Comparative Transcriptome Analysis of Two Cucumber Cultivars with Different Sensitivity to Cucumber Mosaic Virus Infection
Abstract
1. Introduction
2. Results and Discussion
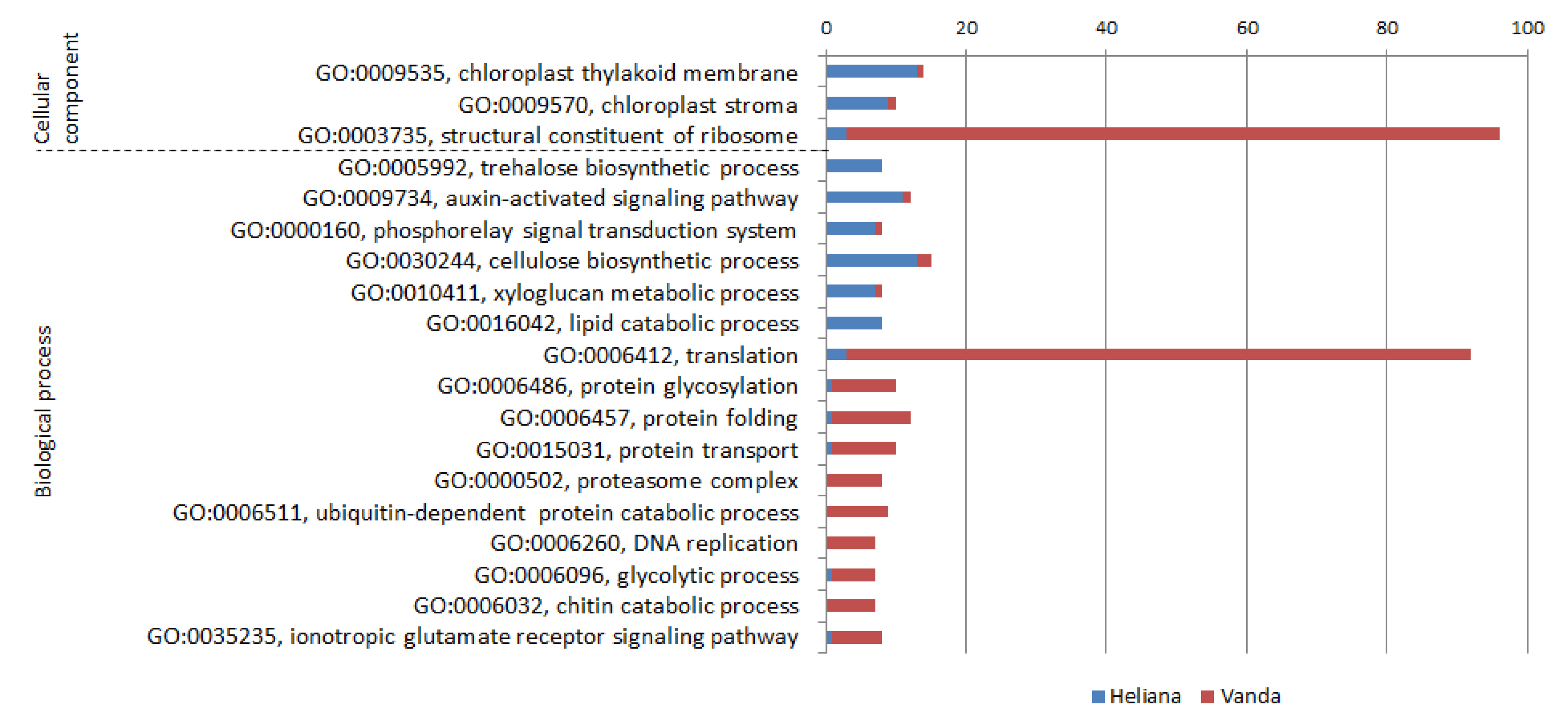
2.1. Gene Ontology Categorization of DEGs
2.2. Mock- and CMV-Inoculated cv. Heliana (Comparison H-/H+)
2.3. Mock- and CMV-Inoculated cv. Vanda (Comparison V-/V+)
2.3.1. Down-Regulation by CMV Infection
2.3.2. Up-Regulation by CMV Infection
2.4. Mock-Inoculated Cultivars (Comparison H-/V-)
2.5. CMV-Inoculated Cultivars (Comparison H+/V+)
2.6. Context with Other Published Data
3. Materials and Methods
3.1. Virus and Plants
3.2. RNA Isolation, cDNA Library Preparation and Sequence Analysis
3.3. Data Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- García-Arenal, F.; Palukaitis, P. Cucumber mosaic virus. In Desk Encyclopedia of Plant and Fungal Virology; Mahy, B.W.J., van Regenmortel, M.H.V., Eds.; Academic Press: Oxford, UK, 2009; pp. 171–176. [Google Scholar]
- Scholthof, K.-B.G.; Adkins, S.; Czosnek, H.; Palukaitis, P.; Jacquot, E.; Hohn, T.; Hohn, B.; Saunders, K.; Candresse, T.; Ahlquist, P.; et al. Top 10 plant viruses in molecular plant pathology. Mol. Plant Pathol. 2011, 12, 938–954. [Google Scholar] [CrossRef] [PubMed]
- Abad, J.; Anastasio, G.; Fraile, A.; García-Arenal, F. A search for resistance to Cucumber mosaic virus in the genus Lycopersicon. J. Plant Pathol. 2000, 82, 39–48. [Google Scholar]
- Kang, W.-H.; Hoang, N.H.; Yang, H.-B.; Kwon, J.-K.; Jo, S.-H.; Seo, J.-K.; Kim, K.-H.; Choi, I.; Kang, B.-C. Molecular mapping and characterization of a single dominant gene controlling CMV resistance in peppers (Capsicum annuum L.). Theor. Appl. Genet. 2010, 120, 1587–1596. [Google Scholar] [CrossRef] [PubMed]
- Song, E.G.; Ryu, K.H. Engineering resistance to a resistance-breaking strain of Cucumber mosaic virus in plants utilizing viral dsRNA. Plant Biotechnol. Rep. 2017, 11, 429–438. [Google Scholar] [CrossRef]
- Whitham, S.; Yang, C.; Goodin, M.M. Global Impact: Elucidating Plant Responses to Viral Infection. Mol. Plant Microbe Interactions 2006, 19, 1207–1215. [Google Scholar] [CrossRef] [PubMed]
- Carr, J.P.; Murphy, A.M. Host Responses—Susceptibility. In Cucumber Mosaic Virus; Palukaitis, P., García-Arenal, F., Eds.; APS Press: St Paul, MN, USA, 2019; pp. 47–58. [Google Scholar]
- Carr, J.P.; Murphy, A.M. Suppression of Plant Defense. In Cucumber Mosaic Virus; Palukaitis, P., García-Arenal, F., Eds.; APS Press: St Paul, MN, USA, 2019; pp. 133–144. [Google Scholar]
- Ando, S.; Miyashita, S.; Takahashi, H. Plant defense systems against cucumber mosaic virus: Lessons learned from CMV–Arabidopsis interactions. J. Gen. Plant Pathol. 2019, 85, 174–181. [Google Scholar] [CrossRef]
- Marathe, R.; Guan, Z.; Anandalakshmi, R.; Zhao, H.; Dinesh-Kumar, S. Study of Arabidopsis thaliana resistome in response to Cucumber mosaic virus infection using whole genome microarray. Plant Mol. Biol. 2004, 55, 501–520. [Google Scholar] [CrossRef] [PubMed]
- Šubr, Z.; Glasa, M. The sensitivity of Cucurbitaceae cultivars to viral diseases studied as base for research of molecular virus-host interactions. In Proceedings of the 21st Czech and Slovak Conference on Plant Protection, Brno, Czech Republic, 5–6 September 2018; p. 30. [Google Scholar]
- Nováková, S.; Šubr, Z.; Kováč, A.; Fialová, I.; Beke, G.; Danchenko, M. Cucumber mosaic virus resistance: Comparative proteomics of contrasting Cucumis sativus cultivars after long-term infection. J. Proteom. 2019, 214, 103626. [Google Scholar] [CrossRef]
- Farver, O.; Wherland, S.; Pecht, I. Intramolecular electron transfer in ascorbate oxidase is enhanced in the presence of oxygen. J. Boil. Chem. 1994, 269, 22933–22936. [Google Scholar]
