Distinct Signaling Pathways Between Human Macrophages and Primary Gingival Epithelial Cells by Aggregatibacter actinomycetemcomitans
Abstract
1. Introduction
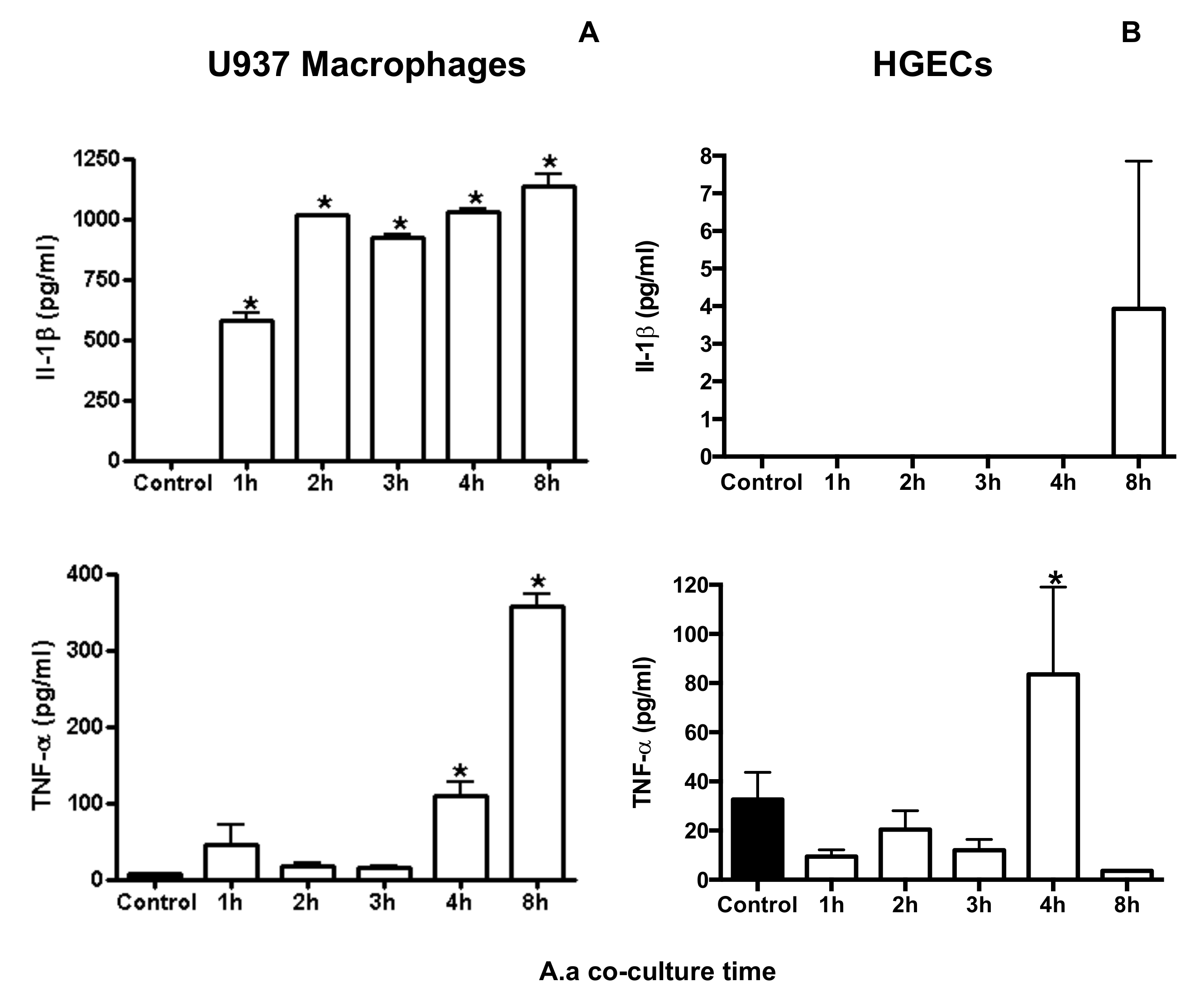
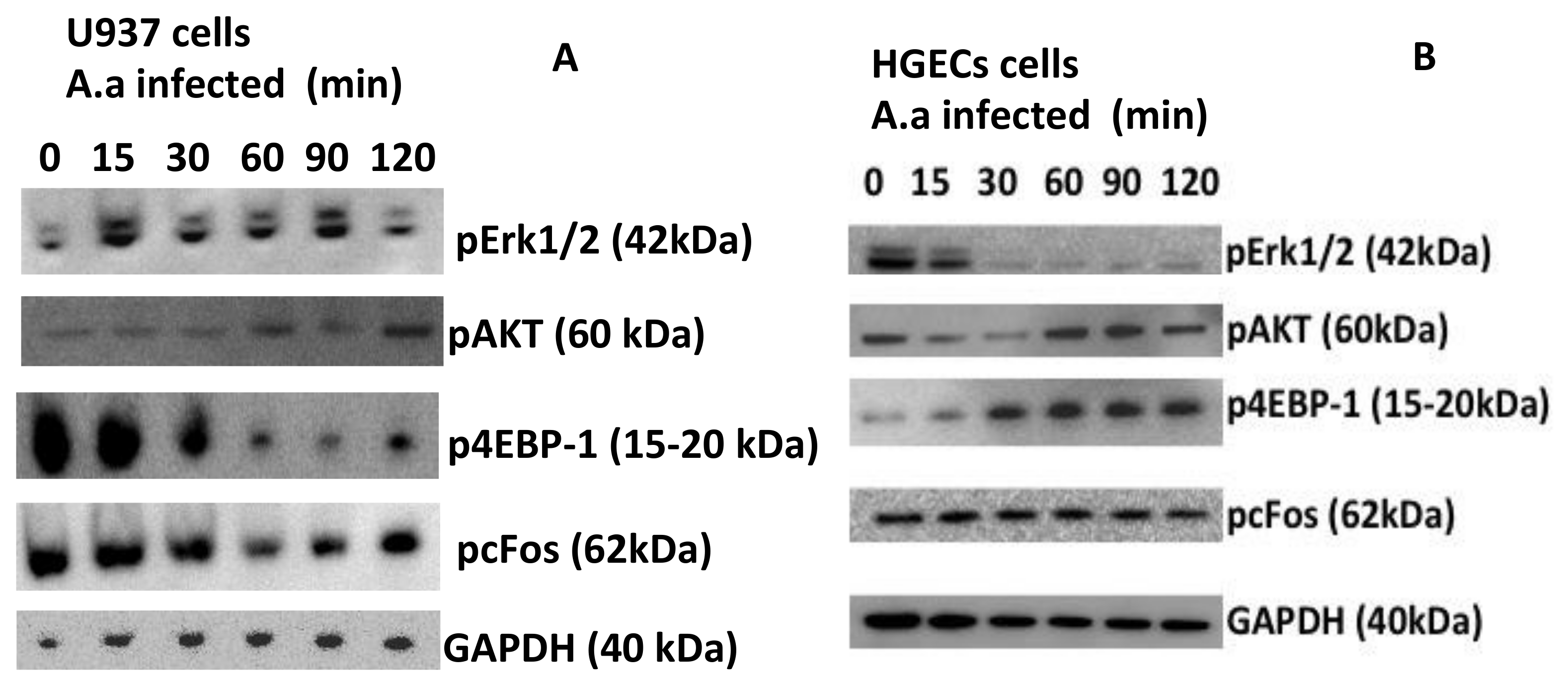
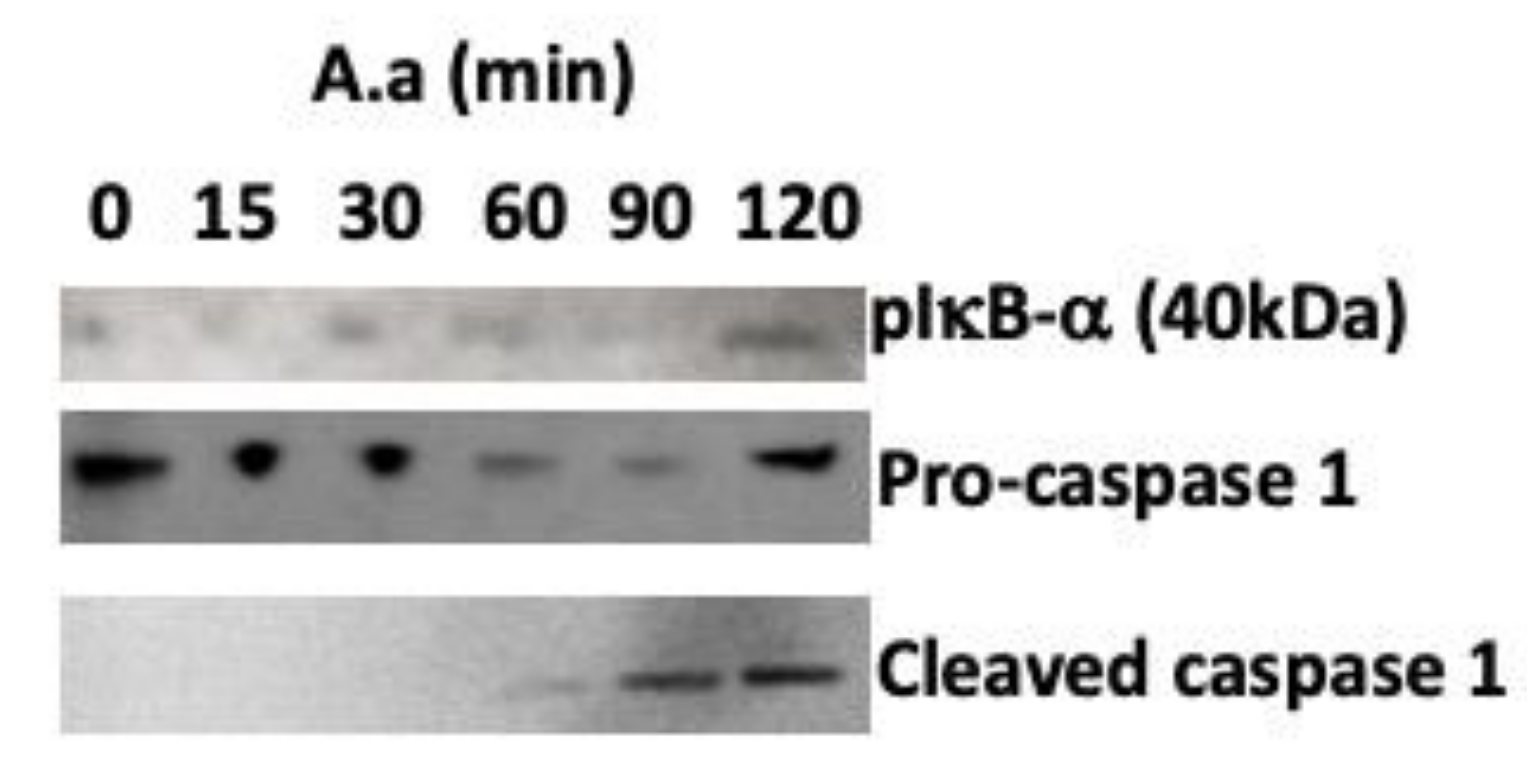
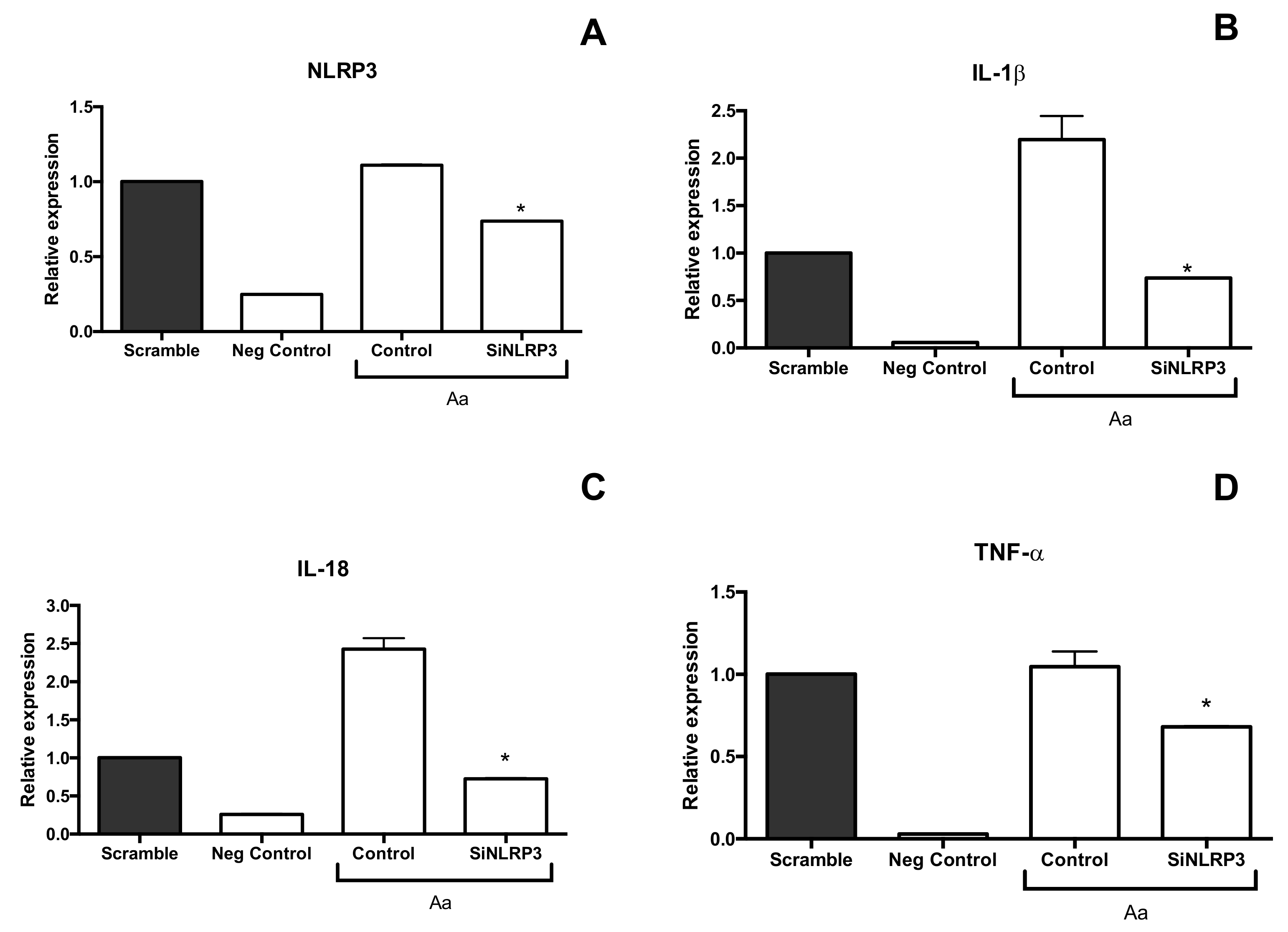
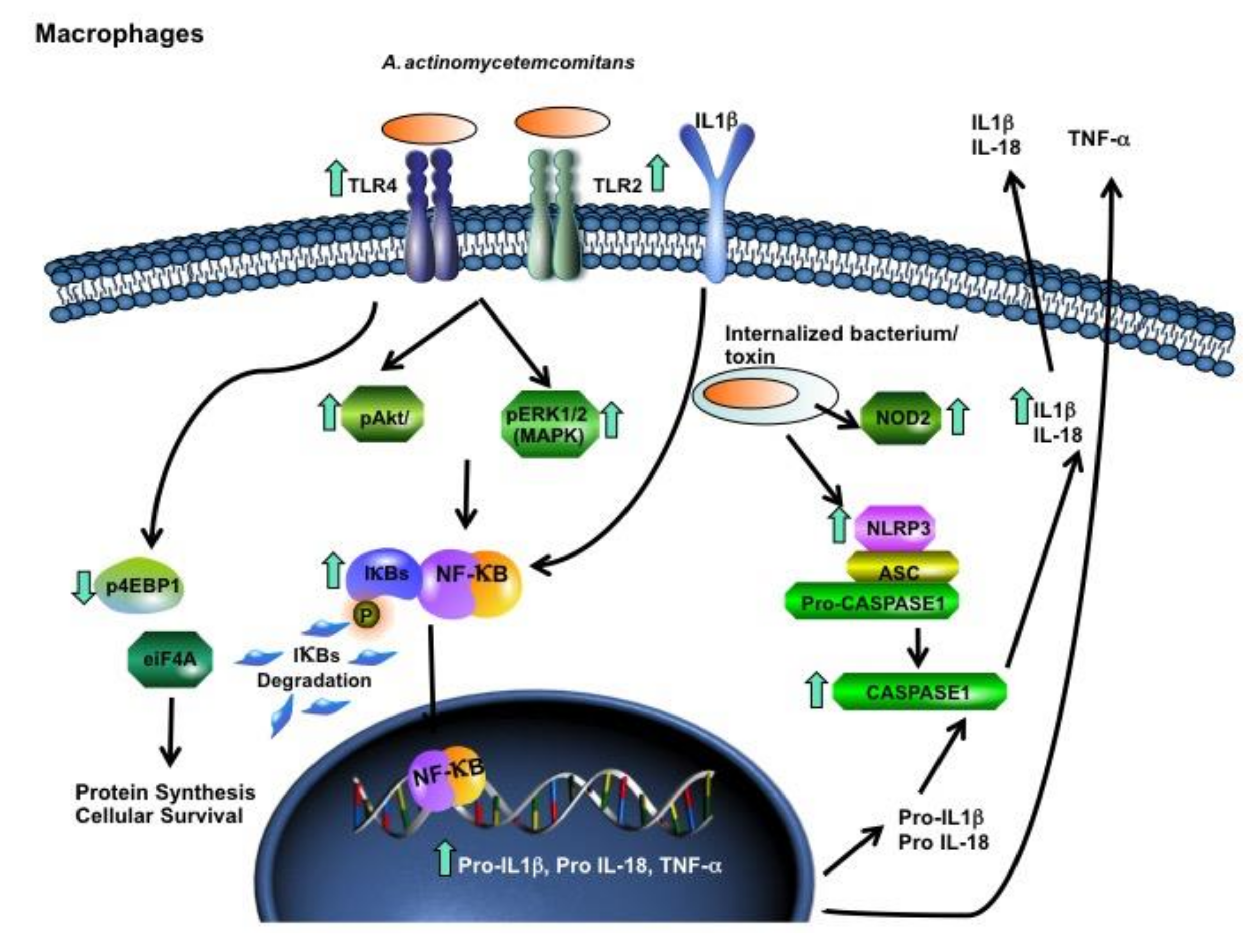
2. Results
3. Discussion
4. Material and Methods
4.1. Eukaryotic Cells and Bacteria Culture Conditions
4.2. Co-culture Assay
4.3. Gene Expression
4.4. Activation of Inflammasome-Related Signaling Pathways
4.5. Cytokines Quantification
4.6. Silencing of NLRP3
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Graves, D. Cytokines that promote periodontal tissue destruction. J. Periodontol. 2008, 79, 1585–1591. [Google Scholar] [CrossRef]
- Kinane, D.F.; Bartold, P.M. Clinical relevance of the host responses of periodontitis. Periodontology 2000 2007, 43, 278–293. [Google Scholar] [CrossRef] [PubMed]
- Benakanakere, M.; Kinane, D.F. Innate cellular responses to the periodontal biofilm. Front. Oral Biol. 2012, 15, 41–55. [Google Scholar] [PubMed]
- Hajishengallis, G. Periodontitis: From microbial immune subversion to systemic inflammation. Nat. Rev. Immunol. 2015, 15, 30–44. [Google Scholar] [CrossRef] [PubMed]
- Benakanakere, M.; Abdolhosseini, M.; Hosur, K.; Finoti, L.S.; Kinane, D.F. TLR2 promoter hypermethylation creates innate immune dysbiosis. J. Dent. Res. 2015, 94, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Fine, D.H.; Markowitz, K.; Furgang, D.; Fairlie, K.; Ferrandiz, J.; Nasri, C.; McKiernan, M.; Gunsolley, J. Aggregatibacter actinomycetemcomitans and its relationship to initiation of localized aggressive periodontitis: Longitudinal cohort study of initially healthy adolescents. J. Clin. Microbiol. 2007, 45, 3859–3869. [Google Scholar] [CrossRef] [PubMed]
- Norskov-Lauritsen, N.; Claesson, R.; Birkeholm Jensen, A.; Aberg, C.H.; Haubek, D. Aggregatibacter Actinomycetemcomitans: Clinical Significance of a Pathobiont Subjected to Ample Changes in Classification and Nomenclature. Pathogens 2019, 8, 243. [Google Scholar] [CrossRef]
- Paturel, L.; Casalta, J.P.; Habib, G.; Nezri, M.; Raoult, D. Actinobacillus actinomycetemcomitans endocarditis. Clin. Microbiol. Infect. 2004, 10, 98–118. [Google Scholar] [CrossRef]
