Lineage, Antimicrobial Resistance and Virulence of Citrobacter spp
Abstract
:1. Background
2. Results
2.1. Multilocus Sequence Typing of Citrobacter Isolates
2.2. Prevalence of Antimicrobial Resistance
2.3. Prevalence of ESBLs and Fluoroquinolone Resistance
2.4. Prevalence of qnrB Genes
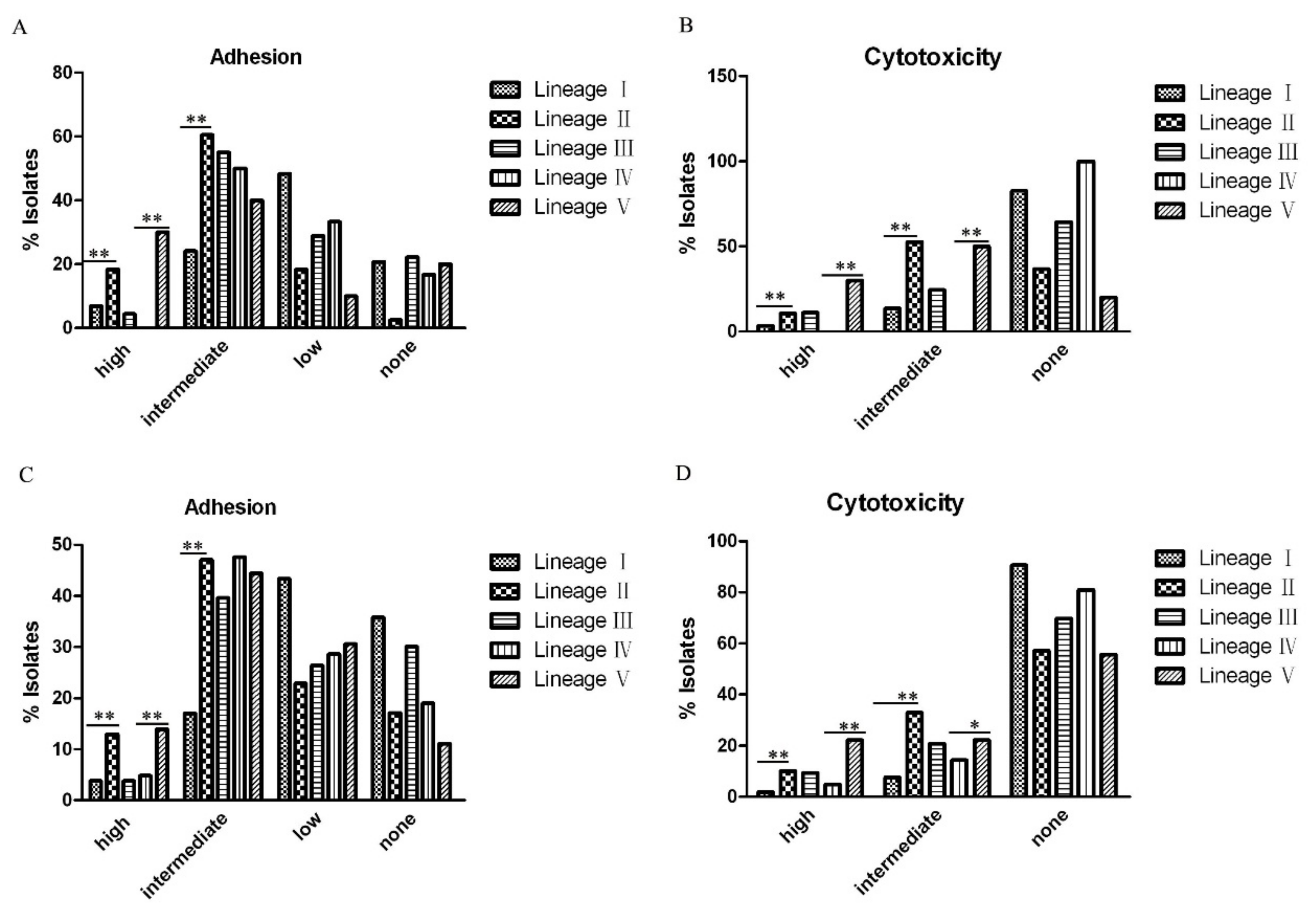
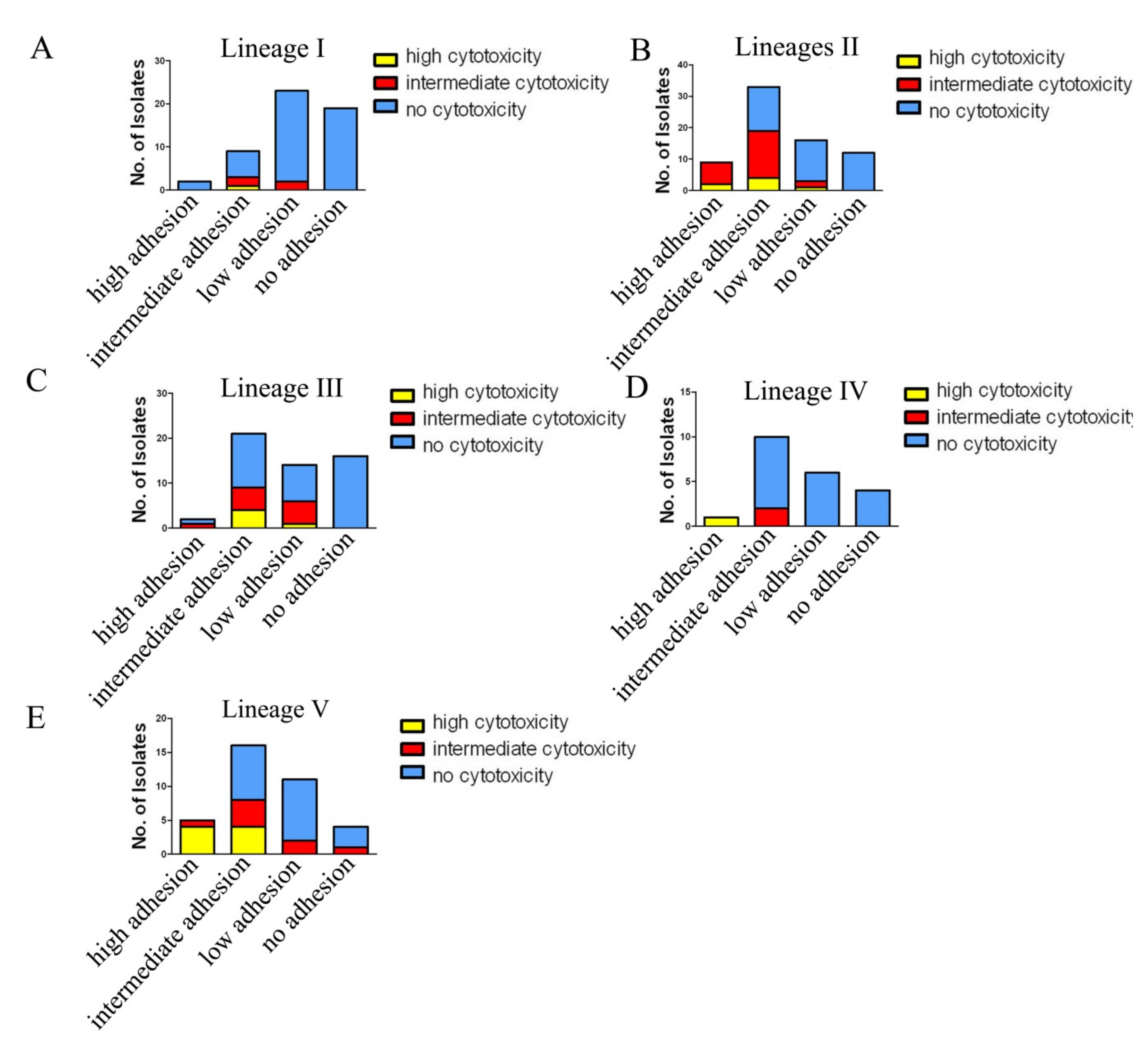
2.5. Adherence and Cytotoxicity of Citrobacter Isolates
3. Discussion
3.1. High Genetic Diversity of Citrobacter spp. Across China and Internationally.
3.2. Association of C. freundii Lineage II and C. youngae Lineage V with Higher Adhesion and Cytotoxicity
3.3. Higher Prevalence of Multidrug Resistance in C. braakii Isolates and Citrobacter Isolates from Food Sources
3.4. Carriage of ESBL Genes by Citrobacter spp. was Relatively Low
3.5. Higher Prevalence of Quinolone Resistance in Lineages I and III of Citrobacter spp with Multiple Mechanism of Resistance Detected
3.6. Citrobacter spp. Carried Many Variants of the qnrB Gene with C. freundii Lineage I as the Main Reservoir
4. Conclusions
5. Methods
5.1. Citrobacter Isolates
5.2. MLST and Phylogenetic Analysis
5.3. Antimicrobial Susceptibility Testing
5.4. PCR Amplification and Sequencing.
5.5. In Vitro Adhesion and Cytotoxicity Assays.
5.6. Statistical Analysis.
5.7. Ethics Approval and Consent to Participate
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Park, Y.-J.; Yu, J.K.; Lee, S.; Oh, E.J.; Woo, G.-J. Prevalence and diversity of qnr alleles in AmpC-producing Enterobacter cloacae, Enterobacter aerogenes, Citrobacter freundii and Serratia marcescens: A multicentre study from Korea. J. Antimicrob. Chemother. 2007, 60, 868–871. [Google Scholar] [CrossRef] [PubMed]
- Guerrant, R.L.; Dickens, M.D.; Wenzel, R.P.; Kapikian, A.Z. Toxigenic bacterial diarrhea: Nursery outbreak involving multiple bacterial strains. J. Pediatr. 1976, 89, 885–891. [Google Scholar] [CrossRef]
- Tschäpe, H.; Prager, R.; Streckel, W.; Fruth, A.; Tietze, E.; Böhme, G. Verotoxinogenic Citrobacter freundii associated with severe gastroenteritis and cases of haemolytic uraemic syndrome in a nursery school: Green butter as the infection source. Epidemiol. Infect. 1995, 114, 441–450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warner, R.D. A large nontypical outbreak of Norwalk virus. Gastroenteritis associated with exposing celery to nonpotable water and with Citrobacter freundii. Arch. Intern. Med. 1991, 151, 2419–2424. [Google Scholar] [CrossRef] [PubMed]
- Doulgeraki, A.; Paramithiotis, S.; Nychas, G.-J.E. Characterization of the Enterobacteriaceae community that developed during storage of minced beef under aerobic or modified atmosphere packaging conditions. Int. J. Food Microbiol. 2011, 145, 77–83. [Google Scholar] [CrossRef]
- Giammanco, G.M.; Aleo, A.; Guida, I.; Mammina, C. Molecular Epidemiological Survey of Citrobacter freundii Misidentified as Cronobacter spp. (Enterobacter sakazakii) and Enterobacter hormaechei Isolated from Powdered Infant Milk Formula. Foodborne Pathog. Dis. 2011, 8, 517–525. [Google Scholar] [CrossRef] [Green Version]
- Samonis, G.; Karageorgopoulos, D.; Kofteridis, D.P.; Matthaiou, D.; Sidiropoulou, V.; Maraki, S.; Falagas, M.E. Citrobacter infections in a general hospital: Characteristics and outcomes. Eur. J. Clin. Microbiol. Infect. Dis. 2008, 28, 61–68. [Google Scholar] [CrossRef]
- Mohanty, S.; Singhal, R.; Sood, S.; Dhawan, B.; Kapil, A.; Das, B.K. Citrobacter infections in a tertiary care hospital in Northern India. J. Infect. 2007, 54, 58–64. [Google Scholar] [CrossRef]
