Pathogenicity and Genetic Characterization of Vietnamese Classical Swine Fever Virus: 2014–2018
Abstract
1. Introduction
2. Results
2.1. Genotype Analysis and Phylogenetic Tree
2.2. Comparison of the Pathogenicity of Different Sub-Genotypes
2.3. Tissue Lesion Finding between Sub-Genotype 2.2 and 2.1c
2.4. Immunohistochemical Staining for Sub-Genotypes 2.2 and 2.1c
3. Discussion
4. Materials and Methods
4.1. Samples, RT-PCR, Phylogenetic Tree, and Virus Isolation
4.2. Animal Experiments
4.3. Immunohistochemical Assay and qRT-PCR
Author Contributions
Funding
Conflicts of Interest
Ethical Approval and Consent to Participate
References
- Meyers, G.; Ruemenapf, T.; Thiel, H.J. Molecular cloning and nucleotide sequence of the genome of hog cholera virus. Virology. 1989, 171, 555–567. [Google Scholar] [CrossRef]
- Moenning, V.; Floegel-Niesmann, G.; Greiser, W. Clinical signs and epidemiology of classical swine fever: A review of new knowledge. Vet. J. 2003, 165, 11–20. [Google Scholar] [CrossRef]
- Brown, V.R.; Bevins, S.N. A review of classical swine fever virus and routes of introduction into the United States and the potential for virus establishment. Front. Vet. Sci. 2018, 5, 31. [Google Scholar] [CrossRef]
- Paton, D.J.; Mcgoldrick, A.; Greiserwilke, A.; Parchariyanon, A.; Song, J.Y.; Liou, P.P.; Stadejek, T.; Lowings, J.P.; Bjorklund, H.; Belak, S. Genetic typing of classical swine fever virus. Vet. Microbiol. 2000, 73, 137–157. [Google Scholar] [CrossRef]
- Tu, C.; Lu, Z.; Li, H.; Yu, X.; Liu, X.; Li, Y.; Zhang, H.; Yin, Z. Phylogenetic comparison of classical swine fever virus in China. Virus Res. 2001, 81, 29–37. [Google Scholar] [CrossRef]
- Holland, W.G.; Do, T.T.; Huong, N.T.; Dung, N.T.; Thanh, N.G.; Vercruysse, J.; Goddeeris, B.M. The effect of Trypanosoma evansi infection on pig performance and vaccination against classical swine fever. Vet. Parasitol. 2003, 111, 115–123. [Google Scholar] [CrossRef]
- Kamakawa, A.; Ho, T.V.; Yamada, S. Epidemiological survey of viral diseases of pigs in the Mekong delta of Vietnam between 1999 and 2003. Vet. Microbiol. 2006, 118, 47–56. [Google Scholar] [CrossRef]
- Parchariyanon, S.; Inui, K.; Damrongwatanapokin, S.; Pinyochon, W.; Lowings, P.; Paton, D. Sequence analysis of E2 glycoprotein genes of classical swine fever viruses: Identification of a novel genogroup in Thailand. Dtsch. Tierarztl. Wochenschr. 2000, 107, 236–238. [Google Scholar]
- Blacksell, S.D.; Khounsy, S.; Boyle, D.B.; Gleeson, L.J.; Westbury, H.A.; Mackenzie, J.S. Genetic typing of classical swine fever viruses from Lao PDR by analysis of the 5’ non-coding region. Virus Genes 2005, 31, 349–355. [Google Scholar] [CrossRef]