- Calandrini, C.A.; Ribeiro, A.C.; Gonnelli, A.C.; Ota-Tsuzuki, C.; Rangel, L.P.; Saba-Chujfi, E.; Mayer, M.P. Microbial composition of atherosclerotic plaques. Oral Dis. 2014, 20, e128–e134. [Google Scholar] [CrossRef]
- Konig, M.F.; Abusleme, L.; Reinholdt, J.; Palmer, R.J.; Teles, R.P.; Sampson, K.; Rosen, A.; Nigrovic, P.A.; Sokolove, J.; Giles, J.T.; et al. Aggregatibacter actinomycetemcomitans-induced hypercitrullination links periodontal infection to autoimmunity in rheumatoid arthritis. Sci. Transl. Med. 2016, 8. [Google Scholar] [CrossRef]
- Fine, D.H.; Markowitz, K.; Fairlie, K.; Tischio-Bereski, D.; Ferrendiz, J.; Furgang, D.; Paster, B.J.; Dewhirst, F.E. A consortium of Aggregatibacter actinomycetemcomitans, Streptococcus parasanguinis, and Filifactor alocis is present in sites prior to bone loss in a longitudinal study of localized aggressive periodontitis. J. Clin. Microbiol. 2013, 51, 2850–2861. [Google Scholar] [CrossRef] [PubMed]
- Shenker, B.J.; Boesze-Battaglia, K.; Scuron, M.D.; Walker, L.P.; Zekavat, A.; Dlakic, M. The toxicity of the Aggregatibacter actinomycetemcomitans cytolethal distending toxin correlates with its phosphatidylinositol-3,4,5-triphosphate phosphatase activity. Cell. Microbiol. 2016, 18, 223–243. [Google Scholar] [CrossRef] [PubMed]
- Kajiya, M.; Komatsuzawa, H.; Papantonakis, A.; Seki, M.; Makihira, S.; Ouhara, K.; Kusumoto, Y.; Murakami, S.; Taubman, M.A.; Kawai, T. Aggregatibacter actinomycetemcomitans Omp29 is associated with bacterial entry to gingival epithelial cells by F-actin rearrangement. PLoS ONE 2011, 6, e18287. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, S.; Sen, G.C. Tyrosine phosphorylation in Toll-like receptor signaling. Cytokine Growth Factor Rev. 2014, 25, 533–541. [Google Scholar] [CrossRef]
- Martinon, F.; Tschopp, J. NLRs join TLRs as innate sensors of pathogens. Trends Immunol. 2005, 26, 447–454. [Google Scholar] [CrossRef]
- Ferrand, J.; Ferrero, R.L. Recognition of Extracellular Bacteria by NLRs and Its Role in the Development of Adaptive Immunity. Front. Immunol. 2013, 4, 344. [Google Scholar] [CrossRef]
- Brewer, S.M.; Brubaker, S.W.; Monack, D.M. Host inflammasome defense mechanisms and bacterial pathogen evasion strategies. Curr. Opin. Immunol. 2019, 60, 63–70. [Google Scholar] [CrossRef]