- Bai, L.; Xia, S.; Lan, R.; Liu, L.; Ye, C.; Wang, Y.; Jin, N.; Cui, Z.; Jing, H.; Xiong, Y.; et al. Isolation and Characterization of Cytotoxic, Aggregative Citrobacter freundii. PLoS ONE 2012, 7, e33054. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Lan, R.; Liu, L.; Wang, Y.; Zhang, Y.; Wang, Y.; Xu, J. Antimicrobial Resistance and Cytotoxicity of Citrobacter spp. in Maanshan Anhui Province, China. Front. Microbiol. 2017, 8, 1357. [Google Scholar] [CrossRef]
- Chen, K.J.; Chen, T.H.; Sue, Y.M. Citrobacter Youngae and Pantoea Agglomerans Peritonitis in a Peritoneal Dialysis Patient. Perit. Dial. Int. 2013, 33, 336–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Basra, P.; Koziol, A.; Wong, A.; Carrillo, C. Complete Genome Sequences of Citrobacter braakii Strains GTA-CB01 and GTA-CB04, Isolated from Ground Beef. Genome Announc. 2015, 3, e01307-14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwak, H.L.; Han, S.K.; Park, S.; Park, S.H.; Shim, J.Y.; Oh, M.; Ricke, S.; Kim, H.Y. Development of a rapid and accurate identification method for Citrobacter species isolated from pork products using a Matrix-Assisted Laser-Desorption Ionization Time-of-Flight Mass Spectrometry (MALDI-TOFMS). J. Microbiol. Biotechnol. 2015, 25, 1537–1541. [Google Scholar] [CrossRef] [PubMed]
- Arens, S.; Verhaegen, J.; Verbist, L. Differentiation and susceptibility of Citrobacter isolates from patients in a university hospital. Clin. Microbiol. Infect. 1997, 3, 53–57. [Google Scholar] [CrossRef] [Green Version]
- Chao, C.-T.; Lee, S.-Y.; Yang, W.-S.; Chen, H.-W.; Fang, C.-C.; Yen, C.-J.; Chiang, C.-K.; Hung, K.-Y.; Huang, J.-W. Citrobacter Peritoneal Dialysis Peritonitis: Rare Occurrence with Poor Outcomes. Int. J. Med Sci. 2013, 10, 1092–1098. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Chen, D.; Liu, L.; Lan, R.; Hao, S.; Jin, W.; Sun, H.; Wang, Y.; Liang, Y.; Xu, J. Genetic Diversity, Multidrug Resistance, and Virulence of Citrobacter freundii From Diarrheal Patients and Healthy Individuals. Front. Microbiol. 2018, 8, 233. [Google Scholar] [CrossRef] [Green Version]
- Akya, A.; Jafari, S.; Ahmadi, K.; Elahi, A. Frequency Of blaCTX-M, blaTEM and blaSHV Genes in Citrobacters Isolated from Imam Reza Hospital in Kermanshah. J. Mazand. Univ. Med. Sci. 2015, 25, 65–73. [Google Scholar]
- Oliveira, H.; Pinto, G.; Oliveira, A.; Oliveira, C.; Faustino, M.A.; Briers, Y.; Domingues, L.; Azeredo, J. Characterization and genome sequencing of a Citrobacter freundii phage CfP1 harboring a lysin active against multidrug-resistant isolates. Appl. Microbiol. Biotechnol. 2016, 100, 10543–10553. [Google Scholar] [CrossRef] [Green Version]
- Moland, E.S.; Hanson, N.D.; Black, J.A.; Hossain, A.; Song, W.; Thomson, K.S. Prevalence of Newer β-Lactamases in Gram-Negative Clinical Isolates Collected in the United States from 2001 to 2002. J. Clin. Microbiol. 2006, 44, 3318–3324. [Google Scholar] [CrossRef] [Green Version]
- Choi, S.-H.; Lee, J.E.; Park, S.J.; Kim, M.-N.; Choo, E.J.; Kwak, Y.G.; Jeong, J.-Y.; Woo, J.H.; Kim, N.J.; Kim, Y.S. Prevalence, microbiology, and clinical characteristics of extended-spectrum β-lactamase-producing Enterobacter spp., Serratia marcescens, Citrobacter freundii, and Morganella morganii in Korea. Eur. J. Clin. Microbiol. Infect. Dis. 2007, 26, 557–561. [Google Scholar] [CrossRef]