- Wensvoort, G.; Terpstra, C. Swine fever: A changing clinical picture. Tijdschr Diergeneeskd. 1985, 110, 263–269. [Google Scholar]
- Koenen, F.; Van Caenegem, G.; Vermeersch, J.P.; Vandenheede, J.; Deluyker, H. Epidemiological characteristic of an outbreak of classical swine fever in an area of high pig density. Vet. Rec. 1996, 12, 367–371. [Google Scholar] [CrossRef]
- Mayer, D.; Thayer, T.M.; Hofmann, M.A.; Tratschin, J.D. Establishment and characterization of two cDNA-derived strains of classical swine fever virus, one highly virulent and one avirulent. Viurs Res. 2003, 98, 105–116. [Google Scholar] [CrossRef]
- Weesendrop, E.; Stegeman, A.; Loeffen, W. Dynamics of virus excretion via different routes in pigs experimentally infected with classical swine fever virus strain of high, moderated, or low virulence. Vet. Microbiol. 2009, 133, 9–22. [Google Scholar] [CrossRef]
- Yoo, S.J.; Kwon, T.; Kang, K.; Kim, H.; Kang, S.D.; Richt, J.A.; Lyoo, Y.S. Genetic evolution of classical swine fever virus under immune environments conditioned by genotype 1-based modified live virus vaccine. Transbound Emerg. Dis. 2018, 65, 735–745. [Google Scholar] [CrossRef]
- Kim, K.S.; Le, V.P.; Choe, S.; Cha, R.M.; Shin, J.; Cho, I.S.; Hung, N.P.; Thach, P.N.; Hang, V.T.T.; An, D.J. Complete genome sequences of classical swine fever virus subgenotype 2.1 and 2.2 strains isolated from Vietnamese pigs. Microbiol. Resour. Announc. 2019, 8, e01634-18. [Google Scholar] [CrossRef]
- An, D.J.; Lim, S.I.; Choe, S.; Kim, K.S.; Cha, R.M.; Cho, I.S.; Song, J.Y.; Hyun, B.H.; Park, B.K. Evolutionary dynamics of classical swine fever virus in South Korea: 1987–2017. Vet. Microbiol. 2018, 225, 79–88. [Google Scholar] [CrossRef]
- Lim, S.I.; Kim, Y.K.; Lim, J.A.; Han, S.H.; Hyun, H.S.; Kim, K.S.; Hyun, B.H.; Kim, J.J.; Cho, I.S.; Song, J.Y.; et al. Antigenic characterization of classical swine fever virus YC11WB isolates from wild boar. J. Vet. Sci. 2017, 18, 201–207. [Google Scholar] [CrossRef]
- Postel, A.; Jha, V.C.; Schmeiser, S.; Becher, P. First molecular identification and characterization of classical swine fever virus isolates from Nepal. Arch. Virol. 2013, 158, 207–210. [Google Scholar] [CrossRef]
- Pan, C.H.; Jong, M.H.; Huang, T.S.; Liu, H.F.; Lin, S.Y.; Lai, S.S. Phylogenetic analysis of classical swine fever virus in Taiwan. Arch. Virol. 2005, 150, 1101–1119. [Google Scholar] [CrossRef]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Bio. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef]
- Rambaut, A. FigFree. Tree Figure dra3wing Tool. 2012. Available online: https://www.semanticscholar.org/paper/Transatlantic-disjunction-in-fleshy-fungi.-II.-The-Petersen-Borovi%C4%8Dka/f8aeaf51c17f62a3c281961756b0c01b408340ba/figure/2 (accessed on 27 February 2020).