- Abderrazak, A.; Syrovets, T.; Couchie, D.; El Hadri, K.; Friguet, B.; Simmet, T.; Rouis, M. NLRP3 inflammasome: From a danger signal sensor to a regulatory node of oxidative stress and inflammatory diseases. Redox Biol. 2015, 4, 296–307. [Google Scholar] [CrossRef]
- Franchi, L.; Muñoz-Planillo, R.; Núñez, G. Sensing and reacting to microbes through the inflammasomes. Nat. Immunol. 2012, 13, 325–332. [Google Scholar] [CrossRef]
- Tsuchiya, K. Inflammasome-associated cell death: Pyroptosis, apoptosis, and physiological implications. Microbiol. Immunol. 2020. [Google Scholar] [CrossRef]
- Kikkert, R.; Laine, M.L.; Aarden, L.A.; van Winkelhoff, A.J. Activation of toll-like receptors 2 and 4 by gram-negative periodontal bacteria. Oral Microbiol. Immunol. 2007, 22, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Umeda, J.E.; Demuth, D.R.; Ando, E.S.; Faveri, M.; Mayer, M.P. Signaling transduction analysis in gingival epithelial cells after infection with Aggregatibacter actinomycetemcomitans. Mol. Oral Microbiol. 2012, 27, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, B.C.; Moffatt, C.E.; Hagerty, D.; Whitmore, S.E.; Brown, T.A.; Graves, D.T.; Lamont, R.J. Interaction of oral bacteria with gingival epithelial cell multilayers. Mol. Oral Microbiol. 2011, 26, 210–220. [Google Scholar] [CrossRef] [PubMed]
- Kato, S.; Nakashima, K.; Nagasawa, T.; Abiko, Y.; Furuichi, Y. Involvement of Toll-like receptor 2 in apoptosis of Aggregatibacter actinomycetemcomitans-infected THP-1 cells. J. Microbiol. Immunol. Infect. 2013, 46, 164–170. [Google Scholar] [CrossRef]
- Kelk, P.; Claesson, R.; Chen, C.; Sjöstedt, A.; Johansson, A. IL-1beta secretion induced by Aggregatibacter (Actinobacillus) actinomycetemcomitans is mainly caused by the leukotoxin. Int. J. Med Microbiol. 2008, 298, 529–541. [Google Scholar] [CrossRef]
- Kelk, P.; Claesson, R.; Hänström, L.; Lerner, U.H.; Kalfas, S.; Johansson, A. Abundant secretion of bioactive interleukin-1beta by human macrophages induced by Actinobacillus actinomycetemcomitans leukotoxin. Infect. Immun. 2005, 73, 453–458. [Google Scholar] [CrossRef]
- Kelk, P.; Abd, H.; Claesson, R.; Sandstrom, G.; Sjostedt, A.; Johansson, A. Cellular and molecular response of human macrophages exposed to Aggregatibacter actinomycetemcomitans leukotoxin. Cell Death Dis. 2011, 2, e126. [Google Scholar] [CrossRef]
