556–567. [Google Scholar] [CrossRef] [PubMed]
- Anagnostakis, S.L. Population Dynamics of Cryphonectria parasitica in a Mixed-Hardwood Forest in Connecticut. Phytopathology 1987, 77, 751. [Google Scholar] [CrossRef]
- Kedera, C.J. Genetic Diversity of Fusarium Section Liseola (Gibberella fujikuroi) in Individual Maize Stalks. Phytopathology 1994, 84, 603. [Google Scholar] [CrossRef]
- McDonald, B.A. DNA Restriction Fragment Length Polymorphisms among Mycosphaerella graminicola (Anamorph Septoria tritici) Isolates Collected from a Single Wheat Field. Phytopathology 1990, 80, 1368. [Google Scholar] [CrossRef]
- López-Villavicencio, M.; Jonot, O.; Coantic, A.; Hood, M.E.; Enjalbert, J.; Giraud, T. Multiple Infections by the Anther Smut Pathogen Are Frequent and Involve Related Strains. PLoS Pathog. 2007, 3, e176. [Google Scholar] [CrossRef]
- Maltby, A.D.; Mihail, J.D. Competition among Sclerotinia sclerotiorum Genotypes within Canola Stems. Can. J. Bot. 1997, 75, 462–468. [Google Scholar] [CrossRef]
- Ekins, M.; Hayden, H.L.; Aitken, E.; Goulter, K.C. Population Structure of Sclerotinia sclerotiorum on Sunflower in Australia. Australas. Plant Pathol. 2010, 40, 99–108. [Google Scholar] [CrossRef]
- Attanayake, R.N.; Carter, P.A.; Jiang, D.; Del Río-Mendoza, L.; Chen, W. Sclerotinia sclerotiorum Populations Infecting Canola from China and the United States are Genetically and Phenotypically Distinct. Phytopathology 2013, 103, 750–761. [Google Scholar] [CrossRef]
- Sirjusingh, C.; Kohn, L.M. Characterization of Microsatellites in the Fungal Plant Pathogen, Sclerotinia sclerotiorum. Mol. Ecol. Notes 2001, 1, 267–269. [Google Scholar] [CrossRef]
- Chitrampalam, P.; Inderbitzin, P.; Maruthachalam, K.; Wu, B.-M.; Subbarao, K.V. The Sclerotinia sclerotiorum Mating Type Locus (MAT) Contains a 3.6-kb Region That Is Inverted in Every Meiotic Generation. PLoS ONE 2013, 8, e56895. [Google Scholar] [CrossRef] [PubMed]