- Kato, N.; Esaka, M. Changes in ascorbate oxidase gene expression and ascorbate levels in cell division and cell elongation in tobacco cells. Physiol. Plant. 1999, 105, 321–329. [Google Scholar] [CrossRef]
- De Tullio, M.; Liso, R.; Arrigoni, O.; De Tullio, M. Ascorbic Acid Oxidase: An Enzyme in Search of a Role. Boil. Plant. 2004, 48, 161–166. [Google Scholar] [CrossRef]
- De Tullio, M.; Guether, M.; Balestrini, R. Ascorbate oxidase is the potential conductor of a symphony of signaling pathways. Plant Signal. Behav. 2013, 8, 23213. [Google Scholar] [CrossRef]
- Sanmartin, M.; Pateraki, I.; Chatzopoulou, F.; Kanellis, A. Differential expression of the ascorbate oxidase multigene family during fruit development and in response to stress. Planta 2006, 225, 873–885. [Google Scholar] [CrossRef] [PubMed]
- García-Pineda, E.; Ernesto, G.-P.; Castro-Mercado, E.; Lozoya-Gloria, E. Gene expression and enzyme activity of pepper (Capsicum annuum L.) ascorbate oxidase during elicitor and wounding stress. Plant Sci. 2004, 166, 237–243. [Google Scholar] [CrossRef]
- Zarrillo, A.; Minutolo, M.; Alioto, D.; Errico, A. Ascorbic acid regulation in leaves and fruits of tomato ecotypes infected by Eggplant Mottled Dwarf Virus. Sci. Hortic. 2017, 225, 512–524. [Google Scholar] [CrossRef]
- Kumari, R.; Kumar, S.; Singh, L.; Hallan, V. Movement Protein of Cucumber Mosaic Virus Associates with Apoplastic Ascorbate Oxidase. PLoS ONE 2016, 11, e0163320. [Google Scholar] [CrossRef]
- Wu, J.; Yang, R.; Yang, Z.; Yao, S.; Zhao, S.; Wang, Y.; Li, P.; Song, X.; Jin, L.; Zhou, T.; et al. ROS accumulation and antiviral defence control by microRNA528 in rice. Nat. Plants 2017, 3, 16203. [Google Scholar] [CrossRef]
- Cândido, E.D.S.; Pinto, M.F.S.; Pelegrini, P.B.; Lima, T.B.; Silva, O.; Pogue, R.; Grossi-De-Sa, M.F.; Franco, O.L. Plant storage proteins with antimicrobial activity: Novel insights into plant defense mechanisms. FASEB J. 2011, 25, 3290–3305. [Google Scholar] [CrossRef]
- Holk, A.; Rietz, S.; Zahn, M.; Quader, H.; Scherer, G.F. Molecular Identification of Cytosolic, Patatin-Related Phospholipases A from Arabidopsis with Potential Functions in Plant Signal Transduction. Plant Physiol. 2002, 130, 90–101. [Google Scholar] [CrossRef]
- Xu, Y.; Huang, B. Comparative transcriptomic analysis reveals common molecular factors responsive to heat and drought stress in Agrostis stolonifera. Sci. Rep. 2018, 8, 15181. [Google Scholar] [CrossRef]
- Rai, A.; Upadhyay, A.R.P. Differential Expression of Pathogenesis Related Protein Genes in Tomato during Inoculation with A. solani. J. Plant Pathol. Microbiol. 2014, 5, 1000217. [Google Scholar]
- Folgado, R.; Panis, B.; Sergeant, K.; Renaut, J.; Swennen, R.; Hausman, J.-F. Differential Protein Expression in Response to Abiotic Stress in Two Potato Species: Solanum commersonii Dun and Solanum tuberosum L. Int. J. Mol. Sci. 2013, 14, 4912–4933. [Google Scholar] [CrossRef] [PubMed]
- Dhondt-Cordelier, S.; Geoffroy, P.; Stelmach, B.A.; Legrand, M.; Heitz, T. Soluble phospholipase A2 activity is induced before oxylipin accumulation in tobacco mosaic virus-infected tobacco leaves and is contributed by patatin-like enzymes. Plant J. 2000, 23, 431–440. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Song, N.; Wu, J. A patatin-like protein synergistically regulated by jasmonate and ethylene signaling pathways plays a negative role in Nicotiana attenuata resistance to Alternaria alternata. Plant Divers. 2018, 41, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, A.; Takahashi, A.; Kakimoto, Y.; Miyazawa, Y.; Fujii, N.; Higashitani, A.; Takahashi, H. A gene essential for hydrotropism in roots. Proc. Natl. Acad. Sci. USA 2007, 104, 4724–4729. [Google Scholar] [CrossRef]
