Molecular Detection and Genetic Characteristics of Equine Herpesvirus in Korea
Abstract
1. Introduction
2. Results
2.1. PCR Detection
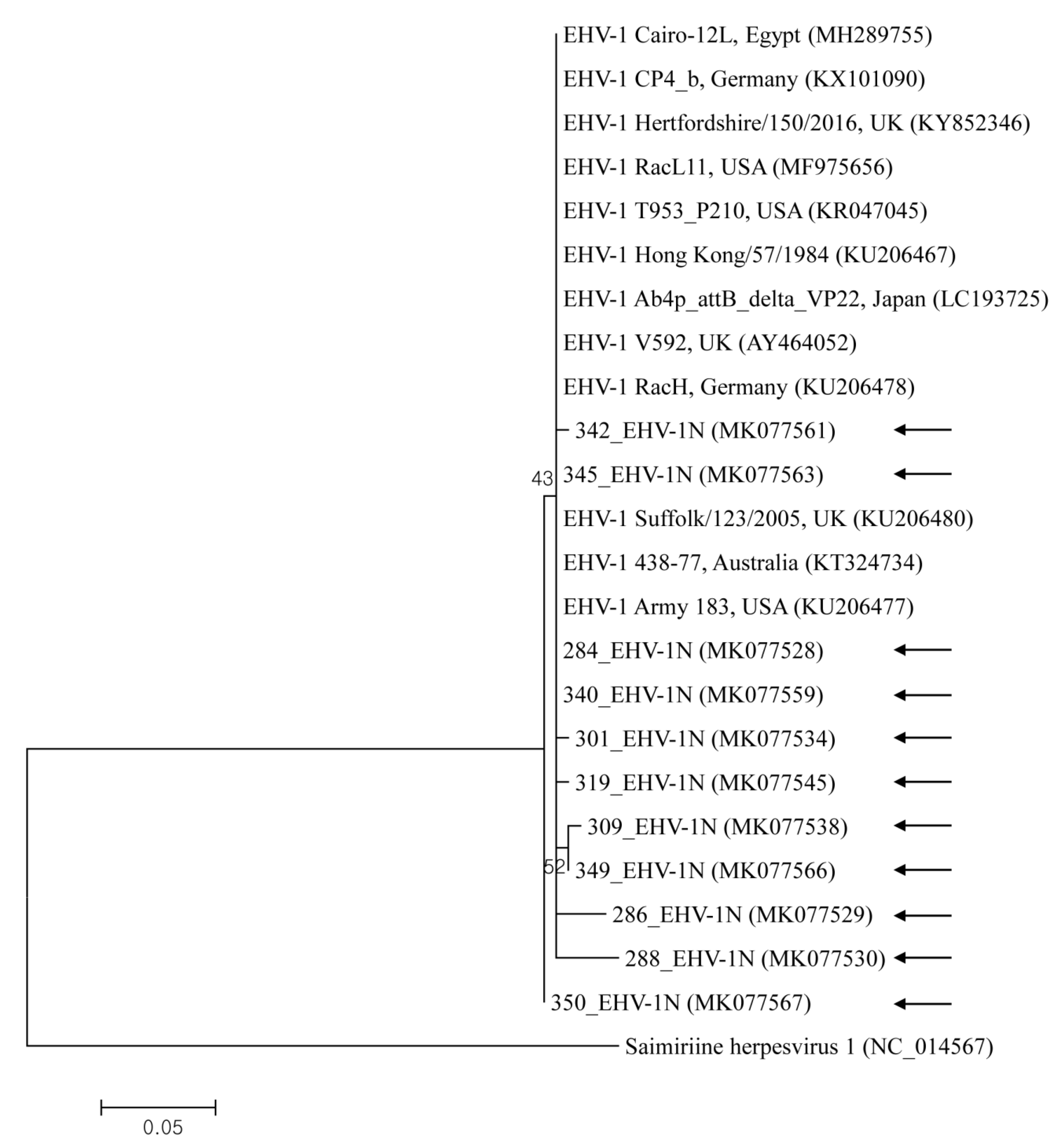
2.2. Molecular and Phylogenetic Analysis
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
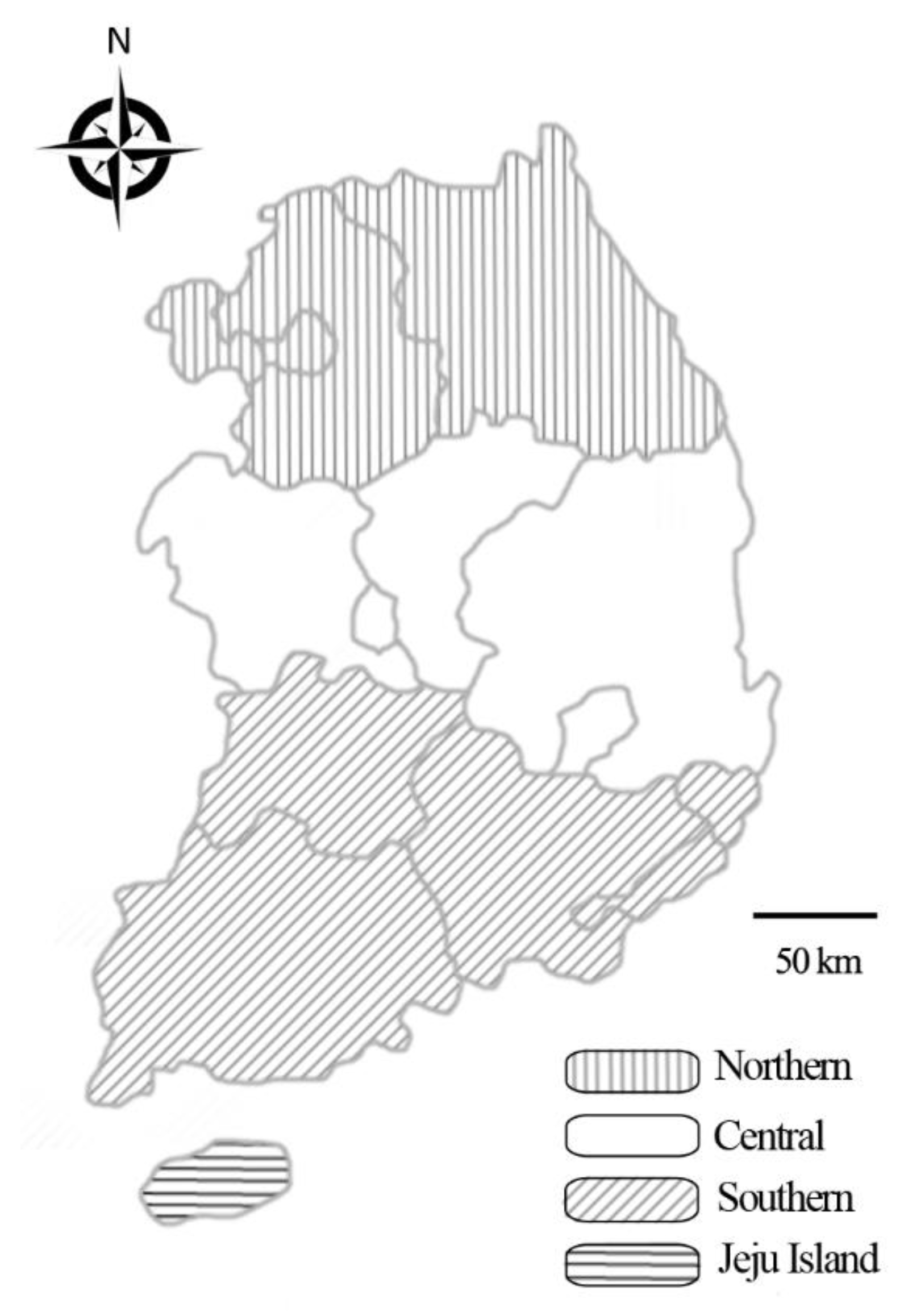
4.2. Sample Size Determination and Sample Collection
4.3. DNA Extraction and PCR
4.4. Cloning
4.5. Sequencing and Phylogeny
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dynon, K.; Black, W.D.; Ficorilli, N.; Hartley, C.A.; Studdert, M.J. Detection of viruses in nasal swab samples from horses with acute, febrile, respiratory disease using virus isolation, polymerase chain reaction and serology. Aust. Vet. J. 2007, 85, 46–50. [Google Scholar] [CrossRef]
- Davison, A.J. Herpesvirus systematics. Vet. Microbiol. 2010, 143, 52–69. [Google Scholar] [CrossRef] [PubMed]
- Allen, G.P.; Kydd, J.H.; Slater, J.D.; Smith, K.C. Equid herpesvirus 1 and equid herpesvirus 4 infections. In Infectious Disease of Livestock; Coetzer, J.A.W., Tustin, R.C., Eds.; Oxford Press: Cape Town, UK, 2004; pp. 829–859. [Google Scholar]
