High Human Papillomavirus DNA loads in Inflammatory Middle Ear Diseases
Abstract
1. Introduction
2. Results
2.1. HPV DNA Detection
2.2. HPV DNA Load
2.3. HPV Genotyping
2.4. Association between HPV and Co-Factors
3. Discussion
4. Materials and Methods
4.1. Patients and Specimens
4.2. DNA Isolation
4.3. Viral DNA Load Quantification
4.4. HPV Genotyping
4.5. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Geißler, C.; Tahtali, A.; Diensthuber, M.; Gassner, D.; Stöver, T.; Wagenblast, J. The role of p16 expression as a predictive marker in HPV-positive oral SCCHN—A retrospective single-center study. Anticancer Res. 2013, 33, 913–916. [Google Scholar] [PubMed]
- Gillison, M.L.; Alemany, L.; Snijders, P.J.F.; Chaturvedi, A.; Steinberg, B.M.; Schwartz, S.; Castellsagué, X. Human papillomavirus and diseases of the upper airway: Head and neck cancer and respiratory papillomatosis. Vaccine 2012, 30 (Suppl. 5), F34–F54. [Google Scholar] [CrossRef] [PubMed]
- Rydzewski, B.; Goździcka-Józefiak, A.; Sokalski, J.; Matusiak, M.; Durzyński, L. Identification of human papilloma viruses (HPV) in inflammatory states and ear neoplasms. Otolaryngol. Pol. Pol. Otolaryngol. 2007, 61, 137–141. [Google Scholar] [CrossRef]
- Jin, Y.T.; Tsai, S.T.; Li, C.; Chang, K.C.; Yan, J.J.; Chao, W.Y.; Eng, H.L.; Chou, T.Y.; Wu, T.C.; Su, I.J. Prevalence of human papillomavirus in middle ear carcinoma associated with chronic otitis media. Am. J. Pathol. 1997, 150, 1327–1333. [Google Scholar]
- Tsai, S.T.; Li, C.; Jin, Y.T.; Chao, W.Y.; Su, I.J. High prevalence of human papillomavirus types 16 and 18 in middle-ear carcinomas. Int. J. Cancer 1997, 71, 208–212. [Google Scholar] [CrossRef]
- Gurgel, R.K.; Karnell, L.H.; Hansen, M.R. Middle ear cancer: A population-based study. Laryngoscope 2009, 119, 1913–1917. [Google Scholar] [CrossRef]
- Gidley, P.W.; Roberts, D.B.; Sturgis, E.M. Squamous cell carcinoma of the temporal bone. Laryngoscope 2010, 120, 1144–1151. [Google Scholar] [CrossRef]
- Shu, M.-T.; Lee, J.-C.; Yang, C.-C.; Wu, K.-C. Squamous cell carcinoma of the middle ear. Ear. Nose. Throat J. 2012, 91, 14. [Google Scholar] [CrossRef]
- Surono, A.; Hariwiyanto, B.; Samodra, E. Detection of Epstein-Barr and Human Papilloma Viruses in the Middle Ear Squamous Cell Carcinoma. Indian J. Otolaryngol. Head Neck Surg. 2018, 70, 66–70. [Google Scholar] [CrossRef] [PubMed]