- Moriwaki, T.; Miyazawa, Y.; Kobayashi, A.; Uchida, M.; Watanabe, C.; Fujii, N.; Takahashi, H. Hormonal regulation of lateral root development in Arabidopsis modulated by MIZ1 and requirement of GNOM activity for MIZ1 function. Plant Physiol. 2011, 157, 1209–1220. [Google Scholar] [CrossRef]
- Shah, A.; Ganguli, S.; Sen, J.; Bhandari, R. Inositol Pyrophosphates: Energetic, Omnipresent and Versatile Signalling Molecules. J. Indian Inst. Sci. 2017, 97, 23–40. [Google Scholar] [CrossRef]
- Miozzi, L.; Napoli, C.; Sardo, L.; Accotto, G.P. Transcriptomics of the Interaction between the Monopartite Phloem-Limited Geminivirus Tomato Yellow Leaf Curl Sardinia Virus and Solanum lycopersicum Highlights a Role for Plant Hormones, Autophagy and Plant Immune System Fine Tuning during Infection. PLoS ONE 2014, 9, e89951. [Google Scholar] [CrossRef]
- Lee, M.O.; Hwang, J.H.; Lee, N.H.; Hong, C.B. Gene expression profile for Nicotiana tabacum in the early phase of flooding stress. J. Plant Boil. 2007, 50, 496–503. [Google Scholar] [CrossRef]
- Li, S.; Tan, Q.; Sun, M.; Xu, G.; Li, C.; Fu, X.; Li, L.; Gao, D.; Li, N. Protein changes in response to photoperiod during dormancy induction in peach leaves and flower buds. Sci. Hortic. 2018, 239, 114–122. [Google Scholar] [CrossRef]
- O’Toole, N.; Hattori, M.; Andres, C.; Iida, K.; Lurin, C.; Schmitz-Linneweber, C.; Sugita, M.; Small, I. On the Expansion of the Pentatricopeptide Repeat Gene Family in Plants. Mol. Boil. Evol. 2008, 25, 1120–1128. [Google Scholar] [CrossRef] [PubMed]
- Barkan, A.; Small, I. Pentatricopeptide Repeat Proteins in Plants. Annu. Rev. Plant Boil. 2014, 65, 415–442. [Google Scholar] [CrossRef] [PubMed]
- Bolwell, G.P.; Bozak, K.; Zimmerlin, A. Plant cytochrome p450. Phytochemistry 1994, 37, 1491–1506. [Google Scholar] [CrossRef]
- Narusaka, Y.; Narusaka, M.; Seki, M.; Umezawa, T.; Ishida, J.; Nakajima, M.; Enju, A.; Shinozaki, K. Crosstalk in the responses to abiotic and biotic stresses in Arabidopsis: Analysis of gene expression in cytochrome P450 gene superfamily by cDNA microarray. Plant Mol. Boil. 2004, 55, 327–342. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Bai, X.; Luo, X.; Chen, Q.; Cai, H.; Ji, W.; Zhu, Y. Identification of wild soybean (Glycine soja) TIFY family genes and their expression profiling analysis under bicarbonate stress. Plant Cell Rep. 2012, 32, 263–272. [Google Scholar] [CrossRef]
- Ye, H.; Du, H.; Tang, N.; Li, X.; Xiong, L. Identification and expression profiling analysis of TIFY family genes involved in stress and phytohormone responses in rice. Plant Mol. Boil. 2009, 71, 291–305. [Google Scholar] [CrossRef]
- Zheng, W.; Ma, L.; Zhao, J.; Li, Z.; Sun, F.; Lu, X. Comparative Transcriptome Analysis of Two Rice Varieties in Response to Rice Stripe Virus and Small Brown Planthoppers during Early Interaction. PLoS ONE 2013, 8, e82126. [Google Scholar] [CrossRef]
- Saha, G.; Park, J.-I.; Kayum, A.; Nou, I.-S. A Genome-Wide Analysis Reveals Stress and Hormone Responsive Patterns of TIFY Family Genes in Brassica rapa. Front. Plant Sci. 2016, 7, 26. [Google Scholar] [CrossRef]
- Vozárová, Z.; Žilová, M.; Šubr, Z. Differentially expressed genes in healthy and plum pox virus-infected Nicotiana benthamiana plants. Acta Virol. 2015, 59, 389–397. [Google Scholar] [CrossRef][Green Version]
- Lacatus, G.; Sunter, G. The Arabidopsis PEAPOD2 transcription factor interacts with geminivirus AL2 protein and the coat protein promoter. Virology 2009, 392, 196–202. [Google Scholar] [CrossRef]