- Mittelholzer, C.; Moser, C.; Tratschin, J.; Hofmann, M.A. Analysis of classical swine fever virus replication kinetics allows differentiation of highly virulent from avirulent strains. Vet. Microbiol. 2000, 74, 293–308. [Google Scholar] [CrossRef]
- Choe, S.; Kim, J.H.; Kim, K.S.; Song, S.; Kang, W.C.; Kim, H.J.; Park, G.N.; Cha, R.M.; Cho, I.S.; Hyun, B.H.; et al. Impact of a Live Attenuated Classical Swine Fever Virus Introduced to Jeju Island, a CSF-Free Area. Pathogens 2019, 20, 8. [Google Scholar] [CrossRef]
- Choe, S.; Kim, J.H.; Kim, K.S.; Song, S.; Cha, R.M.; Kang, W.C.; Kim, H.J.; Park, G.N.; Shin, J.; Jo, H.N.; et al. Adverse Effects of Classical Swine Fever Virus LOM Vaccine and Jeju LOM Strains in Pregnant Sows and Specific Pathogen-Free Pigs. Pathogens 2019, 23, 9. [Google Scholar] [CrossRef]
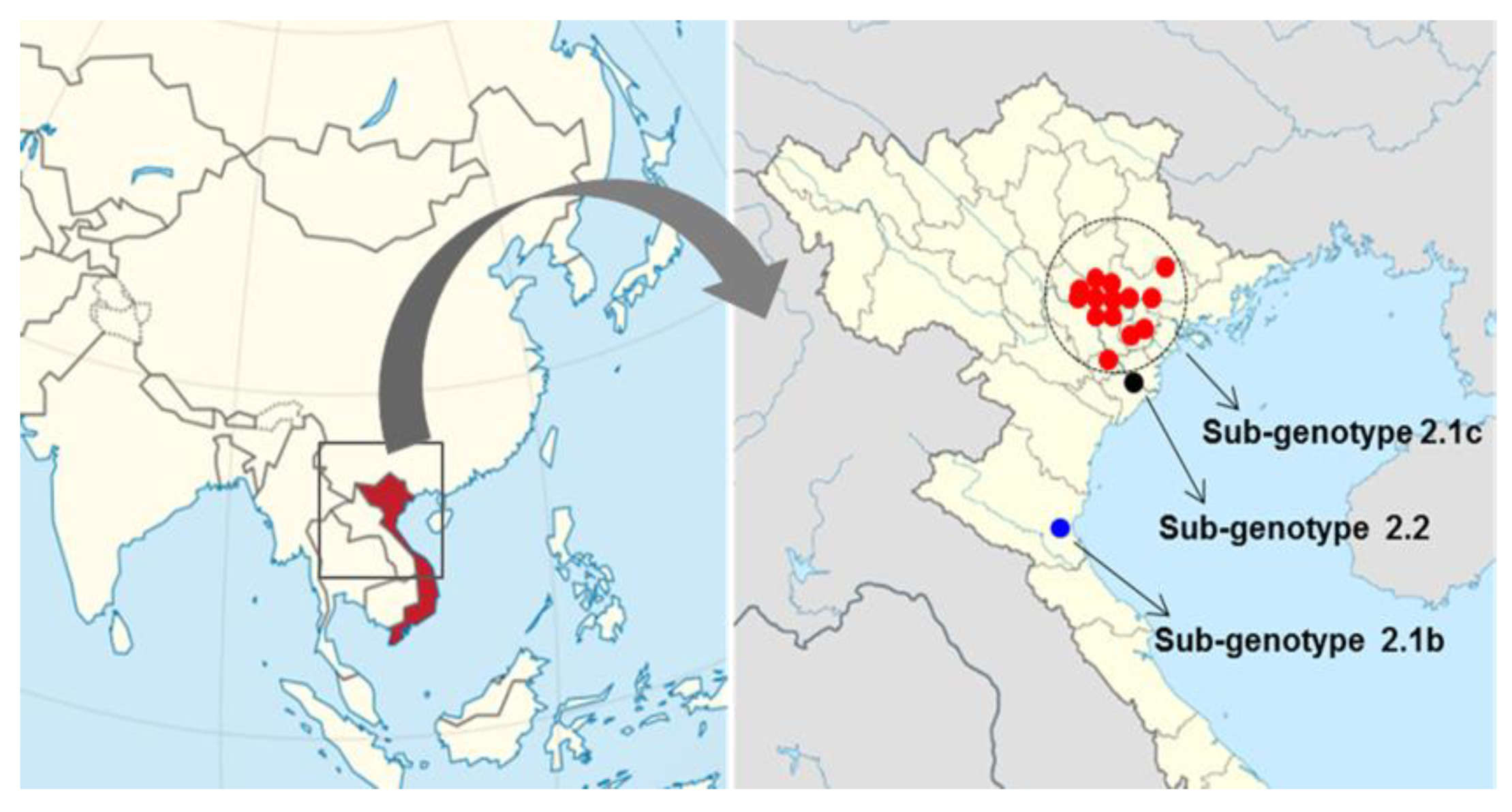
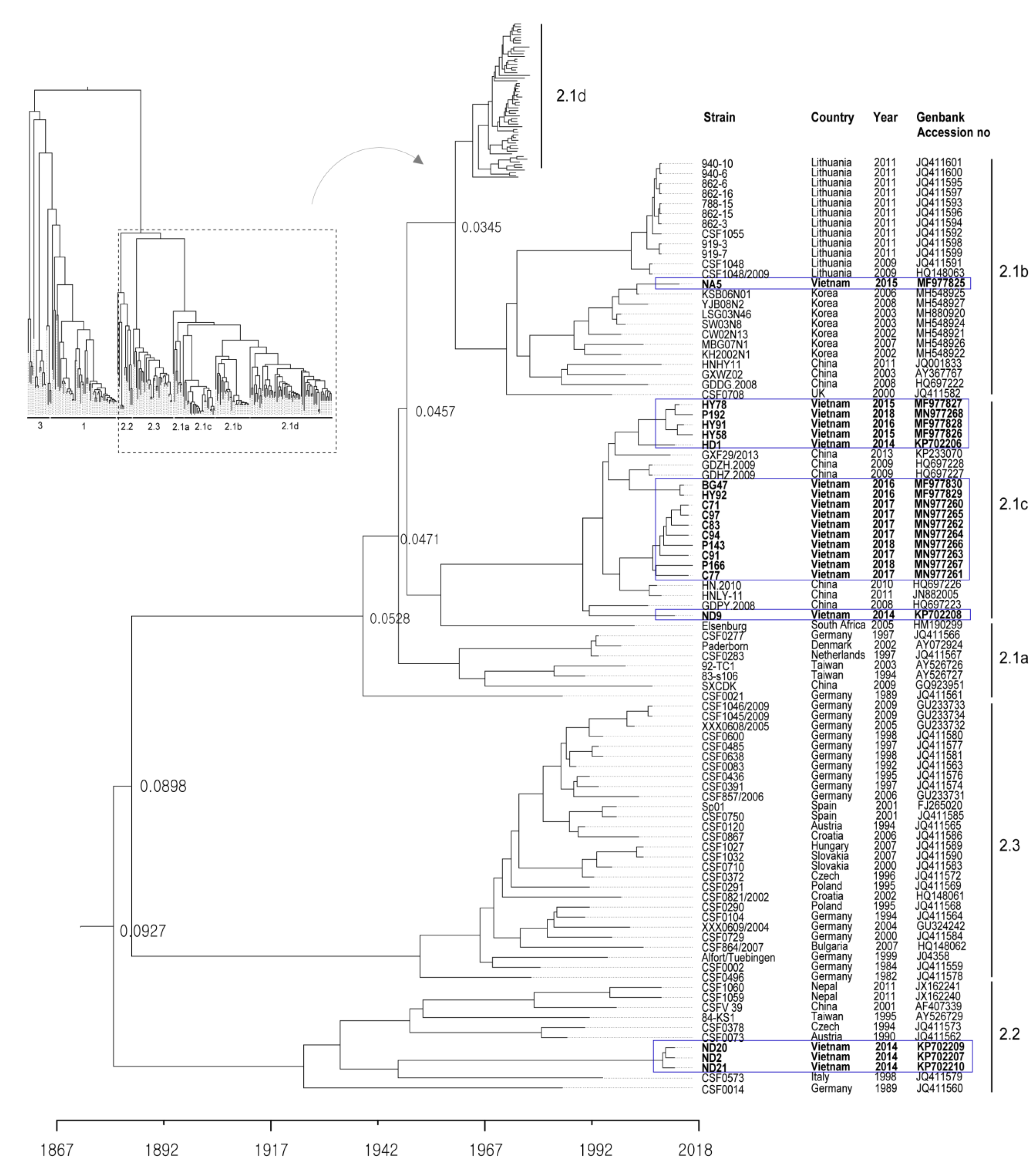

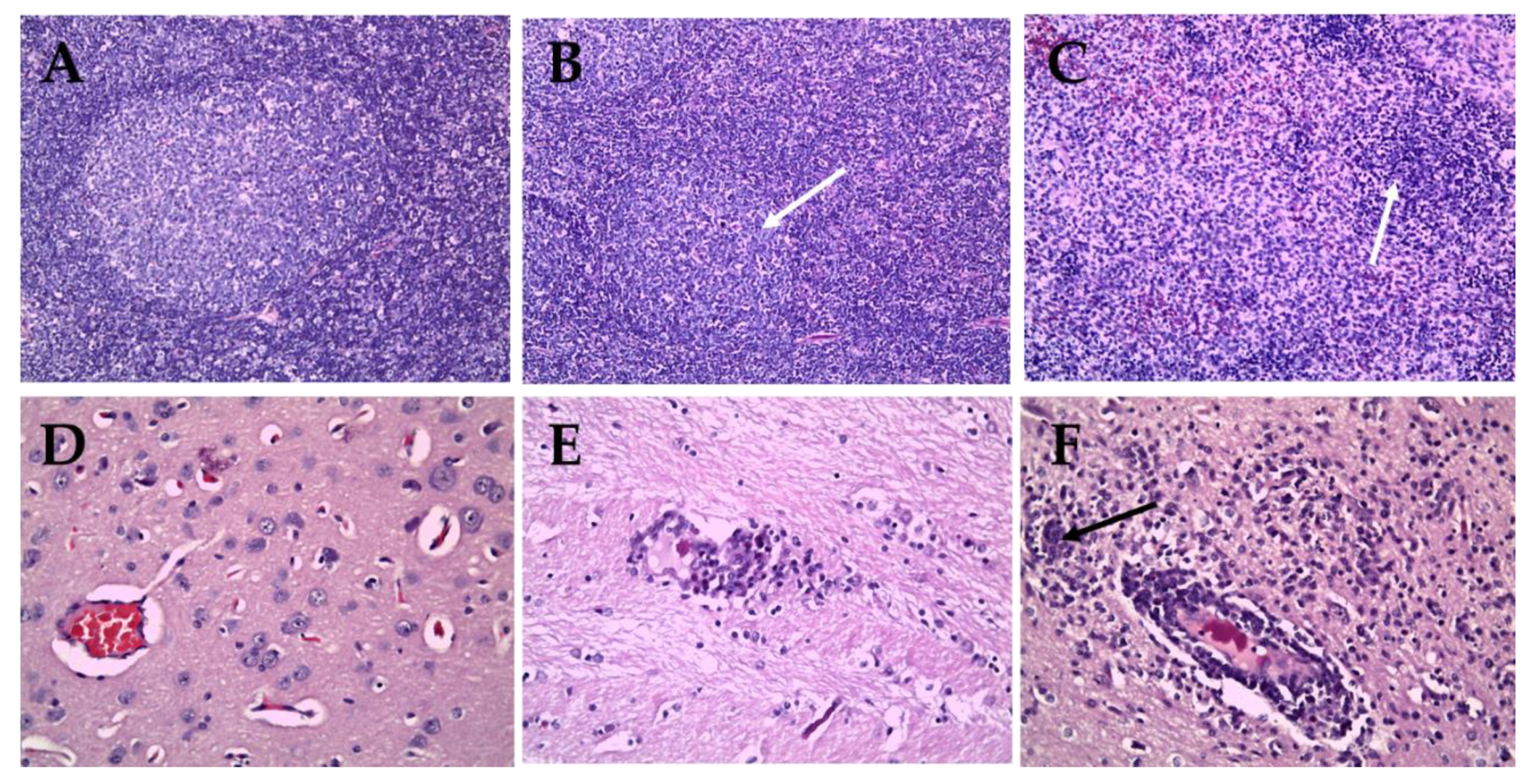
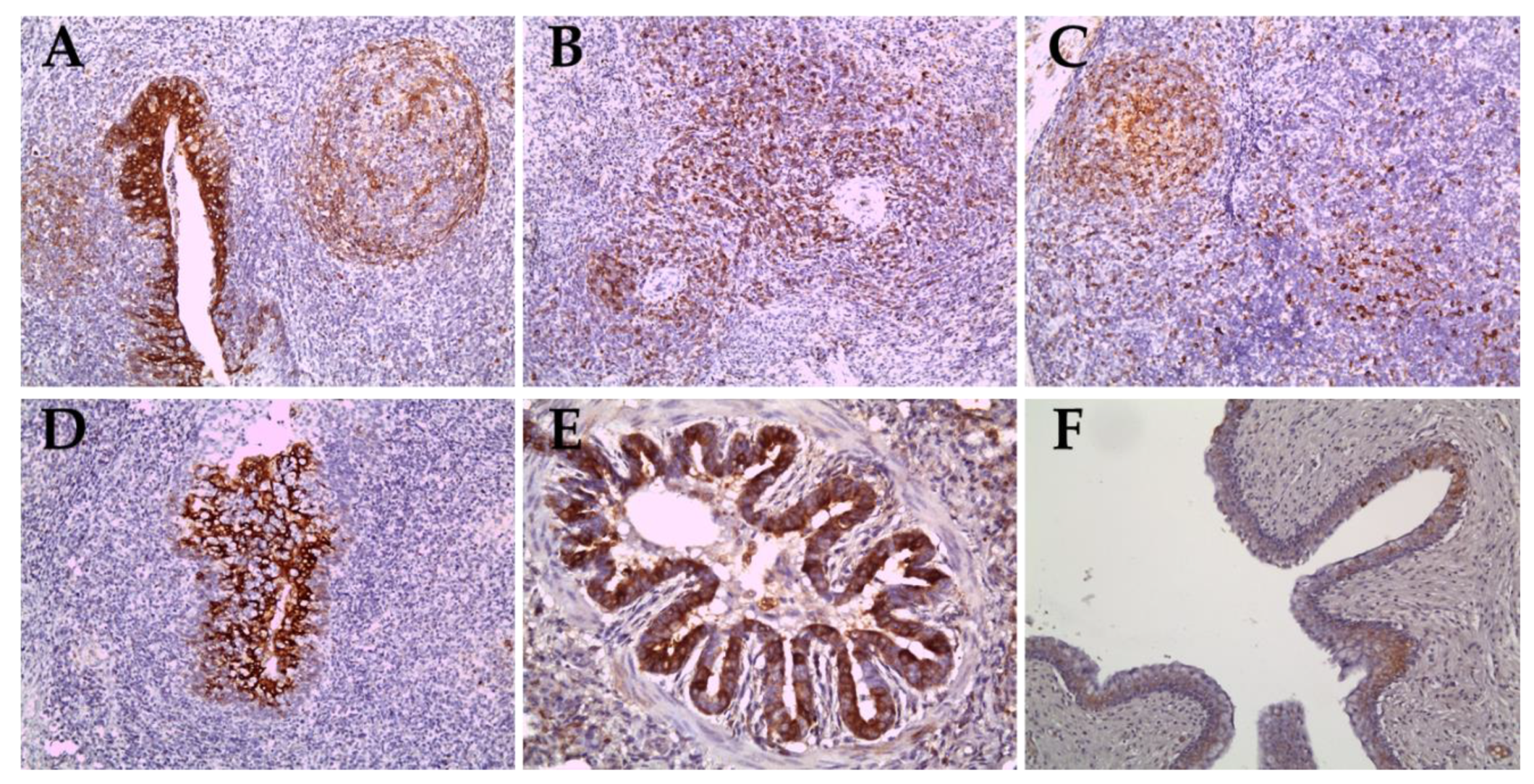
| Date of Collection | Place | Farm Name | Age of Sampled Pig | Strain | CSFV Genotype | Accession Number |