- Kohli, Y.; Brunner, L.J.; Yoell, H.; Milgroom, M.G.; Anderson, J.B.; Morrall, R.A.A.; Kohn, L.M. Clonal Dispersal and Spatial Mixing in Populations of the Plant Pathogenic fungus, Sclerotinia sclerotiorum. Mol. Ecol. 1995, 4, 69–77. [Google Scholar] [CrossRef]
- Aldrich-Wolfe, L.; Travers, S.; Nelson, B.D. Genetic Variation of Sclerotinia sclerotiorum from Multiple Crops in the North Central United States. PLoS ONE 2015, 10, e0139188. [Google Scholar] [CrossRef]
- Sexton, A.; Howlett, B.J. Microsatellite Markers Reveal Genetic Differentiation among Populations of Sclerotinia sclerotiorum from Australian Canola Fields. Curr. Genet. 2004, 46, 357–365. [Google Scholar] [CrossRef]
- Dunn, A.; Kikkert, J.; Pethybridge, S.J. Genotypic Characteristics in Populations of Sclerotinia sclerotiorum from New York State, USA. Ann. Appl. Boil. 2017, 170, 219–228. [Google Scholar] [CrossRef]
- Koufopanou, V.; Burt, A.; Taylor, J.W. Concordance of Gene Genealogies Reveals Reproductive Isolation in the Pathogenic Fungus Coccidioides immitis. Proc. Natl. Acad. Sci. USA 1997, 94, 5478–5482. [Google Scholar] [CrossRef]
- Campbell, L.T.; Currie, B.J.; Krockenberger, M.; Malik, R.; Meyer, W.; Heitman, J.; Carter, D.A. Clonality and Recombination in Genetically Differentiated Subgroups of Cryptococcus gattii. Eukaryot. Cell 2005, 4, 1403–1409. [Google Scholar] [CrossRef]
- Montarry, J.; Andrivon, D.; Glais, I.; Corbière, R.; Mialdea, G.; Delmotte, F. Microsatellite Markers Reveal two Admixed Genetic Groups and an Ongoing Displacement within the French Population of the Invasive Plant Pathogen Phytophthora infestans. Mol. Ecol. 2010, 19, 1965–1977. [Google Scholar] [CrossRef]
- Stewart, J.E.; Thomas, K.A.; Lawrence, C.B.; Dang, H.; Pryor, B.; Timmer, L.M.P.; Peever, T. Signatures of Recombination in Clonal Lineages of the Citrus Brown Spot Pathogen, Alternaria alternata sensu lato. Phytopathology 2013, 103, 741–749. [Google Scholar] [CrossRef]