- Xue, F.; Shu, R.; Xie, Y. The expression of NLRP3, NLRP1 and AIM2 in the gingival tissue of periodontitis patients: RT-PCR study and immunohistochemistry. Arch. Oral Biol. 2015, 60, 948–958. [Google Scholar] [CrossRef]
- Isaza-Guzman, D.M.; Medina-Piedrahita, V.M.; Gutierrez-Henao, C.; Tobon-Arroyave, S.I. Salivary Levels of NLRP3 Inflammasome-Related Proteins as Potential Biomarkers of Periodontal Clinical Status. J. Periodontol. 2017, 88, 1329–1338. [Google Scholar] [CrossRef]
- Shenker, B.J.; Ojcius, D.M.; Walker, L.P.; Zekavat, A.; Scuron, M.D.; Boesze-Battaglia, K. Aggregatibacter actinomycetemcomitans cytolethal distending toxin activates the NLRP3 inflammasome in human macrophages, leading to the release of proinflammatory cytokines. Infect. Immun. 2015, 83, 1487–1496. [Google Scholar] [CrossRef]
- Belibasakis, G.N.; Johansson, A. Aggregatibacter actinomycetemcomitans targets NLRP3 and NLRP6 inflammasome expression in human mononuclear leukocytes. Cytokine 2012, 59, 124–130. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Liu, J.; Pan, C.; Pan, Y. NLRP3 inflammasome is required for apoptosis of Aggregatibacter actinomycetemcomitans-infected human osteoblastic MG63 cells. Acta Histochem. 2014, 116, 1119–1124. [Google Scholar] [CrossRef] [PubMed]
- Thay, B.; Damm, A.; Kufer, T.A.; Wai, S.N.; Oscarsson, J. Aggregatibacter actinomycetemcomitans outer membrane vesicles are internalized in human host cells and trigger NOD1- and NOD2-dependent NF-kappaB activation. Infect. Immun. 2014, 82, 4034–4046. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, O.; Lee, K.L. The inflammasome and danger molecule signaling: At the crossroads of inflammation and pathogen persistence in the oral cavity. Periodontology 2000 2015, 69, 83–95. [Google Scholar] [CrossRef]
- Fukushima, H.; Jimi, E.; Okamoto, F.; Motokawa, W.; Okabe, K. IL-1-induced receptor activator of NF-kappa B ligand in human periodontal ligament cells involves ERK-dependent PGE2 production. Bone 2005, 36, 267–275. [Google Scholar] [CrossRef]
- Stathopoulou, P.G.; Benakanakere, M.R.; Galicia, J.C.; Kinane, D.F. Epithelial cell pro-inflammatory cytokine response differs across dental plaque bacterial species. J. Clin. Periodontol. 2010, 37, 24–29. [Google Scholar] [CrossRef]