- Lunn, D.P.; Davis-Poynter, N.; Flaminio, M.J.; Horohov, D.W.; Osterrieder, K.; Pusterla, N.; Townsend, H.G. Equine herpesvirus-1 consensus statement. J. Vet. Intern. Med. 2009, 23, 450–461. [Google Scholar] [CrossRef] [PubMed]
- Hussey, G.S.; Goehring, L.S.; Lunn, D.P.; Hussey, S.B.; Huang, T.; Osterrieder, N.; Powell, C.; Hand, J.; Holz, C.; Slater, J. Experimental infection with equine herpesvirus type 1 (EHV-1) induces chorioretinal lesions. Vet. Res. 2013, 44, 118. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Raidal, S.L.; Pizzirani, A.; Wilcox, G.E. Detection of respiratory herpesviruses in foals and adult horses determined by nested multiplex PCR. Vet. Microbiol. 2007, 121, 18–28. [Google Scholar] [CrossRef] [PubMed]
- Fortier, G.; van Erck, E.; Pronost, S.; Lekeux, P.; Thiry, E. Equine gammaherpesviruses: Pathogenesis, epidemiology and diagnosis. Vet. J. 2010, 186, 148–156. [Google Scholar] [CrossRef]
- Bell, S.A.; Balasuriya, U.B.; Gardner, I.A.; Barry, P.A.; Wilson, W.D.; Ferraro, G.L.; MacLachlan, N.J. Temporal detection of equine herpesvirus infections of a cohort of mares and their foals. Vet. Microbiol. 2006, 116, 249–257. [Google Scholar] [CrossRef]
- Craig, M.I.; Barrandeguy, M.E.; Fernández, F.M. Equine herpesvirus 2 (EHV-2) infection in thoroughbred horses in Argentina. BMC. Vet. Res. 2005, 1, 9. [Google Scholar] [CrossRef]