- Chao, W.Y.; Chang, S.J.; Jin, Y.T. Detection of human papillomavirus in cholesteatomas. Eur. Arch. Oto-Rhino-Laryngol. 2000, 257, 120–123. [Google Scholar] [CrossRef] [PubMed]
- Si, Y.; Zhang, Z.G.; Chen, S.J.; Zheng, Y.Q.; Chen, Y.B.; Liu, Y.; Jiang, H.; Feng, L.Q.; Huang, X. Attenuated TLRs in middle ear mucosa contributes to susceptibility of chronic suppurative otitis media. Hum. Immunol. 2014, 75, 771–776. [Google Scholar] [CrossRef] [PubMed]
- de Visser, K.E.; Korets, L.V.; Coussens, L.M. De novo carcinogenesis promoted by chronic inflammation is B lymphocyte dependent. Cancer Cell 2005, 7, 411–423. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, J.V.; DE Medeiros Fernandes, T.A.A.; DE Azevedo, J.C.V.; Cobucci, R.N.O.; DE Carvalho, M.G.F.; Andrade, V.S.; DE Araújo, J.M.G. Link between chronic inflammation and human papillomavirus-induced carcinogenesis (Review). Oncol. Lett. 2015, 9, 1015–1026. [Google Scholar] [CrossRef] [PubMed]
- Adefuye, A.; Sales, K. Regulation of inflammatory pathways in cancer and infectious disease of the cervix. Scientifica 2012, 2012, 548150. [Google Scholar] [CrossRef] [PubMed]
- Ongaro, A.; Varani, K.; Masieri, F.F.; Pellati, A.; Massari, L.; Cadossi, R.; Vincenzi, F.; Borea, P.A.; Fini, M.; Caruso, A.; et al. Electromagnetic fields (EMFs) and adenosine receptors modulate prostaglandin E(2) and cytokine release in human osteoarthritic synovial fibroblasts. J. Cell. Physiol. 2012, 227, 2461–2469. [Google Scholar] [CrossRef]
- Kuo, C.-L. Etiopathogenesis of acquired cholesteatoma: Prominent theories and recent advances in biomolecular research. Laryngoscope 2015, 125, 234–240. [Google Scholar] [CrossRef]
- Gheit, T. Mucosal and Cutaneous Human Papillomavirus Infections and Cancer Biology. Front. Oncol. 2019, 9, 355. [Google Scholar] [CrossRef]
- Rotondo, J.C.; Bosi, S.; Bassi, C.; Ferracin, M.; Lanza, G.; Gafà, R.; Magri, E.; Selvatici, R.; Torresani, S.; Marci, R.; et al. Gene expression changes in progression of cervical neoplasia revealed by microarray analysis of cervical neoplastic keratinocytes. J. Cell. Physiol. 2015, 230, 806–812. [Google Scholar] [CrossRef]
- Bergmann, K.; Hoppe, F.; He, Y.; Helms, J.; Müller-Hermelink, H.K.; Stremlau, A.; de Villiers, E.M. Human-papillomavirus DNA in cholesteatomas. Int. J. Cancer 1994, 59, 463–466. [Google Scholar] [CrossRef]
- Franz, P.; Teschendorf, M.; Wohlschlager, J.; Fischer, M. Prevalence of human papillomavirus DNA in cholesteatomas. ORL J. Oto-Rhino-Laryngol. Relat. Spec. 2007, 69, 251–255. [Google Scholar] [CrossRef]