- Stacey, N.J.; Kuromori, T.; Azumi, Y.; Roberts, G.; Breuer, C.; Wada, T.; Maxwell, A.; Roberts, K.; Sugimoto-Shirasu, K. Arabidopsis SPO11-2 functions with SPO11-1 in meiotic recombination. Plant J. 2006, 48, 206–216. [Google Scholar] [CrossRef] [PubMed]
- Kovalchuk, I.; Kovalchuk, O.; Kalck, V.; Boyko, V.; Filkowski, J.; Heinlein, M.; Hohn, B. Pathogen-induced systemic plant signal triggers DNA rearrangements. Nature 2003, 423, 760–762. [Google Scholar] [CrossRef] [PubMed]
- Schürmann, D.; Molinier, J.; Fritsch, O.; Hohn, B. The dual nature of homologous recombination in plants. Trends Genet. 2005, 21, 172–181. [Google Scholar] [CrossRef] [PubMed]
- Gullner, G.; Komives, T.; Király, L.; Schröder, P. Glutathione S-Transferase Enzymes in Plant-Pathogen Interactions. Front. Plant Sci. 2018, 9, 9. [Google Scholar] [CrossRef] [PubMed]
- Fodor, J.; Gullner, G.; Adam, A.L.; Barna, B.; Komives, T.; Kiraly, Z. Local and Systemic Responses of Antioxidants to Tobacco Mosaic Virus Infection and to Salicylic Acid in Tobacco (Role in Systemic Acquired Resistance). Plant Physiol. 1997, 114, 1443–1451. [Google Scholar] [CrossRef] [PubMed]
- Ishihara, T.; Sakurai, N.; Sekine, K.-T.; Hase, S.; Ikegami, M.; Shibata, D.; Takahashi, H. Comparative analysis of expressed sequence tags in resistant and susceptible ecotypes of Arabidopsis thaliana infected with cucumber mosaic virus. Plant Cell Physiol. 2004, 45, 470–480. [Google Scholar] [CrossRef][Green Version]
- Satoh, K.; Kondoh, H.; De Leon, T.B.; Macalalad, R.J.A.; Cabunagan, R.C.; Cabauatan, P.Q.; Mauleon, R.; Kikuchi, S.; Choi, I.-R. Gene expression responses to Rice tungro spherical virus in susceptible and resistant near-isogenic rice plants. Virus Res. 2013, 171, 111–120. [Google Scholar] [CrossRef]
- Love, A.J.; Yun, B.W.; Laval, V.; Loake, G.J.; Milner, J.J. Cauliflower mosaic virus, a Compatible Pathogen of Arabidopsis, Engages Three Distinct Defense-Signaling Pathways and Activates Rapid Systemic Generation of Reactive Oxygen Species. Plant Physiol. 2005, 139, 935–948. [Google Scholar] [CrossRef]
- Kumar, B.K.P.; Kanakala, S.; Malathi, V.G.; Gopal, P.; Usha, R. Transcriptomic and proteomic analysis of yellow mosaic diseased soybean. J. Plant Biochem. Biotechnol. 2016, 26, 224–234. [Google Scholar] [CrossRef]
- Chen, I.-H.; Chiu, M.-H.; Cheng, S.-F.; Hsu, Y.-H.; Tsai, C.-H. The glutathione transferase of Nicotiana benthamiana NbGSTU4 plays a role in regulating the early replication of Bamboo mosaic virus. New Phytol. 2013, 199, 749–757. [Google Scholar] [CrossRef]
- Fanata, W.I.D.; Lee, S.Y.; Lee, K.O. The unfolded protein response in plants: A fundamental adaptive cellular response to internal and external stresses. J. Proteom. 2013, 93, 356–368. [Google Scholar] [CrossRef]
- Qian, D.; Chen, G.; Tian, L.; Qu, L.Q. OsDER1 Is an ER-Associated Protein Degradation Factor That Responds to ER Stress. Plant Physiol. 2018, 178, 402–412. [Google Scholar] [CrossRef] [PubMed]
- Schelhaas, M.; Malmström, J.; Pelkmans, L.; Haugstetter, J.; Ellgaard, L.; Grünewald, K.; Helenius, A. Simian Virus 40 Depends on ER Protein Folding and Quality Control Factors for Entry into Host Cells. Cell 2007, 131, 516–529. [Google Scholar] [CrossRef] [PubMed]
- Lilley, B.N.; Gilbert, J.M.; Ploegh, H.L.; Benjamin, T. Murine Polyomavirus Requires the Endoplasmic Reticulum Protein Derlin-2 To Initiate Infection. J. Virol. 2006, 80, 8739–8744. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekhar, B.; Umesha, S.; Kumar, H.N. Proteomic analysis of salicylic acid enhanced disease resistance in bacterial wilt affected chilli (Capsicum annuum) crop. Physiol. Mol. Plant Pathol. 2017, 98, 85–96. [Google Scholar] [CrossRef]