|---|---|---|---|---|---|---|
| June, 2014 | a Nam Dinh | XTND | 60 | ND2 | 2.2 | KP702207 |
| June, 2014 | a Nam Dinh | XTND | 60 | ND20 | 2.2 | KP702209 |
| June, 2014 | a Nam Dinh | XTND | 60 | ND21 | 2.2 | KP702210 |
| June, 2014 | b Nam Dinh | NDB | 90 | ND9 | 2.1c | KP702208 |
| July, 2014 | c Hai Duong | NGHD | 30 | HD1 | 2.1c | KP702206 |
| Jan, 2015 | Nghe An | NAN | 30 | NA5 | 2.1b | MF977825 |
| June, 2015 | Hung Yen | HYen | 70 | HY58 | 2.1c | MF977826 |
| Sep, 2015 | Hung Yen | HYen | 70 | HY78 | 2.1c | MF977827 |
| Feb, 2016 | Hung Yen | HYN | 50 | HY91 | 2.1c | MF977828 |
| Mar, 2016 | Hung Yen | HYY | 60 | HY92 | 2.1c | MF977829 |
| Mar, 2016 | Bac Giang | BGiang | 61 | BG47 | 2.1c | MF977830 |
| Jul, 2017 | Hung Yen | HYenI | 30 | C71 | 2.1c | MN977260 |
| Aug, 2017 | Hung Yen | HYenJ | 30 | C77 | 2.1c | MN977261 |
| Aug, 2017 | Hung Yen | HYenK | 40 | C83 | 2.1c | MN977262 |
| Sep, 2017 | Hung Yen | HYenJ | 30 | C91 | 2.1c | MN977263 |
| Sep, 2017 | Hung Yen | HYenL | 60 | C94 | 2.1c | MN977264 |