- Li, X.-P.; Hao, C.-L.; Wang, Q.; Yi, X.-M.; Jiang, Z.-S. H19 gene methylation status is associated with male infertility. Exp. Ther. Med. 2016, 12, 451–456. [Google Scholar] [CrossRef] [PubMed]
- Chow, L.T.; Broker, T.R.; Steinberg, B.M. The natural history of human papillomavirus infections of the mucosal epithelia. APMIS Acta Pathol. Microbiol. Immunol. Scand. 2010, 118, 422–449. [Google Scholar] [CrossRef]
- Griffin, N.R.; Bevan, I.S.; Lewis, F.A.; Wells, M.; Young, L.S. Demonstration of multiple HPV types in normal cervix and in cervical squamous cell carcinoma using the polymerase chain reaction on paraffin wax embedded material. J. Clin. Pathol. 1990, 43, 52–56. [Google Scholar] [CrossRef]
- Ayer, B.; Fischer, A.; Spurrett, B.; Houghton, R. Symptoms and HPV infection of the vulva: Clinical manifestations or mere coincidence? Aust. N. Z. J. Obstet. Gynaecol. 2001, 41, 443–446. [Google Scholar] [CrossRef] [PubMed]
- Rieth, K.K.S.; Gill, S.R.; Lott-Limbach, A.A.; Merkley, M.A.; Botero, N.; Allen, P.D.; Miller, M.C. Prevalence of High-Risk Human Papillomavirus in Tonsil Tissue in Healthy Adults and Colocalization in Biofilm of Tonsillar Crypts. JAMA Otolaryngol.-Head Neck Surg. 2018, 144, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Nadji, S.A.; Mokhtari-Azad, T.; Mahmoodi, M.; Yahyapour, Y.; Naghshvar, F.; Torabizadeh, J.; Ziaee, A.A.; Nategh, R. Relationship between lung cancer and human papillomavirus in north of Iran, Mazandaran province. Cancer Lett. 2007, 248, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.-Y.; Sdek, P.; Cao, J.; Chen, W.-T. Human papillomavirus type 16 and 18 DNA in oral squamous cell carcinoma and normal mucosa. Int. J. Oral Maxillofac. Surg. 2004, 33, 71–74. [Google Scholar] [CrossRef] [PubMed]
- Morshed, K.; Polz-Dacewicz, M.; Rajtar, B.; Szymański, M.; Ziaja-Sołtys, M.; Gołabek, W. The prevalence of E6/E7 HPV type 16 in laryngeal cancer and in normal mucosa. Pol. Merkur. Lek. Organ Pol. Tow. Lek. 2005, 19, 291–293. [Google Scholar]
- Buchwald, C.; Franzmann, M.B.; Jacobsen, G.K.; Lindeberg, H. Human papillomavirus and normal nasal mucosa: Detection of human papillomavirus DNA in normal nasal mucosa biopsies by polymerase chain reaction and in situ hybridization. Laryngoscope 1994, 104, 755–757. [Google Scholar] [CrossRef]