- Uchida, Y.; Shiba, H.; Komatsuzawa, H.; Takemoto, T.; Sakata, M.; Fujita, T.; Kawaguchi, H.; Sugai, M.; Kurihara, H. Expression of IL-1 beta and IL-8 by human gingival epithelial cells in response to Actinobacillus actinomycetemcomitans. Cytokine 2001, 14, 152–161. [Google Scholar] [CrossRef]
- Gelani, V.; Fernandes, A.P.; Gasparoto, T.H.; Garlet, T.P.; Cestari, T.M.; Lima, H.R.; Ramos, E.S.; de Souza Malaspina, T.S.; Santos, C.F.; Garlet, G.P.; et al. The role of toll-like receptor 2 in the recognition of Aggregatibacter actinomycetemcomitans. J. Periodontol. 2009, 80, 2010–2019. [Google Scholar] [CrossRef]
- Park, S.R.; Kim, D.J.; Han, S.H.; Kang, M.J.; Lee, J.Y.; Jeong, Y.J.; Lee, S.J.; Kim, T.H.; Ahn, S.G.; Yoon, J.H.; et al. Diverse Toll-like receptors mediate cytokine production by Fusobacterium nucleatum and Aggregatibacter actinomycetemcomitans in macrophages. Infect. Immun. 2014, 82, 1914–1920. [Google Scholar] [CrossRef]
- Diaz-Zuniga, J.; Monasterio, G.; Alvarez, C.; Melgar-Rodriguez, S.; Benitez, A.; Ciuchi, P.; Garcia, M.; Arias, J.; Sanz, M.; Vernal, R. Variability of the dendritic cell response triggered by different serotypes of Aggregatibacter actinomycetemcomitans or Porphyromonas gingivalis is toll-like receptor 2 (TLR2) or TLR4 dependent. J. Periodontol. 2015, 86, 108–119. [Google Scholar] [CrossRef]
- Ando-Suguimoto, E.S.; da Silva, M.P.; Kawamoto, D.; Chen, C.; DiRienzo, J.M.; Mayer, M.P. The cytolethal distending toxin of Aggregatibacter actinomycetemcomitans inhibits macrophage phagocytosis and subverts cytokine production. Cytokine 2014, 66, 46–53. [Google Scholar] [CrossRef]
- Tsai, W.H.; Huang, D.Y.; Yu, Y.H.; Chen, C.Y.; Lin, W.W. Dual roles of NOD2 in TLR4-mediated signal transduction and -induced inflammatory gene expression in macrophages. Cell. Microbiol. 2011, 13, 717–730. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Jiang, B.; Zhang, Y. 4E-BP1, a multifactor regulated multifunctional protein. Cell Cycle 2016, 15, 781–786. [Google Scholar] [CrossRef] [PubMed]
- Vanhaesebroeck, B.; Alessi, D.R. The PI3K-PDK1 connection: More than just a road to PKB. Biochem. J. 2000, 346, 561–576. [Google Scholar] [PubMed]