- Ghorbani, A.; Izadpanah, K.; Dietzgen, R.G. Gene expression and population polymorphism of maize Iranian mosaic virus in Zea mays, and intracellular localization and interactions of viral N, P, and M proteins in Nicotiana benthamiana. Virus Genes 2018, 54, 290–296. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, A.; Zhou, X. ER Stress, UPR and Virus Infections in Plants. In Current Research Topics in Plant Virology; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2016; pp. 173–195. [Google Scholar]
- Xie, Y.-D.; Li, W.; Guo, D.; Dong, J.; Zhang, Q.; Fu, Y.; Ren, D.; Peng, M.; Xia, Y. The Arabidopsis gene SIGMA FACTOR-BINDING PROTEIN 1 plays a role in the salicylate- and jasmonate-mediated defence responses. Plant Cell Environ. 2010, 33, 828–839. [Google Scholar]
- Denay, G.; Vachon, G.; Dumas, R.; Zubieta, C.; Parcy, F.; Denay, G.; Parcy, F. Plant SAM-Domain Proteins Start to Reveal Their Roles. Trends Plant Sci. 2017, 22, 718–725. [Google Scholar] [CrossRef]
- Wan, H.; Yuan, W.; Ye, Q.; Wang, R.; Ruan, M.; Li, Z.; Zhou, G.; Yao, Z.; Zhao, J.; Liu, S.; et al. Analysis of TIR- and non-TIR-NBS-LRR disease resistance gene analogous in pepper: Characterization, genetic variation, functional divergence and expression patterns. BMC Genom. 2012, 13, 502. [Google Scholar] [CrossRef]
- Citovsky, V.; Zaltsman, A.; Kozlovsky, S.V.; Gafni, Y.; Krichevsky, A. Proteasomal degradation in plant–pathogen interactions. Semin. Cell Dev. Boil. 2009, 20, 1048–1054. [Google Scholar] [CrossRef]
- Jin, H.; Li, S.; Villegas, A. Down-Regulation of the 26S Proteasome Subunit RPN9 Inhibits Viral Systemic Transport and Alters Plant Vascular Development. Plant Physiol. 2006, 142, 651–661. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.; Kim, J.; Bang, B.; Kim, I.; Lee, H.-H.; Park, J.; Seo, Y.-S. Comparative Analyses of Tomato yellow leaf curl virus C4 Protein-Interacting Host Proteins in Healthy and Infected Tomato Tissues. Plant Pathol. J. 2016, 32, 377–387. [Google Scholar] [CrossRef] [PubMed]
- Hauser, F.; Li, Z.; Waadt, R.; Schroeder, J.I. SnapShot: Abscisic Acid Signaling. Cell 2017, 171, 1708. [Google Scholar] [CrossRef] [PubMed]
- Schuberth, C.; Buchberger, A. UBX domain proteins: Major regulators of the AAA ATPase Cdc48/p97. Cell. Mol. Life Sci. 2008, 65, 2360–2371. [Google Scholar] [CrossRef] [PubMed]
- Bègue, H.; Mounier, A.; Rosnoblet, C.; Wendehenne, D. Toward the understanding of the role of CDC48, a major component of the protein quality control, in plant immunity. Plant Sci. 2019, 279, 34–44. [Google Scholar] [CrossRef]
- Choi, A.G.; Wong, J.; Marchant, D.; Luo, H. The ubiquitin-proteasome system in positive-strand RNA virus infection. Rev. Med Virol. 2012, 23, 85–96. [Google Scholar] [CrossRef]
- El Kasmi, F.; Chung, E.-H.; Anderson, R.G.; Li, J.; Wan, L.; Eitas, T.K.; Gao, Z.; Dangl, J.L. Signaling from the plasma-membrane localized plant immune receptor RPM1 requires self-association of the full-length protein. Proc. Natl. Acad. Sci. USA 2017, 114, E7385–E7394. [Google Scholar] [CrossRef]
- Liu, J.; Elmore, J.M.; Fuglsang, A.T.; Palmgren, M.B.; Staskawicz, B.J.; Coaker, G. RIN4 Functions with Plasma Membrane H+-ATPases to Regulate Stomatal Apertures during Pathogen Attack. PLoS Boil. 2009, 7, e1000139. [Google Scholar] [CrossRef]
- Mandadi, K.; Scholthof, K.-B.G. Plant immune responses against viruses: How does a virus cause disease? Plant Cell 2013, 25, 1489–1505. [Google Scholar] [CrossRef]
- Chong, J.; Baltz, R.; Schmitt-Keichinger, C.; Beffa, R.; Fritig, B.; Saindrenan, P. Downregulation of a Pathogen-Responsive Tobacco UDP-Glc: Phenylpropanoid Glucosyltransferase Reduces Scopoletin Glucoside Accumulation, Enhances Oxidative Stress, and Weakens Virus Resistance. Plant Cell 2002, 14, 1093–1107. [Google Scholar] [CrossRef]