- Hermansson, R.S.; Olovsson, M.; Hoxell, E.; Lindström, A.K. HPV prevalence and HPV-related dysplasia in elderly women. PLoS ONE 2018, 13, e0189300. [Google Scholar] [CrossRef]
- Bai, Y.; Yan, L.; Li, S.; Bai, Q. Expression of human papillomavirus DNA in cholesteatoma of the middle ear. Zhonghua Er Bi Yan Hou Ke Za Zhi 2000, 35, 352–355. [Google Scholar] [PubMed]
- Ferekidis, E.; Nikolopoulos, T.P.; Yiotakis, J.; Ferekidou, E.; Kandiloros, D.; Papadimitriou, K.; Tzangaroulakis, A. Correlation of clinical and surgical findings to histological features (koilocytosis, papillary hyperplasia) suggesting papillomavirus involvement in the pathogenesis of cholesteatoma. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2006, 12, CR368–CR371. [Google Scholar]
- Doorbar, J. Molecular biology of human papillomavirus infection and cervical cancer. Clin. Sci. Lond. Engl. 1979 2006, 110, 525–541. [Google Scholar] [CrossRef]
- Pyeon, D.; Pearce, S.M.; Lank, S.M.; Ahlquist, P.; Lambert, P.F. Establishment of human papillomavirus infection requires cell cycle progression. PLoS Pathog. 2009, 5, e1000318. [Google Scholar] [CrossRef]
- DiMaio, D.; Mattoon, D. Mechanisms of cell transformation by papillomavirus E5 proteins. Oncogene 2001, 20, 7866–7873. [Google Scholar] [CrossRef] [PubMed]
- Venuti, A.; Paolini, F.; Nasir, L.; Corteggio, A.; Roperto, S.; Campo, M.S.; Borzacchiello, G. Papillomavirus E5: The smallest oncoprotein with many functions. Mol. Cancer 2011, 10, 140. [Google Scholar] [CrossRef] [PubMed]
- Cerasuolo, A.; Annunziata, C.; Tortora, M.; Starita, N.; Stellato, G.; Greggi, S.; Maglione, M.G.; Ionna, F.; Losito, S.; Botti, G.; et al. Comparative analysis of HPV16 gene expression profiles in cervical and in oropharyngeal squamous cell carcinoma. Oncotarget 2017, 8, 34070–34081. [Google Scholar] [CrossRef]
- Andersson, S.; Safari, H.; Mints, M.; Lewensohn-Fuchs, I.; Gyllensten, U.; Johansson, B. Type distribution, viral load and integration status of high-risk human papillomaviruses in pre-stages of cervical cancer (CIN). Br. J. Cancer 2005, 92, 2195–2200. [Google Scholar] [CrossRef]
- Neeff, M.; Biswas, K.; Hoggard, M.; Taylor, M.W.; Douglas, R. Molecular Microbiological Profile of Chronic Suppurative Otitis Media. J. Clin. Microbiol. 2016, 54, 2538–2546. [Google Scholar] [CrossRef]
- Yang, J.A.; Kim, J.Y.; Yoon, Y.K.; Kim, S.; Park, D.W.; Sohn, J.W.; Sim, H.S.; Kim, M.J. Epidemiological and Genetic Characterization of Methicillin-Resistant Staphylococcus aureus Isolates from the Ear Discharge of Outpatients with Chronic Otitis Media. J. Korean Med. Sci. 2008, 23, 762. [Google Scholar] [CrossRef]