- Shao, Y.; Xiong, Z.; Li, X.; Hu, L.; Shen, J.; Li, T.; Hu, F.; Chen, S. Prevalence of plasmid-mediated quinolone resistance determinants in Citrobacter freundii isolates from Anhui province, PR China. J. Med Microbiol. 2011, 60, 1801–1805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minarini, L.; Darini, A.L.C. Mutations in the quinolone resistance-determining regions of gyrA and parC in Enterobacteriaceae isolates from Brazil. Braz. J. Microbiol. 2012, 43, 1309–1314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weigel, L.M.; Steward, C.D.; Tenover, F.C. gyrA Mutations Associated with Fluoroquinolone Resistance in Eight Species of Enterobacteriaceae. Antimicrob. Agents Chemother. 1998, 42, 2661–2667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feil, E.J. Small change: Keeping pace with microevolution. Nat. Rev. Genet. 2004, 2, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Wirth, T.; Falush, D.; Lan, R.; Colles, F.; Mensa, P.; Wieler, L.H.; Karch, H.; Reeves, P.; Maiden, M.C.; Ochman, H.; et al. Sex and virulence in Escherichia coli: An evolutionary perspective. Mol. Microbiol. 2006, 60, 1136–1151. [Google Scholar] [CrossRef] [Green Version]
- Jacoby, G.; Cattoir, V.; Hooper, D.; Martínez-Martínez, L.; Nordmann, P.; Pascual, A.; Poirel, L.; Wang, M. qnr Gene Nomenclature. Antimicrob. Agents Chemother. 2008, 52, 2297–2299. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Huang, Y.; Xu, X.; Zhao, Y.; Sun, Q.; Zhang, Z.; Zhang, X.; Wu, Y.; Wang, J.; Zhou, N.; et al. Complete Genome Sequence of Multidrug-Resistant Citrobacter freundii Strain P10159, Isolated from Urine Samples from a Patient with Esophageal Carcinoma. Genome Announc. 2016, 4, e01754-15. [Google Scholar] [CrossRef] [Green Version]
- Harada, K.; Shimizu, T.; Ozaki, H.; Kimura, Y.; Miyamoto, T.; Tsuyuki, Y. Characterization of Antimicrobial Resistance in Serratia spp. and Citrobacter spp. Isolates from Companion Animals in Japan: Nosocomial Dissemination of Extended-Spectrum Cephalosporin-Resistant Citrobacter freundii. Microorganisms 2019, 7, 64. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.-H.; Wang, N.-Y.; Wu, A.Y.-J.; Lin, C.-C.; Lee, C.-M.; Liu, C.-P. Citrobacter freundii bacteremia: Risk factors of mortality and prevalence of resistance genes. J. Microbiol. Immunol. Infect. 2018, 51, 565–572. [Google Scholar] [CrossRef]
- Zhang, R.; Ichijo, T.; Huang, Y.-L.; Cai, J.; Zhou, H.; Yamaguchi, N.; Nasu, M.; Chen, G.-X. High prevalence of qnr and aac(6’)-Ib-cr genes in both water-borne environmental bacteria and clinical isolates of Citrobacter freundii in China. Microbes Environ. 2012, 27, 158–163. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Chen, H.; Yang, Q.; Chen, M.; Wang, H. High Prevalence of Plasmid-Mediated Quinolone Resistance Genes qnr and aac(6′)-Ib-cr in Clinical Isolates of Enterobacteriaceae from Nine Teaching Hospitals in China . Antimicrob. Agents Chemother. 2008, 52, 4268–4273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacoby, G.A.; Griffin, C.M.; Hooper, D.C. Citrobacter spp. as a Source of qnrB Alleles . Antimicrob. Agents Chemother. 2011, 55, 4979–4984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clinical and Laboratory Standards Institute. M100-S28 Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Eighth Informational Supplement; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018; p. 353. [Google Scholar]
- Poirel, L.; Walsh, T.; Cuvillier, V.; Nordmann, P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 2011, 70, 119–123. [Google Scholar] [CrossRef] [PubMed]