- Diallo, I.S.; Hewitson, G.R.; de Jong, A.; Kelly, M.A.; Wright, D.J.; Corney, B.G.; Rodwell, B.J. Equine herpesvirus infections in yearlings in South-East Queensland. Arch. Virol. 2008, 153, 1643–1649. [Google Scholar] [CrossRef]
- Bak, U.B.; Lim, C.H.; Kang, B.H.; Lee, S.Y. A Pathological Survey on Equine Viral Rhinopneumonitis Occurred in Korea. Korean J. Vet. Res. 1981, 21, 11–23. [Google Scholar]
- Ko, S.; Kang, J.G.; Yeh, J.Y.; Moon, J.S.; Choi, G.C.; Won, S.; Chae, J.S. First report on molecular detection of equine upper respiratory infectious viruses in Republic of Korea. J. Equine Vet. Sci. 2013, 33, 628–636. [Google Scholar] [CrossRef]
- Lee, S.K.; Lee, I. The molecular detection of equine herpesviruses 2 and 5 in genital swabs from clinically normal thoroughbred mares in South Korea. J. Equine Vet. Sci. 2019, 79, 68–72. [Google Scholar] [CrossRef] [PubMed]
- Ministry of Agriculture, Food and Rural Affairs, Korea. Report of “A Fact Finding Survey of Horse Industry in South Korea during 2017”. Available online: http://www.mafra.go.kr/mafra/293/subview.do?enc=Zm5jdDF8QEB8JTJGYmJzJTJGbWFmcmElMkY2OCUyRjMxNjk2NiUyRmFydGNsVmlldy5kbyUzRg%3D%3D/ (accessed on 1 December 2018).