| Sep, 2017 | Hung Yen | HYenM | 50 | C97 | 2.1c | MN977265 |
| Oct, 2018 | Hung Yen | HYenN | 30 | P143 | 2.1c | MN977266 |
| Nov, 2018 | Nam Dinh | NDinhO | 40 | P166 | 2.1c | MN977267 |
| Dec, 2018 | Ha Nam | Ha NamP | 40 | P192 | 2.1c | MN977268 |
| Group | Pig No. | SG | a Strain | OP | DOP | b Clinical Signs | CSFV RNA Copy Number (log 10) in Blood (Days of Post-Infection) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 3 | 5 | 7 | 10 | 14 | 21 | |||||||
| 1 | 11 | 1.1 | GPE− | 30 | alive | None | - | - | - | - | - | - | - |
| 12 | 1.1 | GPE− | 30 | alive | None | - | - | - | - | - | - | - | |
| 2 | 7 | 2.2 | ND20 | 30 | alive | F, A, Di | - | 3.6 | 5.3 | 5.6 | 3.2 | 3.5 | 2.7 |
| 8 | 2.2 | ND20 | 30 | alive | F, A, T, Co | - | 2.7 | 4.1 | 5.3 | 3.8 | 3.1 | 2.4 | |
| 9 | 2.2 | ND20 | 30 | alive | F, A, T, Di | - | 3.0 | 5.5 | 5.1 | 3.3 | 4.2 | 1.8 | |
| 16 | 2.2 | ND20 | 30 | 28 | F, A, Di, T, C | - | 3.4 | 5.8 | 5.4 | 4.5 | 2.5 | 2.9 | |
| 34 | 2.2 | ND20 | 30 | alive | F, A, T, C | - | 2.5 | 4.9 | 4.5 | 2.7 | 2.6 | 1.7 | |