- Voets, G.M.; Fluit, A.C.; Scharringa, J.; Stuart, J.C.; Hall, M.A.L.-V. A set of multiplex PCRs for genotypic detection of extended-spectrum β-lactamases, carbapenemases, plasmid-mediated AmpC β-lactamases and OXA β-lactamases. Int. J. Antimicrob. Agents 2011, 37, 356–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.-Y.; Wang, Y.; Walsh, T.; Yi, L.-X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Xavier, B.B.; Lammens, C.; Ruhal, R.; Kumar-Singh, S.; Butaye, P.; Goossens, H.; Malhotra-Kumar, S. Identification of a novel plasmid-mediated colistin-resistance gene,mcr-2, inEscherichia coli, Belgium, June 2016. Eurosurveillance 2016, 21, 30280. [Google Scholar] [CrossRef]
- Zhao, W.-H.; Hu, Z.-Q. IMP-type metallo-?-lactamases in Gram-negative bacilli: Distribution, phylogeny, and association with integrons. Crit. Rev. Microbiol. 2011, 37, 214–226. [Google Scholar] [CrossRef]
- Frank, J.A.; Reich, C.I.; Sharma, S.; Weisbaum, J.S.; Wilson, B.A.; Olsen, G.J. Critical Evaluation of Two Primers Commonly Used for Amplification of Bacterial 16S rRNA Genes. Appl. Environ. Microbiol. 2008, 74, 2461–2470. [Google Scholar] [CrossRef] [Green Version]
- Mange, J.-P.; Stephan, R.; Borel, N.; Wild, P.; Kim, K.S.; Pospischil, A.; Lehner, A. Adhesive properties of Enterobacter sakazakii to human epithelial and brain microvascular endothelial cells. BMC Microbiol. 2006, 6, 58. [Google Scholar] [CrossRef] [Green Version]
- Dallal, M.M.S.; Validi, M.; Douraghi, M.; Fallah-Mehrabadi, J.; Lormohammadi, L. Evaluation the cytotoxic effect of cytotoxin-producing Klebsiella oxytoca isolates on the HEp-2 cell line by MTT assay. Microb. Pathog. 2017, 113, 416–420. [Google Scholar] [CrossRef]
- Konkel, M.E.; Joens, L.A. Adhesion to and invasion of HEp-2 cells by Campylobacter spp. Infect. Immun. 1989, 57, 2984–2990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Philippe, G. A comprehensive review of Hep-2 cell line in translational research for laryngeal cancer. Am. J. Cancer Res. 2019, 9, 644–649. [Google Scholar]
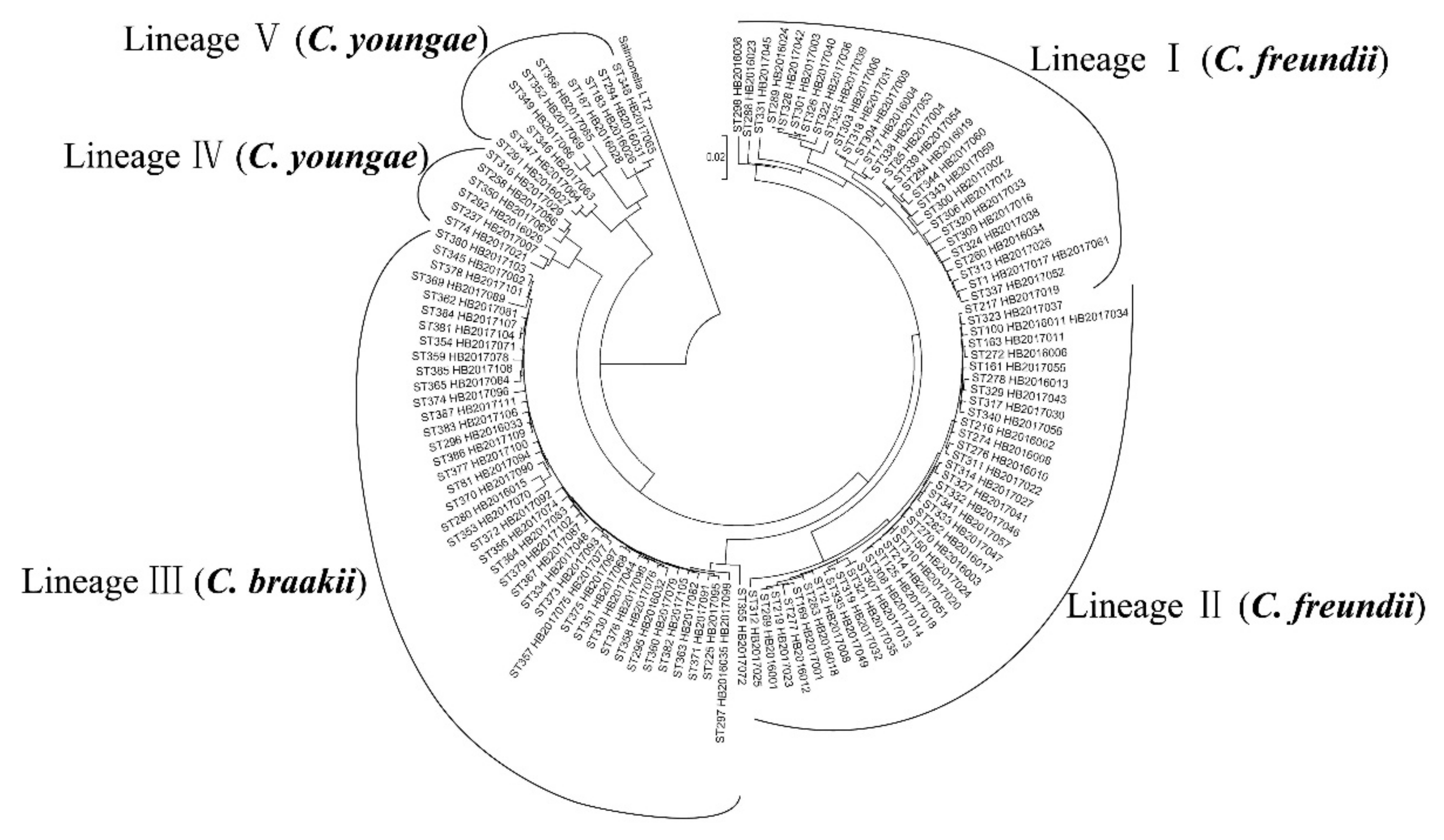
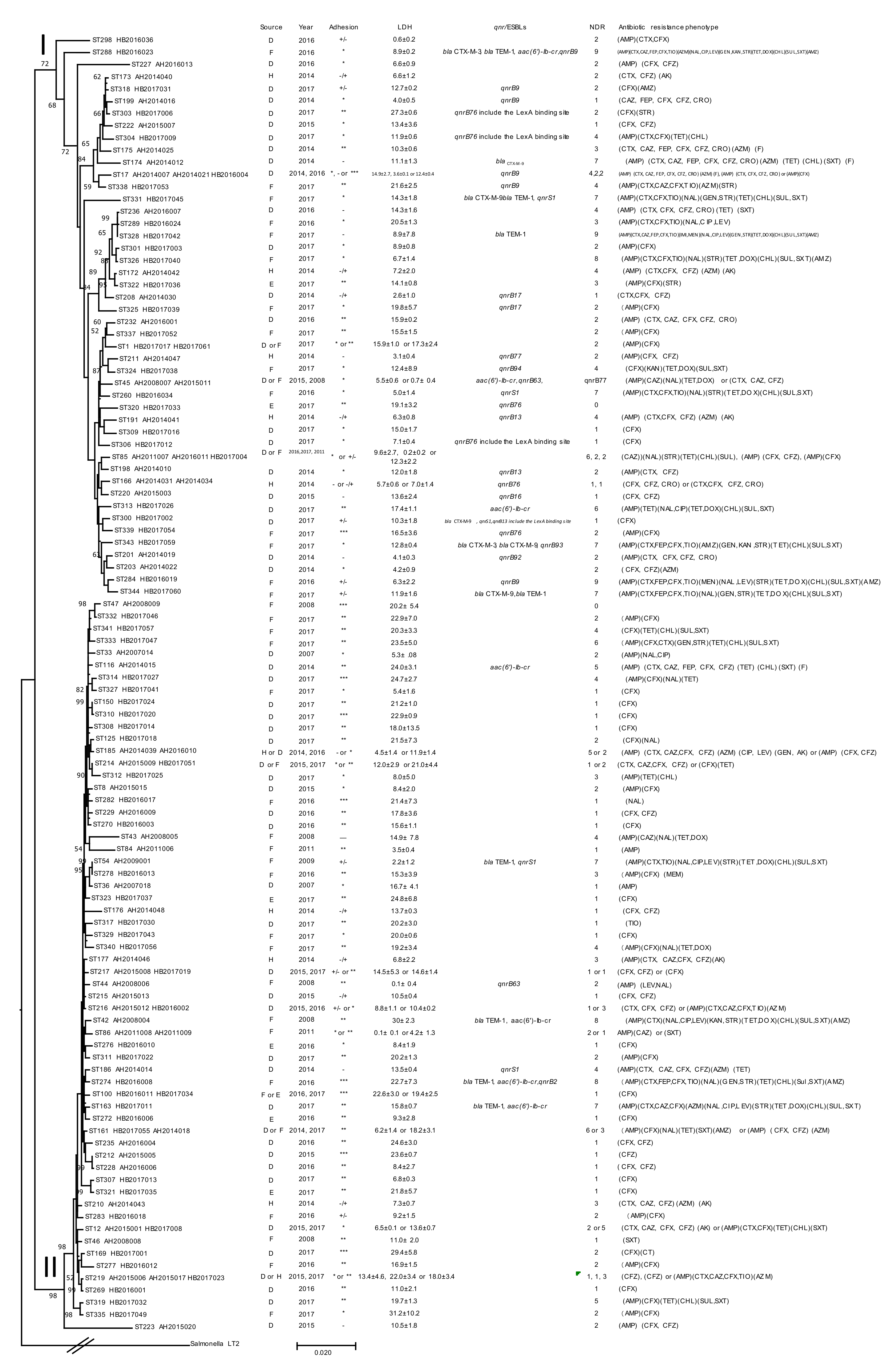
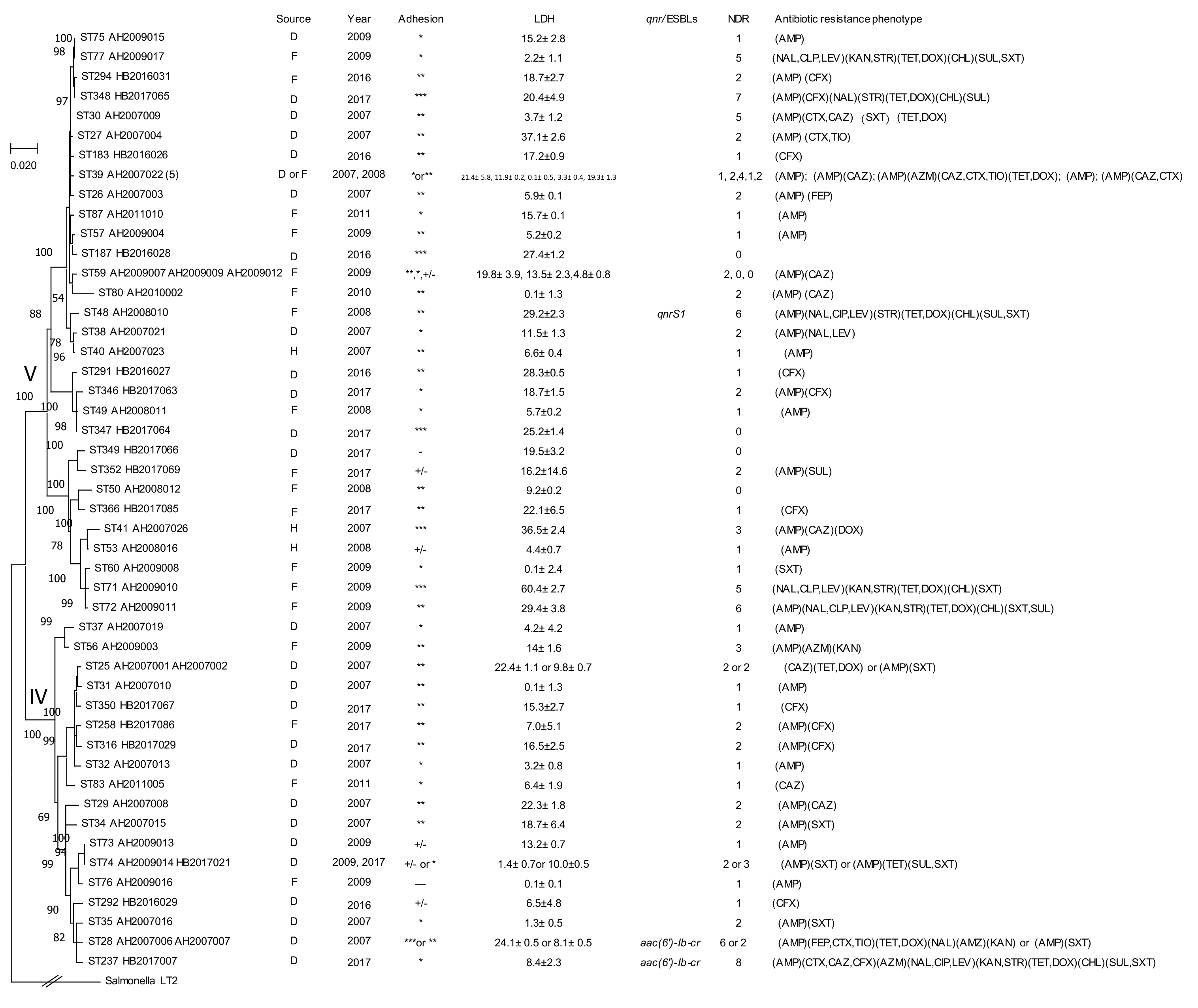


| Clusters and Species | Isolates | Year | Source | STs | Adhesion | LDH | NDR | ESBLs | qnr |
|---|---|---|---|---|---|---|---|---|---|
| Lineage Ⅰ | HB2016004 | 2016 | D | 17 | *** | 12.4 ± 0.4 | 2 | qnrB9 | |
| C. freundii | HB2016019 | 2016 | F | 284 | +/- | 6.3 ± 2.2 | 9 | qnrB9 | |
| HB2016023 | 2016 | F | 288 | * | 8.9 ± 0.2 | 9 | blaCTX-M-3, blaTEM-1 | aac(6')-Ib-cr,qnrB9 | |
| HB2016024 | 2016 | F | 289 | * | 20.5 ± 1.3 | 3 | |||
| HB2016034 | 2016 | F | 260 | * | 5.0 ± 1.4 | 7 | qnrS1 | ||
| HB2016036 | 2016 | D | 298 | +/- | 0.6 ± 0.2 | 2 | |||
| HB2017002 | 2017 | D | 300 | +/- | 10.3 ± 1.8 | 1 | blaCTX-M-9 | qnrS1,qnrB13 include the LexA binding site | |
| HB2017003 | 2017 | D | 301 | * | 8.9 ± 0.8 | 2 | |||
| HB2017004 | 2017 | D | 85 | * | 12.3 ± 2.2 | 2 | |||
| HB2017006 | 2017 | D | 303 | ** | 27.3 ± 0.6 | 2 | qnrB76 include the LexA binding site | ||
| HB2017009 | 2017 | D | 304 | * | 11.9 ± 0.6 | 4 | qnrB76 include the LexA binding site | ||
| HB2017012 | 2017 | D | 306 | * | 7.1 ± 0.4 | 1 | qnrB76 include the LexA binding site | ||
| HB2017016 | 2017 | D | 309 | * | 15.0 ± 1.7 | 1 | |||
| HB2017017 | 2017 | D | 1 | * | 15.9 ± 1.0 | 2 | |||
| HB2017026 | 2017 | D | 313 | ** | 17.4 ± 1.1 | 6 | aac(6')-Ib-cr | ||