- Reed, S.M.; Toribio, R.E. Equine herpesvirus 1 and 4. Vet. Clin. N. Am. Equine Pract. 2004, 20, 631–642. [Google Scholar] [CrossRef] [PubMed]
- Dunowska, M.; Wilks, C.R.; Studdert, M.J.; Meers, J. Viruses associated with outbreaks of equine respiratory disease in New Zealand. N. Z. Vet. J. 2002, 50, 132–139. [Google Scholar] [CrossRef]
- Allen, G.P.; Murray, J.T. Equid herpesvirus 2 and equid herpesvirus 5 infections. In Infectious Disease of Livestock; Coetzer, J.A.W., Thomson, G., Tustin, R.C., Eds.; Oxford Press: Cape Town, UK, 2004; pp. 860–868. [Google Scholar]
- Ataseven, V.S.; Bilge-Dagalp, S.; Oguzoglu, T.C.; Karapinar, Z.; Güzel, M.; Tan, M.T. Detection and sequence analysis of equine Gammaherpesviruses from horses with respiratory tract disease in Turkey. Transbound. Emerg. Dis. 2010, 57, 271–276. [Google Scholar] [CrossRef]
- Stasiak, K.; Dunowska, M.; Rola, J. Prevalence and sequence analysis of equid herpesviruses from the respiratory tract of Polish horses. Virol. J. 2018, 15, 106. [Google Scholar] [CrossRef]
- Negussie, H.; Gizaw, D.; Tesfaw, L.; Li, Y.; Oguma, K.; Sentsui, H.; Tessema, T.S.; Nauwynck, H.J. Detection of equine herpesvirus (EHV) −1, −2, −4 and −5 in Ethiopian equids with and without respiratory problems and genetic characterization of EHV-2 and EHV-5 strains. Transbound. Emerg. Dis. 2017, 64, 1970–1978. [Google Scholar] [CrossRef]
- Ann, N.; Malik, M.; Carlos, R.; Arne, L.; Vilmos, P.; Duncan, H.; Sándor, B. Prevalence of equine herpesvirus types 2 and 5 in horse populations by using type-specific PCR assays. Vet. Res. 2002, 33, 251–259. [Google Scholar]
- Torfason, E.G.; Thorsteinsdóttir, L.; Torsteinsdóttir, S.; Svansson, V. Study of equid herpesviruses 2 and 5 in Iceland with a type-specific polymerase chain reaction. Res. Vet. Sci. 2008, 85, 605–611. [Google Scholar] [CrossRef]
- Rushton, J.; Tichy, A.; Brem, G.; Druml, T.; Nell, B. Ophthalmological findings in a closed herd of Lipizzaners. Equine Vet. J. 2013, 45, 209–213. [Google Scholar] [CrossRef]
- Back, H.; Ullman, K.; Treiberg Berndtsson, L.; Riihimäki, M.; Penell, J.; Ståhl, K.; Valarcher, J.F.; Pringle, J. Viral load of equine herpesviruses 2 and 5 in nasal swabs of actively racing Standardbred trotters: Temporal relationship of shedding to clinical findings and poor performance. Vet. Microbiol. 2015, 179, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Thrusfield, M. Veterinary Epidemiology, 3rd ed.; Blackwell Publishing: Oxford, UK, 2005. [Google Scholar]