- Kim, J.Y.; Paton, J.C.; Briles, D.E.; Rhee, D.K.; Pyo, S. Streptococcus pneumoniae induces pyroptosis through the regulation of autophagy in murine microglia. Oncotarget 2015, 6, 44161–44178. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Shi, J.; He, Q.; Hu, Q.; Wang, Y.Y.; Zhang, L.J.; Chan, W.T.; Chen, W.X. Streptococcus pneumoniae induces autophagy through the inhibition of the PI3K-I/Akt/mTOR pathway and ROS hypergeneration in A549 cells. PLoS ONE 2015, 10, e0122753. [Google Scholar] [CrossRef] [PubMed]
- Owen, K.A.; Meyer, C.B.; Bouton, A.H.; Casanova, J.E. Activation of focal adhesion kinase by Salmonella suppresses autophagy via an Akt/mTOR signaling pathway and promotes bacterial survival in macrophages. PLoS Pathog. 2014, 10, e1004159. [Google Scholar] [CrossRef] [PubMed]
- Monje, P.; Hernandez-Losa, J.; Lyons, R.J.; Castellone, M.D.; Gutkind, J.S. Regulation of the transcriptional activity of c-Fos by ERK. A novel role for the prolyl isomerase PIN1. J. Biol. Chem. 2005, 280, 35081–35084. [Google Scholar] [CrossRef]
- Arai, A.; Mizoguchi, T.; Harada, S.; Kobayashi, Y.; Nakamichi, Y.; Yasuda, H.; Penninger, J.M.; Yamada, K.; Udagawa, N.; Takahashi, N. Fos plays an essential role in the upregulation of RANK expression in osteoclast precursors within the bone microenvironment. J. Cell Sci. 2012, 125, 2910–2917. [Google Scholar] [CrossRef]
- Gutierrez-Venegas, G.; Castillo-Aleman, R. Characterization of the transduction pathway involved in c-fos and c-jun expression induced by Aggregatibacter actinomycetemcomitans lipopolysaccharides in human gingival fibroblasts. Int. Immunopharmacol. 2008, 8, 1513–1523. [Google Scholar] [CrossRef]
- Latz, E.; Xiao, T.S.; Stutz, A. Activation and regulation of the inflammasomes. Nat. Rev. Immunol. 2013, 13, 397–411. [Google Scholar] [CrossRef] [PubMed]
- Hajishengallis, G.; Wang, M.; Liang, S. Induction of distinct TLR2-mediated proinflammatory and proadhesive signaling pathways in response to Porphyromonas gingivalis fimbriae. J. Immunol. 2009, 182, 6690–6696. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, M.; Ceballos-Olvera, I.; del Barrio, L.; Re, F. Role of the inflammasome, IL-1beta, and IL-18 in bacterial infections. Sci. World J. 2011, 11, 2037–2050. [Google Scholar] [CrossRef] [PubMed]