| HB2017031 | 2017 | D | 318 | +/- | 12.7 ± 0.2 | 2 | qnrB9 | ||
| HB2017033 | 2017 | E | 320 | ** | 19.1 ± 3.2 | 0 | qnrB76 | ||
| HB2017036 | 2017 | E | 322 | ** | 14.1 ± 0.8 | 3 | |||
| HB2017038 | 2017 | F | 324 | * | 12.4 ± 8.9 | 4 | qnrB94 | ||
| HB2017039 | 2017 | F | 325 | * | 19.8 ± 5.7 | 2 | qnrB17 | ||
| HB2017040 | 2017 | F | 326 | * | 6.7 ± 1.4 | 8 | |||
| HB2017042 | 2017 | F | 328 | - | 8.9 ± 7.8 | 9 | blaTEM-1 | ||
| HB2017045 | 2017 | F | 331 | * | 14.3 ± 1.8 | 7 | blaCTX-M-9, blaTEM-1 | qnrS1 | |
| HB2017052 | 2017 | F | 337 | ** | 15.5 ± 1.5 | 2 | |||
| HB2017053 | 2017 | F | 338 | ** | 21.6 ± 2.5 | 4 | qnrB9 | ||
| HB2017054 | 2017 | F | 339 | *** | 16.5 ± 3.6 | 2 | qnrB76 | ||
| HB2017059 | 2017 | F | 343 | * | 12.8 ± 0.4 | 7 | blaCTX-M-3, blaCTX-M-9 | qnrB93 | |
| HB2017060 | 2017 | F | 344 | +/- | 11.9 ± 1.6 | 7 | blaCTX-M-9, blaTEM-1 | ||
| HB2017061 | 2017 | F | 1 | ** | 17.3 ± 2.4 | 2 | |||
| Lineage Ⅱ | HB2016001 | 2016 | D | 269 | ** | 11.0 ± 2.1 | 1 | ||
| C. freundii | HB2016002 | 2016 | D | 216 | * | 10.4 ± 0.2 | 3 | ||
| HB2016003 | 2016 | D | 270 | ** | 15.6 ± 1.1 | 1 | |||
| HB2016006 | 2016 | E | 272 | ** | 9.3 ± 2.8 | 1 | |||
| HB2016008 | 2016 | F | 274 | *** | 22.7 ± 7.3 | 8 | blaTEM-1 | aac(6')-Ib-cr,qnrB2 | |
| HB2016010 | 2016 | E | 276 | * | 8.4 ± 1.9 | 1 | |||
| HB2016011 | 2016 | F | 100 | *** | 22.6 ± 3.0 | 1 | |||
| HB2016012 | 2016 | F | 277 | ** | 16.9 ± 1.5 | 2 | |||
| HB2016013 | 2016 | F | 278 | ** | 15.3 ± 3.9 | 3 | |||
| HB2016017 | 2016 | F | 282 | *** | 21.4 ± 7.3 | 1 | |||
| HB2016018 | 2016 | F | 283 | +/- | 9.2 ± 1.5 | 2 | |||
| HB2017001 | 2017 | D | 169 | *** | 29.4 ± 5.8 | 2 | |||
| HB2017008 | 2017 | D | 12 | * | 13.6 ± 0.7 | 5 | |||
| HB2017011 | 2017 | D | 163 | ** | 15.8 ± 0.7 | 7 | blaTEM-1 | aac(6')-Ib-cr | |
| HB2017013 | 2017 | D | 307 | ** | 6.8 ± 0.3 | 1 | |||
| HB2017014 | 2017 | D | 308 | ** | 18.0 ± 13.5 | 1 | |||
| HB2017018 | 2017 | D | 125 | ** | 21.5 ± 7.3 | 2 | |||
| HB2017019 | 2017 | D | 217 | ** | 14.6 ± 1.4 | 1 | |||
| HB2017020 | 2017 | D | 310 | *** | 22.9 ± 0.9 | 1 | |||
| HB2017022 | 2017 | D | 311 | ** | 20.2 ± 1.3 | 2 | |||
| HB2017023 | 2017 | D | 219 | ** | 18.0 ± 3.4 | 3 | |||
| HB2017024 | 2017 | D | 150 | ** | 21.2 ± 1.0 | 1 | |||
| HB2017025 | 2017 | D | 312 | * | 8.0 ± 5.0 | 3 | |||
| HB2017027 | 2017 | D | 314 | *** | 24.7 ± 2.7 | 4 | |||
| HB2017030 | 2017 | D | 317 | ** | 20.2 ± 3.0 | 1 | |||
| HB2017032 | 2017 | D | 319 | ** | 19.7 ± 1.3 | 5 | |||
| HB2017034 | 2017 | E | 100 | *** | 19.4 ± 2.5 | 1 | |||
| HB2017035 | 2017 | E | 321 | ** | 21.8 ± 5.7 | 1 | |||
| HB2017037 | 2017 | E | 323 | ** | 24.8 ± 6.8 | 1 | |||
| HB2017041 | 2017 | F | 327 | * | 5.4 ± 1.6 | 1 | |||
| HB2017043 | 2017 | F | 329 | * | 20.0 ± 0.6 | 1 | |||
| HB2017046 | 2017 | F | 332 | ** | 22.9 ± 7.0 | 2 | |||
| HB2017047 | 2017 | F | 333 | ** | 23.5 ± 5.0 | 6 | |||
| HB2017049 | 2017 | F | 335 | * | 31.2 ± 10.2 | 2 | |||
| HB2017051 | 2017 | F | 214 | ** | 21.0 ± 4.4 | 2 | |||
| HB2017055 | 2017 | F | 161 | ** | 18.2 ± 3.1 | 6 | |||
| HB2017056 | 2017 | F | 340 | ** | 19.2 ± 3.4 | 4 | |||
| HB2017057 | 2017 | F | 341 | ** | 20.3 ± 3.3 | 4 | |||
| Lineage Ⅲ | HB2016015 | 2016 | F | 280 | +/- | 3.4 ± 0.7 | 5 | qnrS1 | |
| C. braakii | HB2016032 | 2016 | E | 295 | ** | 4.2 ± 4.1 | 5 | ||
| HB2016033 | 2016 | F | 296 | ** | 5.3 ± 3.2 | 2 | |||
| HB2016035 | 2016 | F | 297 | * | 11.1 ± 2.2 | 6 | aac(6')-Ib-cr,qnrB2 | ||
| HB2017044 | 2017 | F | 330 | ** | 20.4 ± 5.5 | 2 | |||