- Shilpa, C.; Sandeep, S.; Thanzeemunisa, U.; Prakash, B.G.; Radhika, S.; Virender, S. Current Microbiological Trends of Chronic Suppurative Otitis Media in a Tertiary Care Centre, Mysuru, India. Indian J. Otolaryngol. Head Neck Surg. 2019, 71, 1449–1452. [Google Scholar] [CrossRef]
- Lee, S.K.; Park, D.C.; Kim, M.G.; Boo, S.H.; Choi, Y.J.; Byun, J.Y.; Park, M.S.; Yeo, S.G. Rate of Isolation and Trends of Antimicrobial Resistance of Multidrug Resistant Pseudomonas Aeruginosa from Otorrhea in Chronic Suppurative Otitis Media. Clin. Exp. Otorhinolaryngol. 2012, 5, 17. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.W.; Jung, J.Y.; Pashia, M.E.; Nason, R.; Scholnick, S.; Chole, R.A. Otopathogenic Pseudomonas aeruginosa strains as competent biofilm formers. Arch. Otolaryngol. Head Neck Surg. 2005, 131, 983–989. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Miller, C.S.; Johnstone, B.M. Human papillomavirus as a risk factor for oral squamous cell carcinoma: A meta-analysis, 1982-1997. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 2001, 91, 622–635. [Google Scholar] [CrossRef] [PubMed]
- Rotondo, J.C.; Borghi, A.; Selvatici, R.; Magri, E.; Bianchini, E.; Montinari, E.; Corazza, M.; Virgili, A.; Tognon, M.; Martini, F. Hypermethylation-Induced Inactivation of the IRF6 Gene as a Possible Early Event in Progression of Vulvar Squamous Cell Carcinoma Associated With Lichen Sclerosus. JAMA Dermatol. 2016, 152, 928–933. [Google Scholar] [CrossRef]
- Rotondo, J.C.; Borghi, A.; Selvatici, R.; Mazzoni, E.; Bononi, I.; Corazza, M.; Kussini, J.; Montinari, E.; Gafà, R.; Tognon, M.; et al. Association of Retinoic Acid Receptor β Gene with Onset and Progression of Lichen Sclerosus–Associated Vulvar Squamous Cell Carcinoma. JAMA Dermatol. 2018, 154, 819. [Google Scholar] [CrossRef]
- Contini, C.; Rotondo, J.C.; Magagnoli, F.; Maritati, M.; Seraceni, S.; Graziano, A.; Poggi, A.; Capucci, R.; Vesce, F.; Tognon, M.; et al. Investigation on silent bacterial infections in specimens from pregnant women affected by spontaneous miscarriage. J. Cell. Physiol. 2018, 234, 100–107. [Google Scholar] [CrossRef]
- Tagliapietra, A.; Rotondo, J.C.; Bononi, I.; Mazzoni, E.; Magagnoli, F.; Maritati, M.; Contini, C.; Vesce, F.; Tognon, M.; Martini, F. Footprints of BK and JC polyomaviruses in specimens from females affected by spontaneous abortion. Hum. Reprod. 2019, 34, 433–440. [Google Scholar] [CrossRef]