- Kirisawa, R.; Endo, A.; Iwai, H.; Kawakami, Y. Detection and identification of equine herpesvirus −1 and −4 by polymerase chain reaction. Vet. Microbiol. 1993, 36, 57–67. [Google Scholar] [CrossRef]
- Holloway, S.A.; Lindquester, G.J.; Studdert, M.J.; Drummer, H.E. Identification, sequence analysis and characterisation of equine herpesvirus 5 glycoprotein B. Arch. Virol. 1999, 144, 287–307. [Google Scholar] [CrossRef] [PubMed]

| Sample | Category | No. Tested | EHV-2 | EHV-5 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. Positive (%) | RR | 95% CI | p-Value | No. Positive (%) | RR | 95% CI | p-Value | |||
| Blood | Sex | |||||||||
| Female | 547 | 234 (42.8) | 1.1 | 0.9–1.2 | 0.456 | 129 (23.6) | 1.2 | 1.0–1.6 | 0.1051 | |
| Male | 119 | 49 (41.2) | 1.0 | 0.8–1.2 | 0.921 | 25 (21.0) | 1.0 | 0.7–1.4 | 0.9056 | |
| Castrated | 260 | 103 (39.6) | 0.9 | 0.8–1.1 | 0.4585 | 47 (18.1) | 0.8 | 0.6–1.1 | 0.1101 | |
| Age (years) | ||||||||||
| <5 | 223 | 97 (34.5) | 1.1 | 0.9–1.3 | 0.5339 | 35 (15.7) | 0.7 | 0.5–0.9 | 0.0119 * | |
| 5–10 | 349 | 152 (43.6) | 1.1 | 0.9–1.3 | 0.3723 | 81 (23.2) | 1.1 | 0.9–1.4 | 0.4111 | |
| >10 | 354 | 137 (38.7) | 0.9 | 0.8–1.0 | 0.1505 | 85 (24.0) | 1.2 | 0.9–1.5 | 0.1899 | |
| Region | ||||||||||
| Northern | 248 | 103 (41.5) | 1.0 | 0.8–1.2 | 1 | 54 (21.8) | 1.0 | 0.8–1.3 | 1 | |
| Central | 114 | 31 (27.2) | 0.6 | 0.5–0.8 | 0.0008 * | 11 (9.6) | 0.4 | 0.2–0.7 | 0.0006 * | |
| Southern | 168 | 78 (46.4) | 1.1 | 1.0–1.4 | 0.1945 | 37 (22.0) | 1.0 | 0.7–1.4 | 0.9178 | |
| Jeju Island | 396 | 174 (43.9) | 1.1 | 0.9–1.3 | 0.2522 | 99 (22.7) | 1.3 | 1.0–1.7 | 0.0366 | |
| Breed | ||||||||||
| Thoroughbred | 612 | 256 (41.8) | 1.0 | 0.9–1.2 | 0.9439 | 125 (20.4) | 0.8 | 0.7–1.1 | 0.2066 | |
| Warmblood | 61 | 26 (42.6) | 1.0 | 0.8–1.4 | 0.8937 | 13 (21.3) | 1.0 | 0.6–1.6 | 1 | |
| Native Korean pony | 84 | 41 (48.8) | 1.2 | 0.9–1.5 | 0.1661 | 31 (36.9) | 1.8 | 1.3–2.5 | 0.0008 * | |
| Mixed | 169 | 63 (37.3) | 0.9 | 0.7–1.1 | 0.227 | 32 (18.9) | 0.8 | 0.6–1.2 | 0.3549 | |
| Activity | ||||||||||
| Race | 10 | 4 (40.0) | 1.0 | 0.4–2.1 | 1 | 2 (20.0) | 0.9 | 0.3–3.2 | 1 | |
| Leisure | 613 | 245 (40.0) | 0.9 | 0.8–1.0 | 0.1398 | 122 (19.9) | 0.8 | 0.6–1.0 | 0.0643 | |
| Breeding | 303 | 137 (45.2) | 1.1 | 1.0–1.3 | 0.1361 | 77 (25.4) | 1.3 | 1.0–1.6 | 0.0617 | |
| Sub-total | 926 | 386 (41.7) | 38.5–44.9 | 201 (21.7) | 19.1–24.4 | |||||
| Lung tissue | 187 | 25 (13.4) | 8.5–18.2 | 35 (18.7) | 13.1–24.3 | |||||
| Category | No. Tested | EHV-1 | EHV-2 | EHV-5 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. Positive (%) | RR | 95% CI | p-Value | No. Positive (%) | RR | 95% CI | p-Value | No. Positive (%) | RR | 95% CI | p-Value | ||