| HB2017048 | 2017 | F | 334 | * | 20.6 ± 4.0 | 2 | |||
| HB2017062 | 2017 | F | 345 | * | 14.8 ± 1.9 | 2 | |||
| HB2017068 | 2017 | F | 351 | ** | 12.5 ± 8.9 | 2 | |||
| HB2017070 | 2017 | D | 353 | * | 9.5 ± 0.4 | 2 | |||
| HB2017071 | 2017 | D | 354 | * | 22.2 ± 6.9 | 2 | |||
| HB2017072 | 2017 | D | 355 | ** | 25.2 ± 4.0 | 1 | |||
| HB2017074 | 2017 | D | 356 | *** | 14.9 ± 7.4 | 2 | |||
| HB2017075 | 2017 | D | 357 | ** | 25.2 ± 4.0 | 1 | |||
| HB2017076 | 2017 | D | 358 | ** | 28.9 ± 1.6 | 4 | |||
| HB2017077 | 2017 | D | 357 | ** | 13.6 ± 0.2 | 1 | |||
| HB2017078 | 2017 | D | 359 | +/- | 13.7 ± 5.7 | 1 | |||
| HB2017079 | 2017 | E | 360 | * | 15.5 ± 2.0 | 3 | |||
| HB2017081 | 2017 | E | 362 | ** | 17.7 ± 0.8 | 2 | |||
| HB2017082 | 2017 | F | 363 | ** | 13.1 ± 1.9 | 1 | |||
| HB2017083 | 2017 | F | 364 | ** | 10.8 ± 0.6 | 1 | |||
| HB2017084 | 2017 | F | 365 | ** | 13.9 ± 2.0 | 3 | |||
| HB2017087 | 2017 | F | 367 | * | 3.5 ± 4.0 | 8 | aac(6')-Ib-cr, qnrB2 | ||
| HB2017089 | 2017 | F | 369 | +/- | 11.7 ± 9.7 | 2 | |||
| HB2017090 | 2017 | F | 370 | +/- | 4.5 ± 1.7 | 8 | |||
| HB2017091 | 2017 | F | 371 | - | 4.7 ± 0.1 | 3 | |||
| HB2017092 | 2017 | F | 372 | * | 11.7 ± 10.7 | 4 | |||
| HB2017093 | 2017 | F | 373 | +/- | 6.5 ± 2.0 | 2 | |||
| HB2017094 | 2017 | F | 81 | +/- | 9.8 ± 5.6 | 3 | |||
| HB2017095 | 2017 | F | 225 | ** | 6.2 ± 2.5 | 6 | blaCTX-M-9 | qnrB2 | |
| HB2017096 | 2017 | F | 374 | ** | 31.7 ± 4.8 | 6 | blaCTX-M-3 | ||
| HB2017097 | 2017 | F | 375 | ** | 17.5 ± 1.9 | 0 | |||
| HB2017098 | 2017 | F | 376 | +/- | 14.2 ± 6.5 | 1 | |||
| HB2017099 | 2017 | E | 297 | +/- | 17.1 ± 1.1 | 6 | aac(6')-Ib-cr | ||
| HB2017100 | 2017 | F | 377 | * | 17.9 ± 2.2 | 2 | |||
| HB2017101 | 2017 | F | 378 | *** | 21.1 ± 5.1 | 2 | |||
| HB2017102 | 2017 | F | 379 | * | 25.0 ± 4.2 | 3 | |||
| HB2017103 | 2017 | F | 380 | * | 0.7 ± 0.4 | 4 | |||
| HB2017104 | 2017 | F | 381 | +/- | 14.0 ± 1.7 | 3 | |||
| HB2017105 | 2017 | F | 382 | ** | 13.9 ± 2.9 | 9 | blaTEM-1 | aac(6')-Ib-cr | |
| HB2017106 | 2017 | F | 383 | ** | 18.6 ± 0.3 | 4 | |||
| HB2017107 | 2017 | F | 384 | * | 18.5 ± 1.1 | 7 | |||
| HB2017108 | 2017 | F | 385 | ** | 13.2 ± 3.1 | 6 | blaCTX-M-9 | aac(6')-Ib-cr, qnrB2 | |
| HB2017109 | 2017 | F | 386 | * | 20.4 ± 2.4 | 1 | |||
| HB2017110 | 2017 | F | 375 | ** | 18.0 ± 3.0 | 0 | |||
| HB2017111 | 2017 | F | 387 | ** | 18.0 ± 0.8 | 2 | |||
| Lineage Ⅳ | HB2016029 | 2016 | D | 292 | +/- | 6.5 ± 4.8 | 1 | ||
| C. youngae | HB2017007 | 2017 | D | 237 | * | 8.4 ± 2.3 | 8 | aac(6')-Ib-cr | |
| HB2017021 | 2017 | D | 74 | * | 10.0 ± 0.5 | 3 | |||
| HB2017029 | 2017 | D | 316 | ** | 16.5 ± 2.5 | 2 | |||
| HB2017067 | 2017 | D | 350 | ** | 15.3 ± 2.7 | 1 | |||
| HB2017086 | 2017 | F | 258 | ** | 7.0 ± 5.1 | 2 | |||
| Lineage Ⅴ | HB2016026 | 2016 | D | 183 | ** | 17.2 ± 0.9 | 1 | ||
| C. youngae | HB2016027 | 2016 | D | 291 | ** | 28.3 ± 0.5 | 1 | ||
| HB2016028 | 2016 | D | 187 | *** | 27.4 ± 1.2 | 0 | |||
| HB2016031 | 2016 | F | 294 | ** | 18.7 ± 2.7 | 2 | |||
| HB2017063 | 2017 | D | 346 | * | 18.7 ± 1.5 | 2 | |||
| HB2017064 | 2017 | D | 347 | *** | 25.2 ± 1.4 | 0 | |||
| HB2017065 | 2017 | D | 348 | *** | 20.4 ± 4.9 | 7 | |||
| HB2017066 | 2017 | D | 349 | - | 19.5 ± 3.2 | 0 | |||
| HB2017069 | 2017 | F | 352 | +/- | 16.2 ± 14.6 | 2 | |||
| HB2017085 | 2017 | F | 366 | ** | 22.1 ± 6.5 | 1 |