| 3 | 21 | 2.1c | HY58 | 30 | alive | F, A, De, T | - | 3.2 | 5.7 | 5.9 | 5.2 | 3.5 | 3.7 |
| 24 | 2.1c | HY58 | 30 | 22 | F, A, Di, De, C, T | - | 2.7 | 6.4 | 6.1 | 4.3 | 4.2 | 3.5 | |
| 29 | 2.1c | HY58 | 30 | 23 | F, A, De, C, HP | - | 2.9 | 6.1 | 6.8 | 5.7 | 5.3 | 3.7 | |
| 31 | 2.1c | HY58 | 30 | alive | F, A, De, T, Co | - | 3.3 | 4.7 | 5.6 | 5.8 | 4.2 | 2.6 | |
| 38 | 2.1c | HY58 | 30 | 25 | F, A, Di, T, Co, HP | - | 3.6 | 5.4 | 6.7 | 4.9 | 4.8 | 3.5 | |
| Mean Clinical Sore (21 dpi) | Total Mean Clinical Score | Virulent | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Parameter | *Li | BT | BS | Br | Wa | Sk | Ey | Ap | De | Le | ||
| ND20 strain | 0.8 | 0.2 | 0.5 | 0.8 | 0.8 | 0.9 | 1.0 | 1.5 | 0.5 | 1.4 | 8.4 | moderate |
| HY58 strain | 1.4 | 1.0 | 1.2 | 1.2 | 1.2 | 1.0 | 1.0 | 1.6 | 0.8 | 2.0 | 12.4 | moderate |
| *G | Strain | Pig No. | CSF RNA Copy Number (log 10)/Foci Score | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lung | Heart | Liver | Spleen | Tonsil | LN | SI | Ki | Bl | CNS | |||