- Attanayake, R.N.; Xu, L.; Chen, W. Sclerotinia sclerotiorum Populations: Clonal or recombining? Trop. Plant Pathol. 2019, 44, 23–31. [Google Scholar] [CrossRef]
- Lu, S.; Yun, S.-H.; Lee, T.; Turgeon, B.G. Altering Sexual Reproductive Mode by Interspecific Exchange of MAT loci. Fungal Genet. Boil. 2011, 48, 714–724. [Google Scholar] [CrossRef] [PubMed]
- Sexton, A.; Whitten, A.R.; Howlett, B.J. Population Structure of Sclerotinia sclerotiorum in an Australian Canola Field at Flowering and Stem-infection Stages of the Disease Cycle. Genome 2006, 49, 1408–1415. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the Number of Clusters of Individuals Using the Software Structure: A simulation Study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Earl, D.; Vonholdt, B.M. STRUCTURE HARVESTER: A Website and Program for Visualizing STRUCTURE Output and Implementing the Evanno method. Conserv. Genet. Resour. 2011, 4, 359–361. [Google Scholar] [CrossRef]
- Karunaweera, N.D.; Ferreira, M.U.; Munasinghe, A.; Barnwell, J.W.; Collins, W.E.; King, C.L.; Kawamoto, F.; Hartl, D.L.; Wirth, D. Extensive Microsatellite Diversity in the Human Malaria Parasite Plasmodium vivax. Gene 2008, 410, 105–112. [Google Scholar] [CrossRef]
- Excoffier, L.; Laval, G.; Schneider, S. Arlequin ver. 3.0: An Integrated Software Package for Population Genetics Data Analysis. Evol. Bioinf. Online 2005, 1, 47–50. [Google Scholar] [CrossRef]
- Kamvar, Z.N.; Brooks, J.C.; Grünwald, N.J. Novel R Tools for Analysis of Genome-wide Population Genetic Data with Emphasis on Clonality. Front. Genet. 2015, 6, 208. [Google Scholar] [CrossRef] [PubMed]
- Agapow, P.-M.; Burt, A. Indices of Multilocus Linkage Disequilibrium. Mol. Ecol. Notes 2001, 1, 101–102. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic Analysis in Excel. Population Genetic Software for Teaching and Research–an Update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