| Antibiotic | C. freundii (n = 67) Resistance (%) | C. youngae (n = 16) Resistance (%) | C.braakii (n = 45) Resistance (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| D (n = 30) | F (n = 30) | E (n = 7) | D (n = 12) | F (n = 4) | E (n = 0) | D (n = 8) | F (n = 33) | E (n = 4) | |
| PENICILLINS | |||||||||
| Ampicillin | 15 (50.0) | 23 (76.7) | 1 (14.3) | 5 (41.7) | 3 (75.0) | 0 (0) | 3 (37.5) | 16 (48.5) | 4 (100.0) |
| CEPHALOSPORINS | |||||||||
| Cefotaxime | 6 (20.0) | 12 (40.0) | 0 (0) | 1 (8.3) | 0 (0) | 0 (0) | 0 (0) | 7 (21.2) | 2 (50.0) |
| Ceftazidime | 3 (10.0) | 3 (10.0) | 0 (0) | 1 (8.3) | 0 (0) | 0 (0) | 0 (0) | 1 (3.0) | 0 (0) |
| Cefepime | 0 (0) | 6 (20.0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (3.0) | 0 (0) |
| Cefoxitin | 28 (93.3) | 29 (96.7) | 6 (85.7) | 8 (66.7) | 3 (75.0) | 0 (0) | 7 (87.5) | 29 (87.9) | 4 (100.0) |
| Ceftiofur Sodium | 3 (10.0) | 11 (36.7) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 7 (21.2) | 1 (25.0) |
| MONOBACTAMS | |||||||||
| Aztreonam | 2 (6.7) | 3 (10.0) | 0 (0) | 1 (8.3) | 0 (0) | 0 (0) | 0 (0) | 5 (15.2) | 0 (0) |
| CARBAPENEMS | |||||||||
| Imipenem | 0 (0) | 1 (3.3) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (3.0) | 0 (0) |
| Meropenem | 0 (0) | 3 (10.0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| QUINOLONES | |||||||||
| Nalidixicacid | 4 (13.3) | 12 (40.0) | 0 (0) | 2 (16.7) | 0 (0) | 0 (0) | 3 (37.5) | 15 (45.5) | 2 (50.0) |
| Ciprofloxacin | 2 (6.7) | 4 (13.3) | 0 (0) | 1 (8.3) | 0 (0) | 0 (0) | 0 (0) | 3 (9.1) | 0 (0) |
| Levofloxacin | 1 (3.3) | 4 (13.3) | 0 (0) | 1 (8.3) | 0 (0) | 0 (0) | 0 (0) | 3 (9.1) | 0 (0) |
| AMINOGLYCOSIDES | |||||||||
| Gentamicin | 0 (0) | 7 (23.3) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2 (6.1) | 0 (0) |
| Amikacin | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Streptomycin | 2 (6.7) | 11 (36.7) | 1 (14.3) | 2 (16.7) | 0 (0) | 0 (0) | 0 (0) | 8 (24.2) | 2 (50.0) |
| Kanamycin | 0 (0) | 3 (10.0) | 0 (0) | 1 (8.3) | 0 (0) | 0 (0) | 0 (0) | 1 (3.0) | 0 (0) |
| TETRACYCLINES | |||||||||
| Tetracycline | 7 (23.3) | 15 (50.0) | 0 (0) | 3 (25.0) | 0 (0) | 0 (0) | 1 (12.5) | 17 (51.5) | 1 (25.0) |
| Doxycycline | 2 (6.7) | 9 (30.0) | 0 (0) | 2 (16.7) | 0 (0) | 0 (0) | 1 (12.5) | 12 (36.4) | 1 (25.0) |
| PHENICOLS | |||||||||
| Chloramphenicol | 6 (20.0) | 11 (36.7) | 0 (0) | 2 (16.7) | 0 (0) | 0 (0) | 0 (0) | 7 (21.2) | 2 (50.0) |
| SULFONAMIDES | |||||||||
| Trimethoprim/Sulfamethoxazole | 4 (13.3) | 13 (43.3) | 0 (0) | 2 (16.7) | 1 (25.0) | 0 (0) | 0 (0) | 11 (33.3) | 2 (50.0) |
| Sulfafurazole | 3 (10.0) | 12 (40.0) | 0 (0) | 3 (25.0) | 0 (0) | 0 (0) | 0 (0) | 9 (37.3) | 2 (50.0) |
| MACROLIDES | |||||||||
| Azithromycin | 1 (3.3) | 6 (20.0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 3 (9.1) | 0 (0) |
| MDR | 9 (30.0) | 17 (56.7) | 1 (14.3) | 3 (25.0) | 0 (0) | 0 (0) | 1 (12.5) | 17 (51.5) | 3 (75.0) |
| Isolates | Species | Year | Source | ST | NDR | NAL | CIP | LEV | PMQR | gyrA Position |
|---|---|---|---|---|---|---|---|---|---|---|
| HB2016008 | C. freundii | 2016 | F | 274 | 8 | >128 | 4 | aac(6’)-Ib-cr,qnrB2 | Thr59Ile | |
| HB2016017 | C. freundii | 2016 | F | 282 | 1 | >128 | Thr59Ile | |||
| HB2017011 | C. freundii | 2017 | D | 163 | 7 | >64 | 16 | 8 | aac(6’)-Ib-cr | Thr59Ile |
| HB2017018 | C. freundii | 2017 | D | 125 | 2 | >128 | Thr59Ile | |||
| HB2017027 | C. freundii | 2017 | D | 314 | 4 | >64 | No mutation | |||
| HB2017055 | C. freundii | 2017 | F | 161 | 6 | >128 | Thr59Ile | |||
| HB2017056 | C. freundii | 2017 | F | 340 | 4 | >128 | Thr59Ile | |||