- Matros, A.; Mock, H.-P. Ectopic Expression of a UDP-Glucose:phenylpropanoid Glucosyltransferase Leads to Increased Resistance of Transgenic Tobacco Plants against Infection with Potato Virus Y. Plant Cell Physiol. 2004, 45, 1185–1193. [Google Scholar] [CrossRef] [PubMed]
- Rehman, H.M.; Nawaz, M.A.; Shah, Z.H.; Ludwig-Müller, J.; Chung, G.; Ahmad, M.Q.; Yang, S.H.; Lee, S.I. Comparative genomic and transcriptomic analyses of Family-1 UDP glycosyltransferase in three Brassica species and Arabidopsis indicates stress-responsive regulation. Sci. Rep. 2018, 8, 1875. [Google Scholar] [CrossRef] [PubMed]
- Conti, G.; Rodriguez, M.C.; Manacorda, C.; Asurmendi, S. Transgenic Expression of Tobacco mosaic virus Capsid and Movement Proteins Modulate Plant Basal Defense and Biotic Stress Responses in Nicotiana tabacum. Mol. Plant Microbe Interactions 2012, 25, 1370–1384. [Google Scholar] [CrossRef] [PubMed]
- Sarowar, S.; Kim, Y.J.; Kim, K.D.; Hwang, B.K.; Ok, S.H.; Shin, J.S. Overexpression of lipid transfer protein (LTP) genes enhances resistance to plant pathogens and LTP functions in long-distance systemic signaling in tobacco. Plant Cell Rep. 2008, 28, 419–427. [Google Scholar] [CrossRef] [PubMed]
- Van Ooijen, G.; Mayr, G.; Kasiem, M.M.A.; Albrecht, M.; Cornelissen, B.J.C.; Takken, F.L.W. Structure–function analysis of the NB-ARC domain of plant disease resistance proteins. J. Exp. Bot. 2008, 59, 1383–1397. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.; Jo, Y.; Lian, S.; Jo, K.-M.; Chu, H.; Yoon, J.-Y.; Choi, S.-K.; Kim, K.-H.; Cho, W.K. Comparative analysis of chrysanthemum transcriptome in response to three RNA viruses: Cucumber mosaic virus, Tomato spotted wilt virus and Potato virus X. Plant Mol. Boil. 2015, 88, 233–248. [Google Scholar] [CrossRef]
- Acevedo-Garcia, J.; Kusch, S.; Panstruga, R. Magical mystery tour: MLO proteins in plant immunity and beyond. New Phytol. 2014, 204, 273–281. [Google Scholar] [CrossRef]
- Thiel, H.; Varrelmann, M. Identification of Beet necrotic yellow vein virus P25 Pathogenicity Factor–Interacting Sugar Beet Proteins That Represent Putative Virus Targets or Components of Plant Resistance. Mol. Plant Microbe Interactions 2009, 22, 999–1010. [Google Scholar] [CrossRef]
- Barros, J.; Serk, H.; Granlund, I.; Pesquet, E. The cell biology of lignification in higher plants. Ann. Bot. 2015, 115, 1053–1074. [Google Scholar] [CrossRef]
- Xie, M.; Zhang, J.; Tschaplinski, T.J.; Tuskan, G.A.; Chen, J.; Muchero, W. Regulation of Lignin Biosynthesis and Its Role in Growth-Defense Tradeoffs. Front. Plant Sci. 2018, 9, 1427. [Google Scholar] [CrossRef]
- Gerke, V.; Creutz, C.E.; Moss, S.E. Annexins: Linking Ca2+ signalling to membrane dynamics. Nat. Rev. Mol. Cell Boil. 2005, 6, 449–461. [Google Scholar] [CrossRef]
- Mortimer, J.C.; Laohavisit, A.; MacPherson, N.; Webb, A.A.R.; Brownlee, C.; Battey, N.H.; Davies, J.M. Annexins: Multifunctional components of growth and adaptation. J. Exp. Bot. 2008, 59, 533–544. [Google Scholar] [CrossRef]
- Diab, H.; Limami, A.M. Reconfiguration of N Metabolism upon Hypoxia Stress and Recovery: Roles of Alanine Aminotransferase (AlaAT) and Glutamate Dehydrogenase (GDH). Plants 2016, 5, 25. [Google Scholar] [CrossRef] [PubMed]