- Sfakianakis, A.; Barr, C.E.; Kreutzer, D.L. Actinobacillus actinomycetemcomitans-induced expression of IL-1alpha and IL-1beta in human gingival epithelial cells: Role in IL-8 expression. Eur. J. Oral Sci. 2001, 109, 393–401. [Google Scholar] [CrossRef]
- Handfield, M.; Mans, J.J.; Zheng, G.; Lopez, M.C.; Mao, S.; Progulske-Fox, A.; Narasimhan, G.; Baker, H.V.; Lamont, R.J. Distinct transcriptional profiles characterize oral epithelium-microbiota interactions. Cell. Microbiol. 2005, 7, 811–823. [Google Scholar] [CrossRef]
- Stathopoulou, P.G.; Galicia, J.C.; Benakanakere, M.R.; Garcia, C.A.; Potempa, J.; Kinane, D.F. Porphyromonas gingivalis induce apoptosis in human gingival epithelial cells through a gingipain-dependent mechanism. BMC Microbiol. 2009, 9, 107. [Google Scholar] [CrossRef]
- Kim, S.; Park, M.H.; Song, Y.R.; Na, H.S.; Chung, J. Aggregatibacter actinomycetemcomitans-Induced AIM2 Inflammasome Activation Is Suppressed by Xylitol in Differentiated THP-1 Macrophages. J. Periodontol. 2016, 87, e116–126. [Google Scholar] [CrossRef]
- Palm, N.W.; de Zoete, M.R.; Flavell, R.A. Immune-microbiota interactions in health and disease. Clin. Immunol. 2015. [Google Scholar] [CrossRef]
- Sundstrom, C.; Nilsson, K. Establishment and characterization of a human histiocytic lymphoma cell line (U-937). Int. J. Cancer 1976, 17, 565–577. [Google Scholar] [CrossRef]
- Minta, J.O.; Pambrun, L. In vitro induction of cytologic and functional differentiation of the immature human monocytelike cell line U-937 with phorbol myristate acetate. Am. J. Pathol. 1985, 119, 111–126. [Google Scholar]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
| Gene Expression (HGECs) | Control (0 h) | Aa 1 h | Aa 2 h | Aa 3 h | Aa 4 h | Aa 8 h |
|---|---|---|---|---|---|---|
| Fold-Change (±SD) | ||||||
| tlr-4 | 1.00 | 0.79 (±0.14) | 0.74 (±0.10) | 0.68 (±0.08) | 0.39 (±0.04) | 0.32 (±0.01) |
| tlr-2 | 1.00 | 0.78 (±0.01) | 0.66 (±0.09) | 0.47 (±0.04) | 0.41 (±0.03) | 0.46 (±0.04) |
| nlrp3 | 1.00 | 1.15 (±0.23) | 0.76 (±0.11) | 0.64 (±0.09) | 0.50 (±0.03) | 1.07 (0.05) |
| nod1 | 1.00 | 1.15 (±0.24) | 1.38 (±0.20) | 0.80 (±0.00) | 0.58 (±0.02) | 0.42 (0 05) |
| nod2 | 1.00 | 0.77 (0.09) | 0.77 (±0.09) | 0.72 (±0.08) | 0.77 (±0.01) | 0.76 (0.03) |
| il-1β | 1.00 | 1.22 (±0.06) * | 0.72 (±0.01) | 0.33 (±0.01) | 0.51 (±0.02) | 0.13 (±0.01) |