| 1 | GPE− | 11 | - | - | - | - | - | - | - | - | - | - |
| 12 | - | - | - | - | - | - | - | - | - | - | ||
| 2 | ND20 | 7 | - | - | - | 3.3/* | 4.5/* | - | - | 4.3/* | 3.1/* | - |
| 8 | - | - | - | - | 3.7/* | 3.8/* | - | 2.5/* | - | - | ||
| 9 | - | - | - | 4.1/* | 4.9/* | 3.1/* | - | - | - | - | ||
| 16 | 3.3/+ | 4.1/+ | - | 5.2/+ | 6.4/+++ | 6.2/++ | - | 3.7/- | 3.8/+ | - | ||
| 34 | - | - | - | - | 3.6/- | 5.4/+ | 4.5/+ | - | - | - | ||
| 3 | HY58 | 21 | - | 2.1/* | - | 5.2/* | 5.7/* | 4.2/* | 3.4/* | - | - | - |
| 24 | 4.4/* | - | - | 4.1/* | 5.9/* | 4.6/* | - | 3.8/* | 4.8/* | - | ||
| 29 | 5.1/++ | 4.2/+ | 4.6/+ | 5.5/+ | 6.6/++ | 6.1/++ | 5.3/++ | 3.5/+ | 5.2/++ | 5.8/++ | ||
| 31 | 4.7/+ | - | - | 3.7/- | 6.2/++ | 5.9/+ | 4.8+ | 3.6/- | 4.7/+ | - | ||
| 38 | 3.5/* | - | 3.4/* | 4.8/* | 5.2/* | - | 3.9/* | 4.6/* | 3.1/* | 4.5/* | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choe, S.; Le, V.P.; Shin, J.; Kim, J.-H.; Kim, K.-S.; Song, S.; Cha, R.M.; Park, G.-N.; Nguyen, T.L.; Hyun, B.-H.; et al. Pathogenicity and Genetic Characterization of Vietnamese Classical Swine Fever Virus: 2014–2018. Pathogens 2020, 9, 169. https://doi.org/10.3390/pathogens9030169