- Marsh, E.; Alvarez, S.; Hicks, L.M.; Barbazuk, W.; Qiu, W.; Kovács, L.; Schachtman, D.P. Changes in protein abundance during powdery mildew infection of leaf tissues of Cabernet Sauvignon grapevine (Vitis vinifera L.). Proteomics 2010, 10, 2057–2064. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.-J.; Park, C.-J.; An, J.-M.; Ham, B.-K.; Lee, B.-J.; Paek, K.-H. CaAlaAT1 catalyzes the alanine: 2-oxoglutarate aminotransferase reaction during the resistance response against Tobacco mosaic virus in hot pepper. Planta 2005, 221, 857–867. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Calvino, L.; Osorio, S.; Hernandez, L.; Hamada, I.B.; Del Toro, F.J.; Donaire, L.; Yu, A.; Bustos, R.; Fernie, A.R.; Martínez-Rivas, J.M.; et al. Virus-Induced Alterations in Primary Metabolism Modulate Susceptibility to Tobacco rattle virus in Arabidopsis. Plant Physiol. 2014, 166, 1821–1838. [Google Scholar] [CrossRef]
- Manova, V.; Gruszka, D. DNA damage and repair in plants—From models to crops. Front. Plant Sci. 2015, 6, 187. [Google Scholar] [CrossRef]
- Li, Y.; Shi, X.; Gao, X. In vivo effects of piperonyl butoxide on the activities of 7-ethoxycoumarin O-deethylase and carboxylesterase in larval Chilo suppressalis (Lepidoptera: Pyralidae). Acta Entomol. Sin. 2016, 59, 1159–1165. [Google Scholar]
- Li, S.-W.; Leng, Y.; Shi, R.-F. Transcriptomic profiling provides molecular insights into hydrogen peroxide-induced adventitious rooting in mung bean seedlings. BMC Genom. 2017, 18, 188. [Google Scholar] [CrossRef]
- Dang, P.; Lamb, M.C.; Bowen, K.L.; Chen, C.Y. Identification of expressed R-genes associated with leaf spot diseases in cultivated peanut. Mol. Boil. Rep. 2018, 46, 225–239. [Google Scholar] [CrossRef]
- Takahashi, H.; Suzuki, M.; Natsuaki, K.; Shigyo, T.; Hino, K.; Teraoka, T.; Hosokawa, D.; Ehara, Y. Mapping the virus and host genes involved in the resistance response in cucumber mosaic virus-Infected Arabidopsis thaliana. Plant Cell Physiol. 2001, 42, 340–347. [Google Scholar] [CrossRef] [PubMed]
- Seo, Y.-S.; Rojas, M.R.; Lee, J.-Y.; Lee, S.-W.; Jeon, J.-S.; Ronald, P.; Lucas, W.J.; Gilbertson, R.L. A viral resistance gene from common bean functions across plant families and is up-regulated in a non-virus-specific manner. Proc. Natl. Acad. Sci. USA 2006, 103, 11856–11861. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Lee, J.-H.; Kang, W.-H.; Kim, J.; Huy, H.N.; Park, S.-W.; Son, E.-H.; Kwon, J.-K.; Kang, B.-C. Identification of Cucumber mosaic resistance 2 (cmr2) That Confers Resistance to a New Cucumber mosaic virus Isolate P1 (CMV-P1) in Pepper (Capsicum spp.). Front. Plant Sci. 2018, 9, 1106. [Google Scholar] [CrossRef] [PubMed]
- Guiu-Aragonés, C.; Monforte, A.; Saladié, M.; Corrêa, R.X.; Garcia-Mas, J.; Martin-Hernandez, A.M. The complex resistance to cucumber mosaic cucumovirus (CMV) in the melon accession PI161375 is governed by one gene and at least two quantitative trait loci. Mol. Breed. 2014, 34, 351–362. [Google Scholar] [CrossRef]
- Jian, W.; Zhang, D.-W.; Zhu, F.; Wang, S.-X.; Zhu, T.; Pu, X.-J.; Zheng, T.; Feng, H.; Lin, H. Nitrate reductase-dependent nitric oxide production is required for regulation alternative oxidase pathway involved in the resistance to Cucumber mosaic virus infection in Arabidopsis. Plant Growth Regul. 2015, 77, 99–107. [Google Scholar] [CrossRef]
- Shi, L.; Yang, Y.; Xie, Q.; Miao, H.; Bo, K.; Song, Z.; Wang, Y.; Xie, B.; Zhang, S.; Gu, X. Inheritance and QTL mapping of cucumber mosaic virus resistance in cucumber (Cucumis Sativus L.). PLoS ONE 2018, 13, e0200571. [Google Scholar] [CrossRef]
- Richards, K.; Jonard, G.; Jacquemond, M.; Lot, H. Nucleotide sequence of cucumber mosaic virus-associated RNA 5. Virology 1978, 89, 395–408. [Google Scholar] [CrossRef]
- Lu, J.; Du, Z.-X.; Kong, J.; Chen, L.-N.; Qiu, Y.-H.; Li, G.-F.; Meng, X.-H.; Zhu, S.-F. Transcriptome Analysis of Nicotiana tabacum Infected by Cucumber mosaic virus during Systemic Symptom Development. PLoS ONE 2012, 7, e43447. [Google Scholar] [CrossRef]