| Il-18 | 1.00 | 1.27 (±0.12) * | 0.93 (±0.09) | 0.70 (±0.05) | 0.64 (±0.09) | 0.43 (±0.02) |
| tnf | 1.00 | 0.88 (±0.05) | 0.98 (±0.06) | 2.00 (±0.21) * | 3.72 (±0.13) * | 0.94 (±0.09) |
| Gene Expression (U937 cells) | Control (0 h) | Aa 1 h | Aa 2 h | Aa 3 h | Aa 4 h | Aa 8 h |
|---|---|---|---|---|---|---|
| Fold-Change (±SD) | ||||||
| tlr-4 | 1.00 | 1.27 (±0.03) * | 1.30 (±0.10) * | 1.15 (±0.01) * | 0.44 (±0.00) | 0.25 (±0.00) |
| tlr-2 | 1.00 | 1.18 (±0.10) | 1.51 (±0.18) * | 1.47 (±0.14) * | 0.75 (±0.04) | 0.68 (±0.01) |
| nlrp3 | 1.00 | 0.92 (±0.01) | 2.03 (±0.17) * | 1.85 (0.18) * | 0.70 (±0.02) | 0.83 (0.04) |
| nod1 | 1.00 | 1.24 (±0.03) | 1.07 (±0.05) | 1.13 (±0.40) | 1.90 (±0.17) | 0.76 (0.09) |
| nod2 | 1.00 | 1.25 (0.07) * | 1.24 (±0.02) * | 1.29 (±0.07) * | 1.67 (±0.13) * | 2.34 (0.06) * |
| il-1b | 1.00 | 10.75 (±0.43) * | 31.79 (±0.30) * | 43.19 (±1.01) * | 26.75 (±2.88) * | 54.56 (±7.20) * |
| Il-18 | 1.00 | 1.62 (±0.05) * | 1.34 (±0.16) * | 1.42 (±0.17) * | 1.23 (±0.13) | 0.83 (±0.01) |
| tnf | 1.00 | 7.18 (±0.41) | 85.13 (±6.84) * | 134.96 (±26.93) * | 85.53 (±1.44) * | 68.86 (±2.54) * |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ando-Suguimoto, E.S.; Benakanakere, M.R.; Mayer, M.P.A.; Kinane, D.F. Distinct Signaling Pathways Between Human Macrophages and Primary Gingival Epithelial Cells by Aggregatibacter actinomycetemcomitans. Pathogens 2020, 9, 248. https://doi.org/10.3390/pathogens9040248
Ando-Suguimoto ES, Benakanakere MR, Mayer MPA, Kinane DF. Distinct Signaling Pathways Between Human Macrophages and Primary Gingival Epithelial Cells by Aggregatibacter actinomycetemcomitans. Pathogens. 2020; 9(4):248. https://doi.org/10.3390/pathogens9040248
Chicago/Turabian StyleAndo-Suguimoto, Ellen S., Manjunatha R. Benakanakere, Marcia P.A. Mayer, and Denis F. Kinane. 2020. "Distinct Signaling Pathways Between Human Macrophages and Primary Gingival Epithelial Cells by Aggregatibacter actinomycetemcomitans" Pathogens 9, no. 4: 248. https://doi.org/10.3390/pathogens9040248
APA StyleAndo-Suguimoto, E. S., Benakanakere, M. R., Mayer, M. P. A., & Kinane, D. F. (2020). Distinct Signaling Pathways Between Human Macrophages and Primary Gingival Epithelial Cells by Aggregatibacter actinomycetemcomitans. Pathogens, 9(4), 248. https://doi.org/10.3390/pathogens9040248