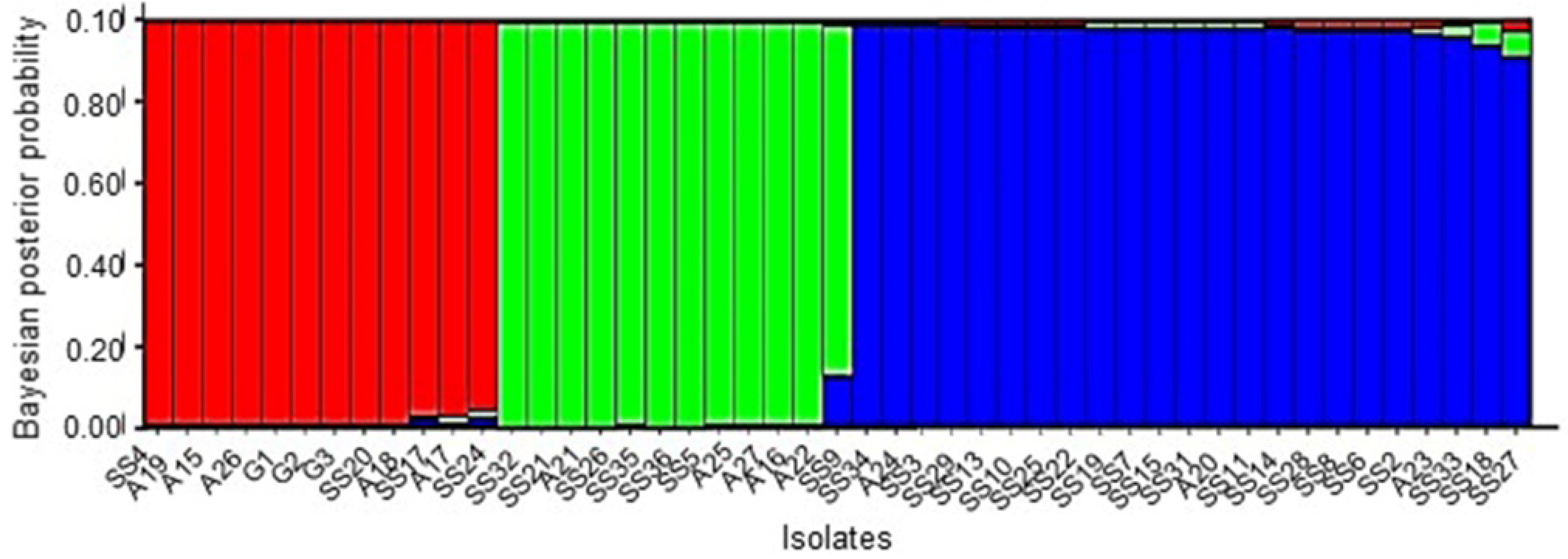
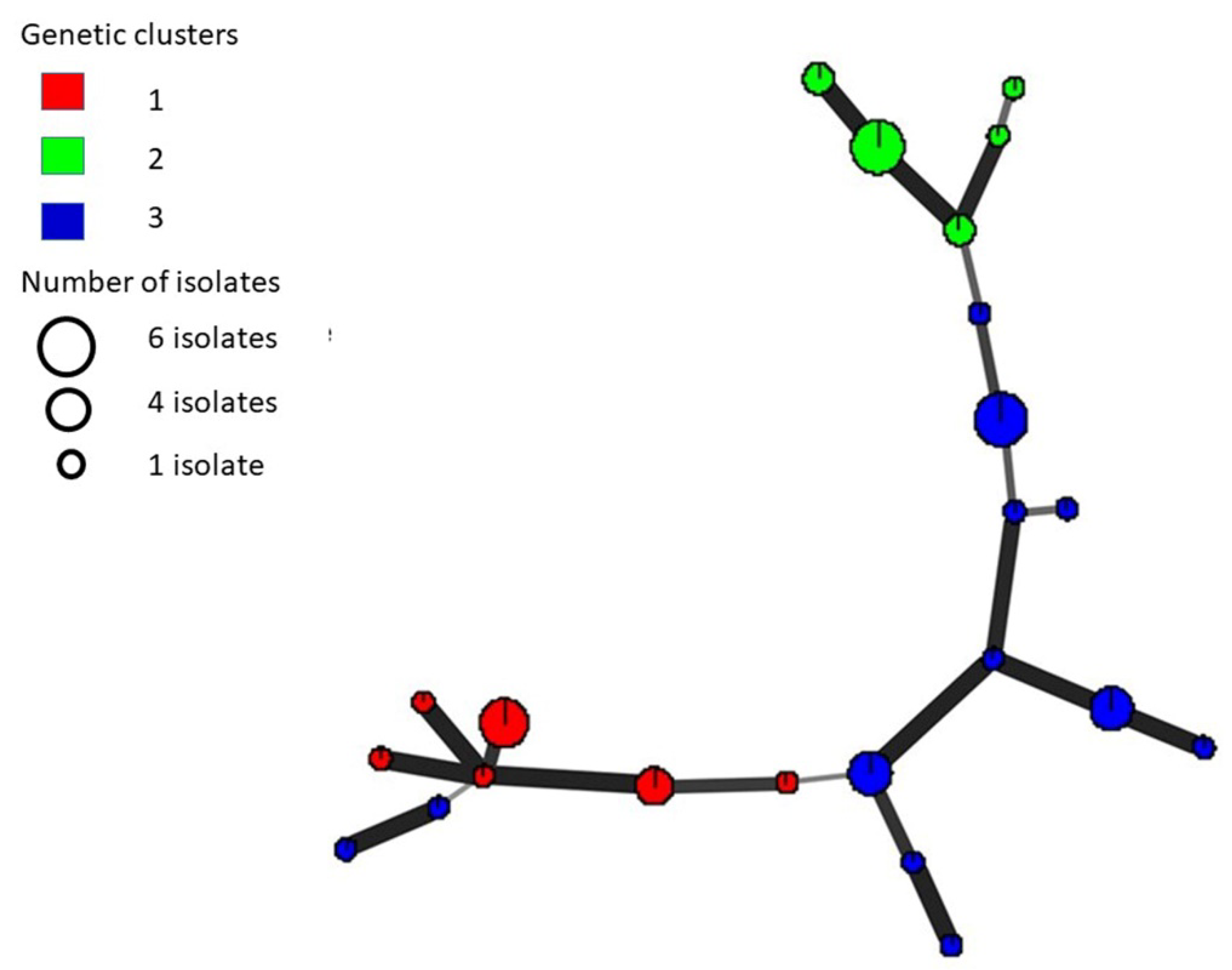
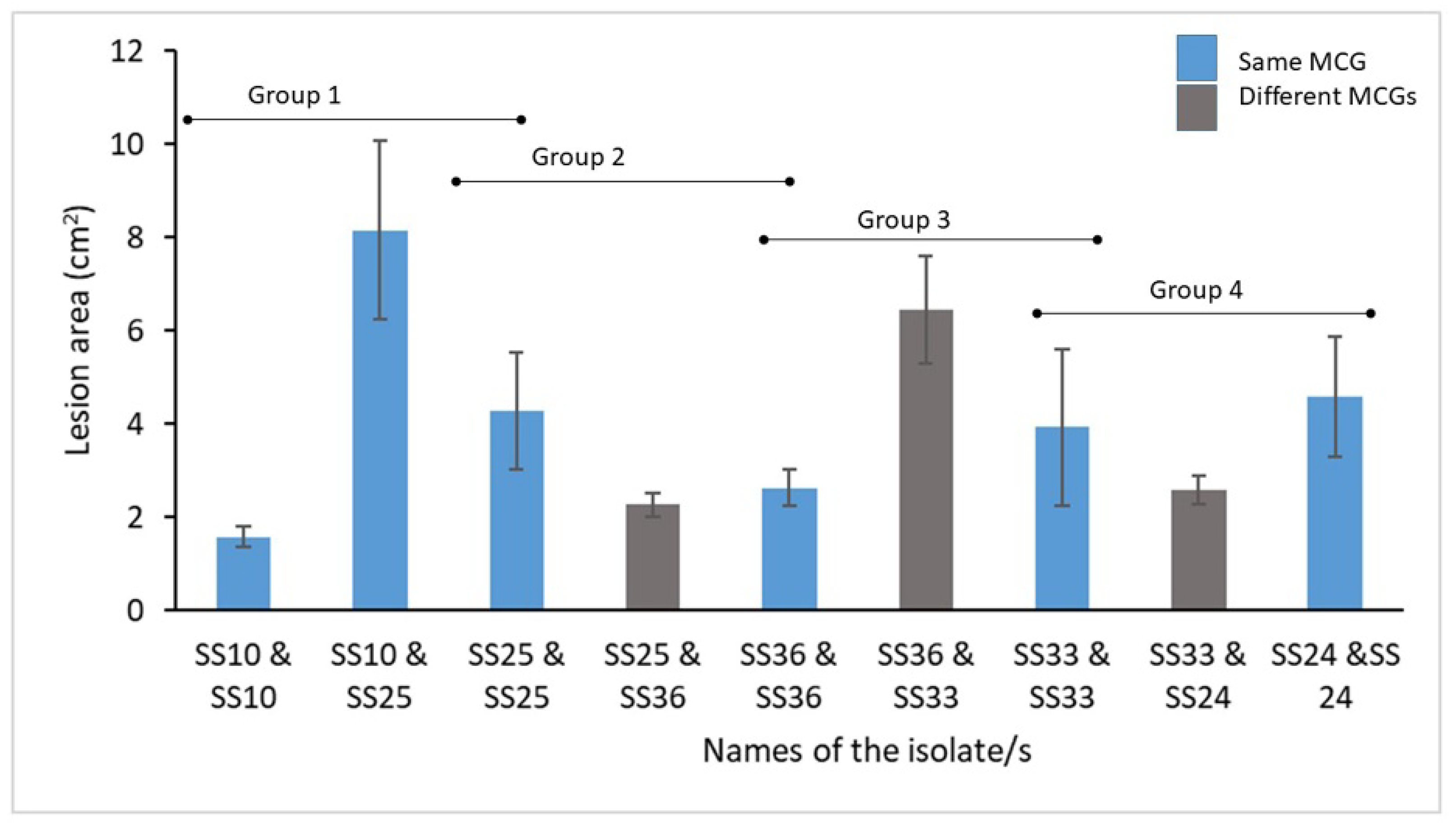

| Isolate Name a | Location a | MCG | MLH | Isolate Name a | Location a | MCG | MLH |
|---|---|---|---|---|---|---|---|
| SS5 | Pattipola Field 2 | 1 | 1 | SS11 | Pattipola Field 4 | 3 | 2 |
| SS26 | Ambewela Field 1 | 1 | 1 | SS15 | Pattipola Field 5 | 3 | 2 |
| SS35 | Ambewela Field 3 | 1 | 1 | SS19 | Pattipola Field 6 | 3 | 2 |
| SS36 | Ambewela Field 4 | 1 | 1 | SS31 | Ambewela Field 1 | 3 | 2 |
| A25 | Seetha Eliya Field 5 | 1 | 1 | A20 | Seetha Eliya Field 3 | 3 | 2 |
| A27 | Seetha Eliya Field 6 | 1 | 1 | SS18 | Pattipola Field 6 | 3 | 15 |
| G2 | Meepilimana Filed 1 | 1 | 3 | A15 | Seetha Eliya Field 1 | 4 | 3 |
| SS6 | Pattipola Field 2 | 1 | 4 | A26 | Seetha Eliya Field 6 | 4 | 3 |
| SS21 | Pattipola Field 7 | 1 | 7 | G1 | Meepilimana Filed 1 | 4 | 3 |
| A21 | Seetha Eliya Field 3 | 1 | 7 | G3 | Meepilimana Filed 1 | 4 | 3 |
| A16 | Seetha Eliya Field 1 | 1 | 8 | SS17 | Pattipola Field 5 | 4 | 14 |
| A22 | Seetha Eliya Field 3 | 1 | 8 | SS20 | Pattipola Field 6 | 4 | 16 |
| SS9 | Pattipola Field 3 | 1 | 12 | SS24 | Pattipola Field 7 | 4 | 17 |
| SS32 | Ambewela Field 2 | 1 | 21 | SS2 | Pattipola Field 1 | 5 | 4 |
| SS8 | Pattipola Field 3 | 2 | 4 | SS4 | Pattipola Field 1 | 6 | 6 |
| A23 | Seetha Eliya Field 4 | 2 | 4 | A18 | Seetha Eliya Field 2 | 7 | 6 |
| SS10 | Pattipola Field 3 | 2 | 5 | A19 | Seetha Eliya Field 2 | 8 | 6 |
| SS13 | Pattipola Field 4 | 2 | 5 | A24 | Seetha Eliya Field 4 | 9 | 9 |
| SS22 | Pattipola Field 7 | 2 | 5 | A17 | Seetha Eliya Field 1 | 10 | 10 |
| SS25 | Pattipola Field 8 | 2 | 5 | SS27 | Ambewela Field 1 | 11 | 18 |
| SS3 | Pattipola Field 1 | 2 | 11 | SS29 | Ambewela Field 2 | 12 | 20 |