- de Araujo, M.R.; De Marco, L.; Santos, C.F.; Rubira-Bullen, I.R.F.; Ronco, G.; Pennini, I.; Vizzini, L.; Merletti, F.; Gillio-Tos, A. GP5+/6+ SYBR Green methodology for simultaneous screening and quantification of human papillomavirus. J. Clin. Virol. 2009, 45, 90–95. [Google Scholar] [CrossRef]
- Tagliapietra, A.; Rotondo, J.C.; Bononi, I.; Mazzoni, E.; Magagnoli, F.; Gonzalez, L.O.; Contini, C.; Vesce, F.; Tognon, M.; Martini, F. Droplet-digital PCR assay to detect Merkel cell polyomavirus sequences in chorionic villi from spontaneous abortion affected females. J. Cell. Physiol. 2020, 235, 1888–1894. [Google Scholar] [CrossRef]
- Rotondo, J.C.; Mazzoni, E.; Bononi, I.; Tognon, M.; Martini, F. Association between Simian Virus 40 and Human Tumors. Front. Oncol. 2019, 9, 670. [Google Scholar] [CrossRef]
- Rotondo, J.C.; Bononi, I.; Puozzo, A.; Govoni, M.; Foschi, V.; Lanza, G.; Gafà, R.; Gaboriaud, P.; Touzé, F.A.; Selvatici, R.; et al. Merkel Cell Carcinomas Arising in Autoimmune Disease Affected Patients Treated with Biologic Drugs, Including Anti-TNF. Clin. Cancer Res. 2017, 23, 3929–3934. [Google Scholar] [CrossRef] [PubMed]
- Rotondo, J.C.; Selvatici, R.; Di Domenico, M.; Marci, R.; Vesce, F.; Tognon, M.; Martini, F. Methylation loss at H19 imprinted gene correlates with methylenetetrahydrofolate reductase gene promoter hypermethylation in semen samples from infertile males. Epigenetics 2013, 8, 990–997. [Google Scholar] [CrossRef] [PubMed]
- Rotondo, J.C.; Candian, T.; Selvatici, R.; Mazzoni, E.; Bonaccorsi, G.; Greco, P.; Tognon, M.; Martini, F. Tracing Males From Different Continents by Genotyping JC Polyomavirus in DNA From Semen Samples. J. Cell. Physiol. 2017, 232, 982–985. [Google Scholar] [CrossRef] [PubMed]
- Mazzoni, E.; Pietrobon, S.; Masini, I.; Rotondo, J.C.; Gentile, M.; Fainardi, E.; Casetta, I.; Castellazzi, M.; Granieri, E.; Caniati, M.L.; et al. Significant low prevalence of antibodies reacting with simian virus 40 mimotopes in serum samples from patients affected by inflammatory neurologic diseases, including multiple sclerosis. PLoS ONE 2014, 9, e110923. [Google Scholar] [CrossRef]
- Rotondo, J.C.; Giari, L.; Guerranti, C.; Tognon, M.; Castaldelli, G.; Fano, E.A.; Martini, F. Environmental doses of perfluorooctanoic acid change the expression of genes in target tissues of common carp. Environ. Toxicol. Chem. 2018, 37, 942–948. [Google Scholar] [CrossRef]