- Glasa, M.; Soltys, K.; Predajna, L.; Sihelská, N.; Nováková, S.; Šubr, Z.; Kraic, J.; Mihalik, D. Molecular and Biological Characterisation of Turnip mosaic virus Isolates Infecting Poppy (Papaver somniferum and P. rhoeas) in Slovakia. Viruses 2018, 10, 430. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Wu, S.; Bai, Y.; Sun, H.; Jiao, C.; Guo, S.; Zhao, K.; Blanca, J.; Zhang, Z.; Huang, S.; et al. Cucurbit Genomics Database (CuGenDB): A central portal for comparative and functional genomics of cucurbit crops. Nucleic Acids Res. 2018, 47, D1128–D1136. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Gene Ontology Consortium. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004, 32, 258D–261D. [Google Scholar] [CrossRef] [PubMed]
- Supek, F.; Bosnjak, M.; Skunca, N.; Šmuc, T. REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef]
- Koster, J.; Rahmann, S. Snakemake—A scalable bioinformatics workflow engine. Bioinformatics 2012, 28, 2520–2522. [Google Scholar] [CrossRef]
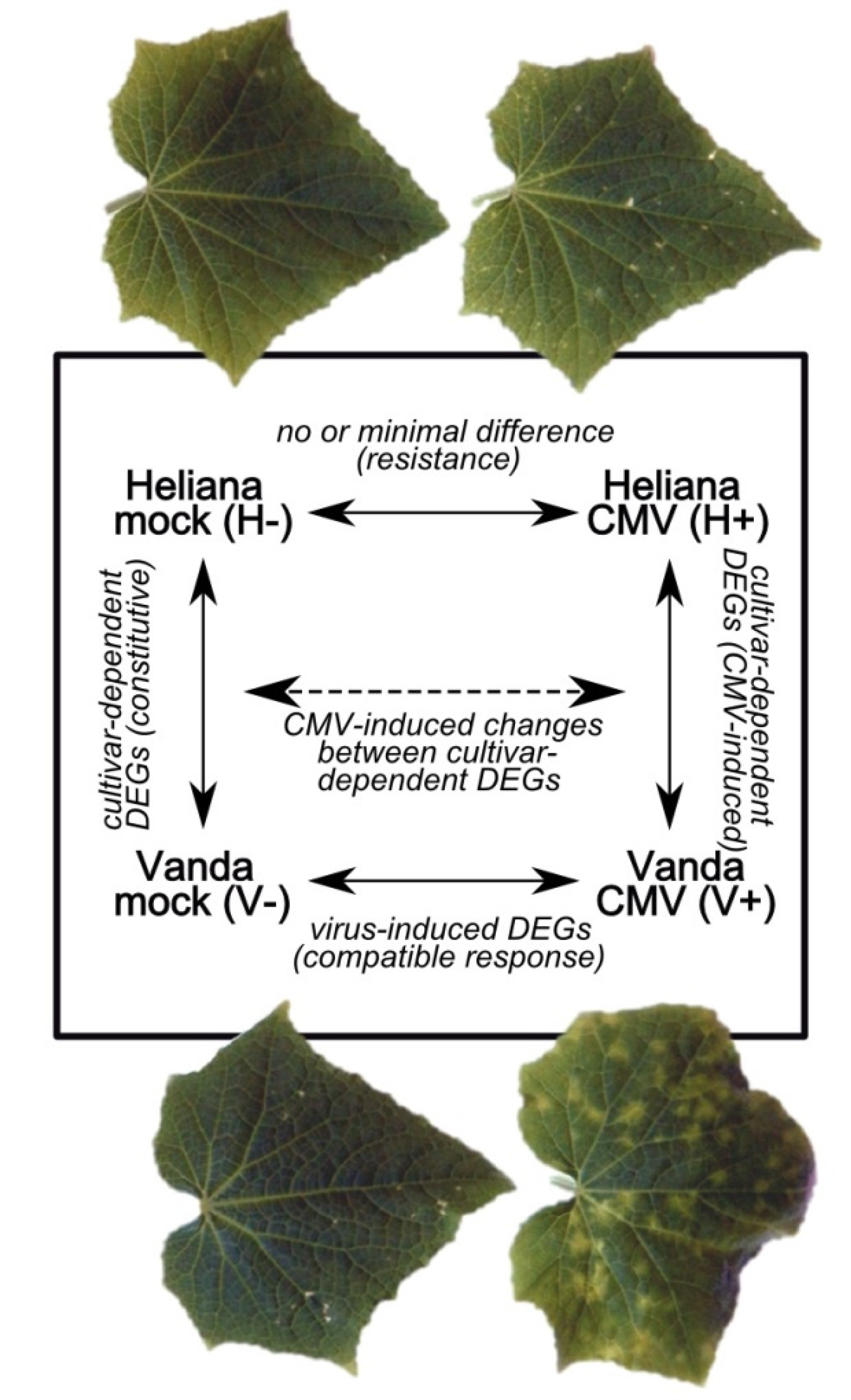


| Comparison 1 | Significant DEGs | Down-Regulated 2 | Up-Regulated 2 |
|---|---|---|---|
| H-/H+ | 9 (1) | 8 (1) | 1 (0) |
| V-/V+ | 3006 (359) | 1379 (168) | 1627 (191) |
| H-/V- | 617 (53) | 427 (32) | 190 (21) |
| H+/V+ | 2456 (219) | 1112 (99) | 1344 (120) |
| ID | Function | Fold Change | |
|---|---|---|---|
| H-/V- | V-/V+ | ||
| CsGy7G013580.1 | sodium/metabolite cotransporter BASS3, chloroplastic | −1.68 | 1.58 |
| CsGy7G019210.1 | pentatricopeptide repeat-containing protein At1g15510, chloroplastic | −1.62 | 1.71 |
| CsGy6G013240.1 | ubiquinone biosynthesis O-methyltransferase, mitochondrial | 1.65 | −2.06 |
| CsGy4G018480.1 | extradiol ring-cleavage dioxygenase | 1.73 | −1.53 |
| CsGy2G023650.1 | U-box domain-containing protein 12-like | 1.83 | −2.62 |
| CsGy6G033990.1 | epoxide hydrolase | 1.93 | −1.74 |
| CsGy1G011590.1 | sugar transporter, putative | 2.29 | −1.58 |
| CsGy2G004500.1 | 26S proteasome regulatory subunit 6A homolog A | 2.48 | −2.11 |
| CsGy7G018670.1 | receptor like protein 9 | 2.57 | −1.83 |
| CsGy7G019140.1 | serine/threonine-protein kinase receptor | 3.35 | −1.87 |
| CsGy4G009730.1 | receptor-like protein kinase FERONIA | 4.02 | −1.88 |
| CsGy1G016880.1 | sugar transporter, putative | 4.88 | −1.83 |
| CsGy1G017330.1 | sterile alpha motif, type 2 | 5.19 | −2.82 |
| CsGy4G004000.1 | derlin | 6.08 | −1.83 |
| CsGy4G018180.1 | sigma factor binding protein 2, chloroplastic | 9.84 | −2.91 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Šubr, Z.; Predajňa, L.; Šoltys, K.; Bokor, B.; Budiš, J.; Glasa, M. Comparative Transcriptome Analysis of Two Cucumber Cultivars with Different Sensitivity to Cucumber Mosaic Virus Infection. Pathogens 2020, 9, 145. https://doi.org/10.3390/pathogens9020145
Šubr Z, Predajňa L, Šoltys K, Bokor B, Budiš J, Glasa M. Comparative Transcriptome Analysis of Two Cucumber Cultivars with Different Sensitivity to Cucumber Mosaic Virus Infection. Pathogens. 2020; 9(2):145. https://doi.org/10.3390/pathogens9020145
Chicago/Turabian StyleŠubr, Zdeno, Lukáš Predajňa, Katarína Šoltys, Boris Bokor, Jaroslav Budiš, and Miroslav Glasa. 2020. "Comparative Transcriptome Analysis of Two Cucumber Cultivars with Different Sensitivity to Cucumber Mosaic Virus Infection" Pathogens 9, no. 2: 145. https://doi.org/10.3390/pathogens9020145
APA StyleŠubr, Z., Predajňa, L., Šoltys, K., Bokor, B., Budiš, J., & Glasa, M. (2020). Comparative Transcriptome Analysis of Two Cucumber Cultivars with Different Sensitivity to Cucumber Mosaic Virus Infection. Pathogens, 9(2), 145. https://doi.org/10.3390/pathogens9020145