| SS14 | Pattipola Field 5 | 2 | 13 | SS33 | Ambewela Field 3 | 13 | 22 |
| SS28 | Ambewela Field 1 | 2 | 19 | SS34 | Ambewela Field 3 | 14 | 23 |
| SS7 | Pattipola Field 2 | 3 | 2 |
| Microsatellite Locus a | Allele (bp) b | Frequency c | Expected Heterozygosity (He) c |
|---|---|---|---|
| 5-2 | 340 | 0.2340 | 0.4947 |
| 342 | 0.6596 | ||
| 344 | 0.0638 | ||
| 346 | 0.0213 | ||
| 7-2 | 180 | 0.0426 | 0.6957 |
| 182 | 0.2553 | ||
| 190 | 0.4043 | ||
| 194 | 0.2979 | ||
| 12-2 | 235 | 0.2340 | 0.6818 |
| 237 | 0.3617 | ||
| 241 | 0.3830 | ||
| 243 | 0.0213 | ||
| 13-2 | 313 | 0.0426 | 0.5402 |
| 315 | 0.6596 | ||
| 329 | 0.1277 | ||
| 331 | 0.1277 | ||
| 333 | 0.0426 | ||
| 17-3 | 375 | 0.0213 | 0.7438 |
| 378 | 0.0426 | ||
| 381 | 0.4043 | ||
| 384 | 0.2553 | ||
| 399 | 0.1915 | ||
| 402 | 0.0638 | ||
| 405 | 0.0213 | ||
| 55-4 | 176 | 0.9574 | 0.0842 |
| 188 | 0.0213 | ||
| 204 | 0.0213 | ||
| 110-4 | 392 | 0.2553 | 0.3885 |
| 396 | 0.7447 | ||
| 114-4 | 359 | 0.0426 | 0.8343 |
| 383 | 0.1489 | ||
| 387 | 0.0213 | ||
| 395 | 0.2553 | ||
| 411 | 0.0213 | ||
| 415 | 0.2128 | ||
| 419 | 0.0213 | ||
| 435 | 0.1702 | ||
| 439 | 0.0638 |
| Host (Number of Samples) | |||
|---|---|---|---|
| Genetic Diversity | Cabbage (N = 47) | Canola in China a (N = 30) | Canola in the USA a (N = 29) |
| Gene diversity | |||
| Number of polymorphic loci | 8 | 7 | 8 |
| Total number of alleles | 38 | 24 | 29 |
| Mean number of alleles per locus | 4.75 | 3 | 3.63 |
| Mean expected heterozygosity (He) | 0.56 | 0.46 | 0.59 |
| Genotypic diversity | |||
| Number of mycelial compatibility groups | 14 | 27 | 19 |
| Number of multilocus haplotypes (g) | 23 | 29 | 19 |
| Genotypic richness (g/N) | 0.49 | 0.97 | 0.66 |
| Population (N = Number of Isolates) | IA | p | |
|---|---|---|---|
| Whole population (N = 47) | 1.524 | 0.224 | <0.001 |
| Clone corrected whole population (N = 23) | 1.126 | 0.162 | <0.001 |