- Mazzoni, E.; Di Stefano, M.; Fiore, J.R.; Destro, F.; Manfrini, M.; Rotondo, J.C.; Casali, M.V.; Vesce, F.; Greco, P.; Scutiero, G.; et al. Serum IgG Antibodies from Pregnant Women Reacting to Mimotopes of Simian Virus 40 Large T Antigen, the Viral Oncoprotein. Front. Immunol. 2017, 8, 411. [Google Scholar] [CrossRef]
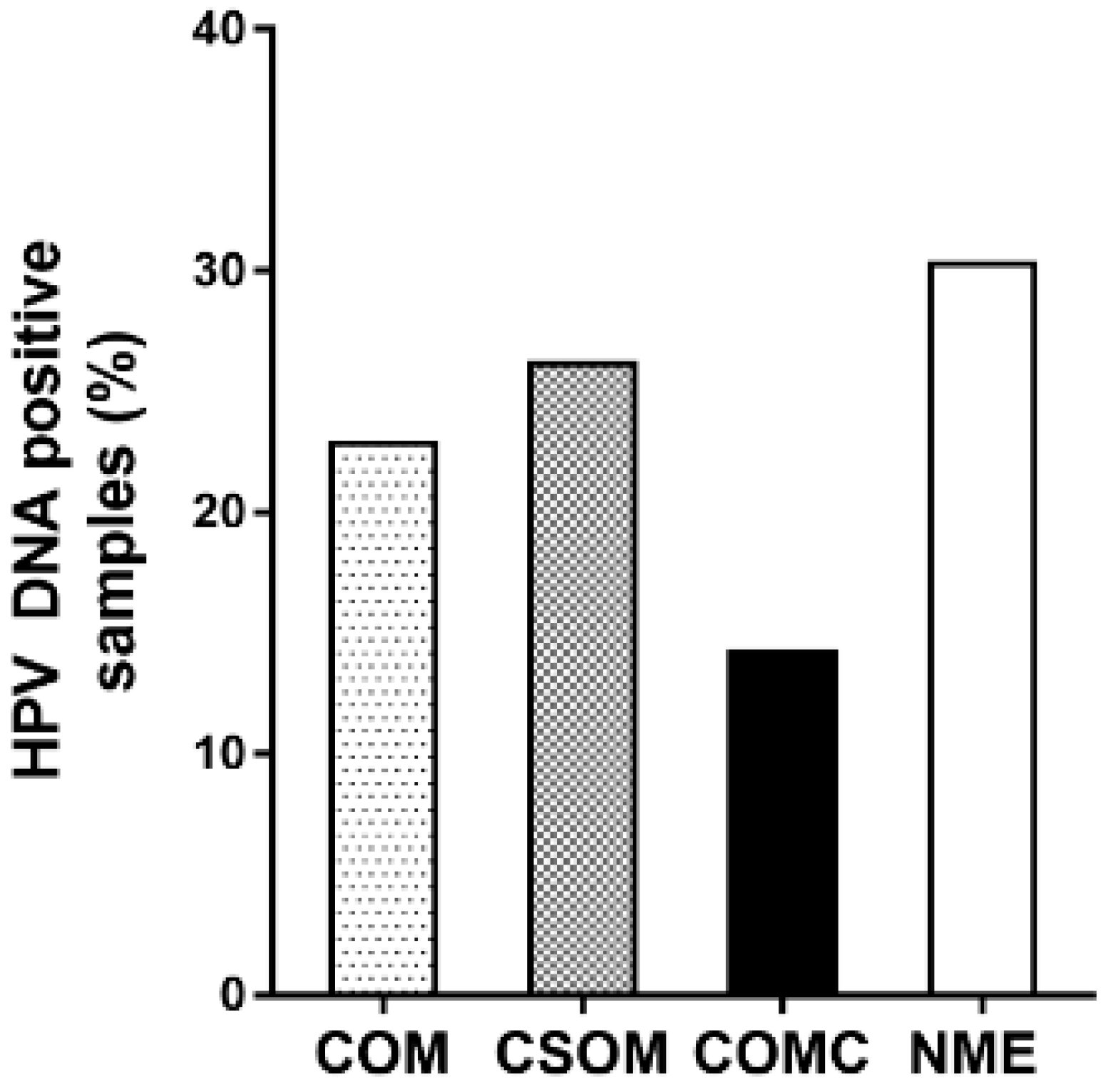
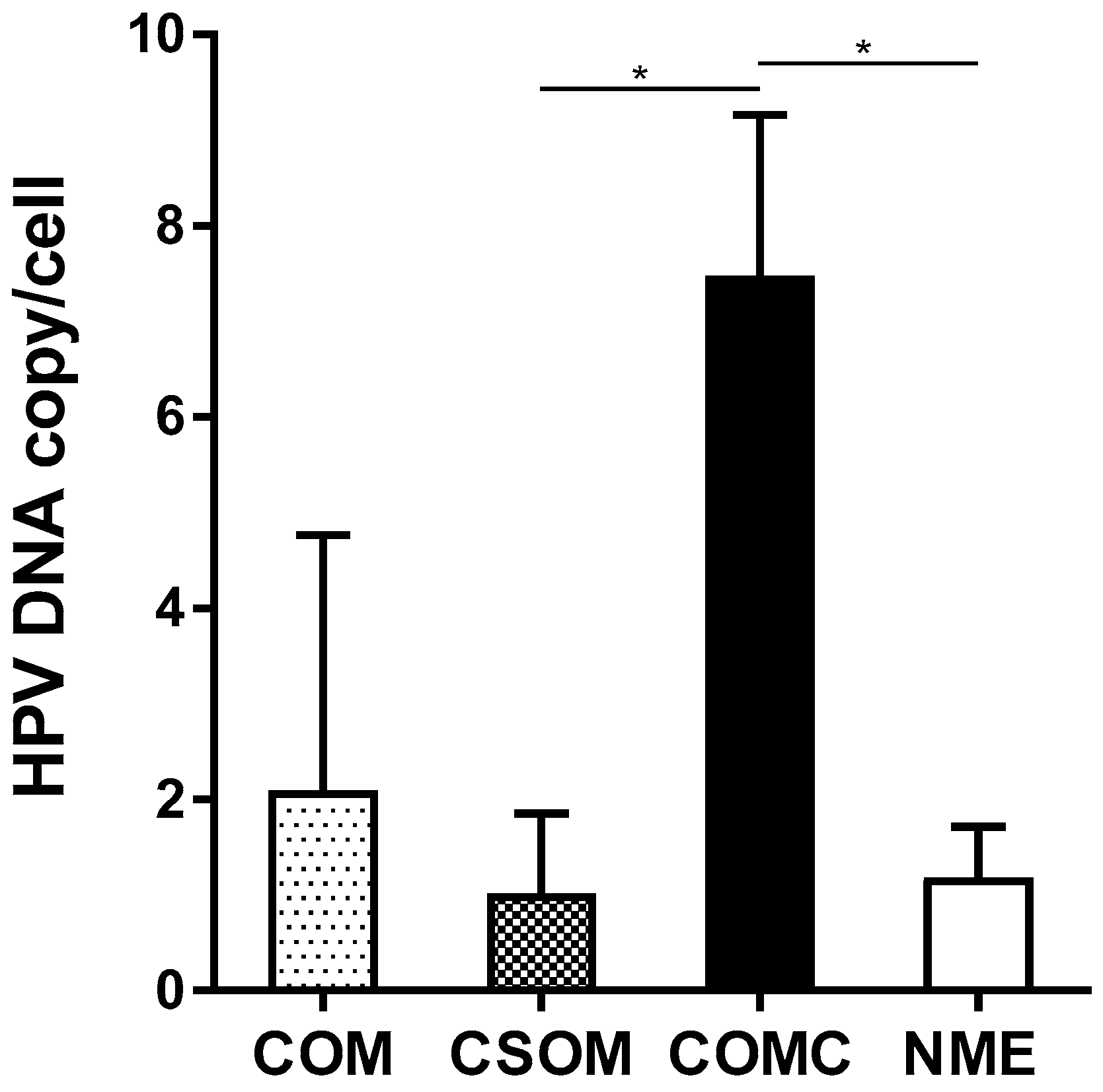
| Middle Ear Mucosa Specimens | Number of Patients | Mean HPV DNA Load (Copy/Cell) | Range (Copy/Cell) |
|---|---|---|---|
| COM | 12 | 2.09 | 0.01–8.67 |
| CSOM | 10 | 1.02 | 0.01–2.36 |
| COMC | 2 | 7.47 †,‡ | 6.28–8.67 |
| NME | 17 | 1.18 | 0.20–1.96 |
| Patients | CSOM | COMC | NME |
|---|---|---|---|
| Age (yrs) | |||
| ≤64 | 5/22 (23) | 1/14 (7) | 16/44 (36) |
| ≥65 | 10/16 (63) †,‡ | 0/0 (0) | 1/12 (8) |
| Smoke | |||
| Yes | 4/10 (40) | 1/6 (17) | 5/7 (71) § |
| No | 6/28 (21) | 0/8 (0) | 12/49 (25) |
| Sex | |||
| M | 5/20 (25) | 1/6 (17) | 6/20 (30) |
| F | 5/18 (28) | 0/8 (0) | 11/36 (31) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Malagutti, N.; Rotondo, J.C.; Cerritelli, L.; Melchiorri, C.; De Mattei, M.; Selvatici, R.; Oton-Gonzalez, L.; Stomeo, F.; Mazzoli, M.; Borin, M.; et al. High Human Papillomavirus DNA loads in Inflammatory Middle Ear Diseases. Pathogens 2020, 9, 224. https://doi.org/10.3390/pathogens9030224
Malagutti N, Rotondo JC, Cerritelli L, Melchiorri C, De Mattei M, Selvatici R, Oton-Gonzalez L, Stomeo F, Mazzoli M, Borin M, et al. High Human Papillomavirus DNA loads in Inflammatory Middle Ear Diseases. Pathogens. 2020; 9(3):224. https://doi.org/10.3390/pathogens9030224
Chicago/Turabian StyleMalagutti, Nicola, John Charles Rotondo, Luca Cerritelli, Claudio Melchiorri, Monica De Mattei, Rita Selvatici, Lucia Oton-Gonzalez, Francesco Stomeo, Manuela Mazzoli, Michela Borin, and et al. 2020. "High Human Papillomavirus DNA loads in Inflammatory Middle Ear Diseases" Pathogens 9, no. 3: 224. https://doi.org/10.3390/pathogens9030224
APA StyleMalagutti, N., Rotondo, J. C., Cerritelli, L., Melchiorri, C., De Mattei, M., Selvatici, R., Oton-Gonzalez, L., Stomeo, F., Mazzoli, M., Borin, M., Mores, B., Ciorba, A., Tognon, M., Pelucchi, S., & Martini, F. (2020). High Human Papillomavirus DNA loads in Inflammatory Middle Ear Diseases. Pathogens, 9(3), 224. https://doi.org/10.3390/pathogens9030224