| HB2016019 | C. freundii | 2016 | F | 284 | 9 | >128 | 8 | qnrB9 | Thr59Ile | |
| HB2016023 | C. freundii | 2016 | F | 288 | 9 | >128 | 32 | 16 | aac(6’)-Ib-cr, qnrB9 | Thr59Ile |
| HB2016024 | C. freundii | 2016 | F | 289 | 3 | >128 | 4 | 8 | Thr59Ile | |
| HB2016034 | C. freundii | 2016 | F | 260 | 7 | 32 | qnrS1 | No mutation | ||
| HB2017026 | C. freundii | 2017 | D | 313 | 6 | >128 | 4 | aac(6’)-Ib-cr | Thr59Ile | |
| HB2017040 | C. freundii | 2017 | F | 326 | 8 | >128 | Thr59Ile | |||
| HB2017042 | C. freundii | 2017 | F | 328 | 9 | >128 | 8 | 16 | Thr59Ile | |
| HB2017045 | C. freundii | 2017 | F | 331 | 7 | 64 | qnrS1 | No mutation | ||
| HB2017060 | C. freundii | 2017 | F | 344 | 7 | >128 | No mutation | |||
| HB2016015 | C. braakii | 2016 | F | 280 | 5 | >128 | qnrS1 | Thr59Ile | ||
| HB2016033 | C. braakii | 2016 | F | 296 | 2 | >128 | Thr59Ile | |||
| HB2016035 | C. braakii | 2016 | F | 297 | 6 | >128 | 8 | 8 | aac(6’)-Ib-cr, qnrB2 | Thr59Ile |
| HB2017070 | C. braakii | 2017 | D | 353 | 2 | >64 | Thr59Ile | |||
| HB2017076 | C. braakii | 2017 | D | 358 | 4 | >128 | Thr59Ile | |||
| HB2017078 | C. braakii | 2017 | D | 359 | 1 | >64 | 2 | No mutation | ||
| HB2017079 | C. braakii | 2017 | E | 360 | 3 | >128 | Thr59Ile | |||
| HB2017084 | C. braakii | 2017 | F | 365 | 3 | >64 | Thr59Ile | |||
| HB2017087 | C. braakii | 2017 | F | 367 | 8 | >128 | aac(6’)-Ib-cr, qnrB2 | Thr59Ile | ||
| HB2017090 | C. braakii | 2017 | F | 370 | 8 | >128 | 8 | >16 | Thr59Ile | |
| HB2017091 | C. braakii | 2017 | F | 371 | 3 | >128 | Thr59Ile | |||
| HB2017092 | C. braakii | 2017 | F | 372 | 4 | 64 | No mutation | |||
| HB2017095 | C. braakii | 2017 | F | 225 | 6 | >128 | qnrB2 | No mutation | ||
| HB2017099 | C. braakii | 2017 | E | 297 | 6 | >128 | 8 | 8 | aac(6’)-Ib-cr | Thr59Ile |
| HB2017102 | C. braakii | 2017 | F | 379 | 3 | 32 | No mutation | |||
| HB2017103 | C. braakii | 2017 | F | 380 | 4 | >128 | Thr59Ile | |||
| HB2017105 | C. braakii | 2017 | F | 382 | 9 | >128 | 8 | 16 | aac(6’)-Ib-cr | No mutation |
| HB2017106 | C. braakii | 2017 | F | 383 | 4 | 32 | No mutation | |||
| HB2017107 | C. braakii | 2017 | F | 384 | 7 | >128 | Thr59Ile | |||
| HB2017108 | C. braakii | 2017 | F | 385 | 6 | >128 | aac(6’)-Ib-cr, qnrB2 | Thr59Ile | ||
| HB2017007 | C. youngae | 2017 | D | 237 | 8 | >64 | 8 | 8 | aac(6’)-Ib-cr | Thr59Ile |
| HB2017065 | C. youngae | 2017 | D | 348 | 7 | >128 | Thr59Ile, Gln111Arg, Ile134Val |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, L.; Qin, L.; Hao, S.; Lan, R.; Xu, B.; Guo, Y.; Jiang, R.; Sun, H.; Chen, X.; LV, X.; et al. Lineage, Antimicrobial Resistance and Virulence of Citrobacter spp. Pathogens 2020, 9, 195. https://doi.org/10.3390/pathogens9030195
Liu L, Qin L, Hao S, Lan R, Xu B, Guo Y, Jiang R, Sun H, Chen X, LV X, et al. Lineage, Antimicrobial Resistance and Virulence of Citrobacter spp. Pathogens. 2020; 9(3):195. https://doi.org/10.3390/pathogens9030195
Chicago/Turabian StyleLiu, Liyun, Liyun Qin, Shuai Hao, Ruiting Lan, Baohong Xu, Yumei Guo, Ruiping Jiang, Hui Sun, Xiaoping Chen, Xinchao LV, and et al. 2020. "Lineage, Antimicrobial Resistance and Virulence of Citrobacter spp" Pathogens 9, no. 3: 195. https://doi.org/10.3390/pathogens9030195
APA StyleLiu, L., Qin, L., Hao, S., Lan, R., Xu, B., Guo, Y., Jiang, R., Sun, H., Chen, X., LV, X., Xu, J., & Zhao, C. (2020). Lineage, Antimicrobial Resistance and Virulence of Citrobacter spp. Pathogens, 9(3), 195. https://doi.org/10.3390/pathogens9030195