| Cluster 1 (N = 12) a | 0.036 | 0.018 | 0.463 |
| Clone corrected cluster 1 (N = 6) a | −0.455 | −0.228 | 1.000 |
| Cluster 2 (N = 12) a | 0.700 | 0.358 | 0.021 |
| Clone corrected cluster 2 (N = 5) a | 0.771 | 0.387 | 0.13 |
| Cluster 3 (N = 23) a | 1.661 | 0.279 | <0.001 |
| Clone corrected cluster 3 (N = 12) a | 1.211 | 0.202 | 0.001 |
| Locus | 5-2 | 7-2 | 12-2 | 13-2 | 17-3 | 55-4 | 110-4 |
|---|---|---|---|---|---|---|---|
| 7-2 | 0.892 | ||||||
| 12-2 | 0.972 | 1.000 | |||||
| 13-2 | 0.811 | 1.000 | 1.000 | ||||
| 17-3 | 0.947 | 1.000 | 1.000 | 1.000 | |||
| 55-4 | 0.738 | 1.000 | 1.000 | 1.000 | 1.000 | ||
| 110-4 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | |
| 114-4 | 0.453 | 0.543 | 0.423 | 0.675 | 0.321 | 0.826 | 0.714 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mahalingam, T.; Chen, W.; Rajapakse, C.S.; Somachandra, K.P.; Attanayake, R.N. Genetic Diversity and Recombination in the Plant Pathogen Sclerotinia sclerotiorum Detected in Sri Lanka. Pathogens 2020, 9, 306. https://doi.org/10.3390/pathogens9040306
Mahalingam T, Chen W, Rajapakse CS, Somachandra KP, Attanayake RN. Genetic Diversity and Recombination in the Plant Pathogen Sclerotinia sclerotiorum Detected in Sri Lanka. Pathogens. 2020; 9(4):306. https://doi.org/10.3390/pathogens9040306
Chicago/Turabian StyleMahalingam, Thirega, Weidong Chen, Chandima Shashikala Rajapakse, Kandangamuwa Pathirannahalage Somachandra, and Renuka Nilmini Attanayake. 2020. "Genetic Diversity and Recombination in the Plant Pathogen Sclerotinia sclerotiorum Detected in Sri Lanka" Pathogens 9, no. 4: 306. https://doi.org/10.3390/pathogens9040306
APA StyleMahalingam, T., Chen, W., Rajapakse, C. S., Somachandra, K. P., & Attanayake, R. N. (2020). Genetic Diversity and Recombination in the Plant Pathogen Sclerotinia sclerotiorum Detected in Sri Lanka. Pathogens, 9(4), 306. https://doi.org/10.3390/pathogens9040306