Choe S, Le VP, Shin J, Kim J-H, Kim K-S, Song S, Cha RM, Park G-N, Nguyen TL, Hyun B-H, et al. Pathogenicity and Genetic Characterization of Vietnamese Classical Swine Fever Virus: 2014–2018. Pathogens. 2020; 9(3):169. https://doi.org/10.3390/pathogens9030169
Chicago/Turabian StyleChoe, SeEun, Van Phan Le, Jihye Shin, Jae-Hoon Kim, Ki-Sun Kim, Sok Song, Ra Mi Cha, Gyu-Nam Park, Thi Lan Nguyen, Bang-Hun Hyun, and et al. 2020. "Pathogenicity and Genetic Characterization of Vietnamese Classical Swine Fever Virus: 2014–2018" Pathogens 9, no. 3: 169. https://doi.org/10.3390/pathogens9030169
APA StyleChoe, S., Le, V. P., Shin, J., Kim, J.-H., Kim, K.-S., Song, S., Cha, R. M., Park, G.-N., Nguyen, T. L., Hyun, B.-H., Park, B.-K., & An, D.-J. (2020). Pathogenicity and Genetic Characterization of Vietnamese Classical Swine Fever Virus: 2014–2018. Pathogens, 9(3), 169. https://doi.org/10.3390/pathogens9030169