| Sex | |||||||||||||
| Female | 130 | 20 (15.4) | 1.5 | 0.9–2.6 | 0.1832 | 10 (7.7) | 1.8 | 0.85–4.1 | 0.2343 | 19 (14.6) | 1.7 | 0.9–3.0 | 0.0821 |
| Male | 128 | 10 (7.8) | 0.6 | 0.3–1.1 | 0.0949 | 3 (2.3) | 0.3 | 0.1–1.1 | 0.0602 | 5 (3.9) | 0.3 | 0.1–0.7 | 0.0024 * |
| Castrated | 126 | 16 (12.7) | 1.1 | 0.6–1.9 | 0.7411 | 8 (6.3) | 1.3 | 0.5–3.0 | 0.6354 | 17 (13.5) | 1.5 | 0.8–2.6 | 0.2214 |
| Age (years) | |||||||||||||
| <5 | 192 | 14 (7.3) | 0.4 | 0.2–0.8 | 0.007 * | 4 (2.1) | 0.2 | 0.1–0.7 | 0.0058 * | 7 (3.6) | 0.2 | 0.1–0.4 | < 0.0001 * |
| 5–10 | 127 | 16 (12.6) | 1.1 | 0.6–1.9 | 0.8675 | 7 (5.5) | 1.0 | 0.4–2.5 | 1 | 15 (11.8) | 1.1 | 0.6–2.0 | 0.8636 |
| >10 | 65 | 16 (24.6) | 2.6 | 1.5–4.5 | 0.0014 * | 10 (15.4) | 4.5 | 2.0–10.1 | 0.0008 * | 21 (32.3) | 4.7 | 2.7–8.0 | < 0.0001 * |
| Region | |||||||||||||
| Northern | 207 | 1 (0.5) | 0.01 | 0–0.1 | < 0.0001 * | 10 (4.8) | 0.8 | 0.3–1.8 | 0.6542 | 15 (7.2) | 0.5 | 0.3–0.8 | 0.0092 * |
| Central | 31 | 8 (25.8) | 2.4 | 1.2–4.7 | 0.0212 | 2 (6.5) | 1.2 | 0.3–4.9 | 0.6825 | 5 (16.1) | 1.5 | 0.6–3.5 | 0.3706 |
| Southern | 86 | 27 (31.4) | 4.9 | 2.9–8.4 | < 0.0001 * | 6 (7.0) | 1.4 | 0.6–3.5 | 0.5893 | 15 (17.4) | 1.9 | 1.0–3.3 | 0.0507 |
| Jeju Island | 60 | 10 (16.7) | 1.5 | 0.8–2.9 | 0.2766 | 3 (5.0) | 0.9 | 0.3–3.0 | 1 | 8 (13.3) | 1.2 | 0.6–2.5 | 0.5122 |
| Activity | |||||||||||||
| Race | 262 | 15 (5.7) | 0.2 | 0.1–0.4 | < 0.0001 * | 3 (1.1) | 0.1 | 0.02–0.3 | < 0.0001 * | 4 (1.5) | 0.05 | 0.02–0.1 | < 0.0001 * |
| Leisure | 98 | 21 (21.4) | 2.5 | 1.4–4.2 | 0.0018 * | 14 (14.3) | 5.8 | 2.4–14.0 | < 0.0001 * | 30 (30.6) | 6.7 | 3.7–12.4 | < 0.0001 * |
| Breeding | 24 | 10 (41.7) | 4.2 | 2.4–7.3 | 0.0001 * | 4 (16.7) | 3.5 | 1.3–9.7 | 0.0343 | 9 (37.5) | 4.0 | 2.2–7.3 | 0.0004 * |
| Total | 384 | 46 (12.0) | 8.7–15.2 | 21 (5.5) | 3.2–7.7 | 43 (11.2) | 8.0–14.4 | ||||||
| Sample Type | Classified Infection | Virus | No. Detected (%) |
|---|---|---|---|
| Blood | Total number positive | EHV-2 | 386 (41.7) |
| EHV-5 | 201 (21.7) | ||
| Unique detection | EHV-2 only | 279 (30.1) | |
| EHV-5 only | 94 (10.2) | ||
| Double detection | EHV-2, EHV-5 | 107 (11.6) | |
| Lung | Total number positive | EHV-2 | 25 (13.4) |
| EHV-5 | 35 (18.7) | ||
| Unique detection | EHV-2 only | 13 (7.0) | |
| EHV-5 only | 23 (12.3) | ||
| Double detection | EHV-2, EHV-5 | 12 (6.4) | |
| Nasal swab | Total number positive | EHV-1 | 46 (12.0) |
| EHV-2 | 21 (5.5) | ||
| EHV-5 | 43 (11.2) | ||
| Unique detection | EHV-1 only | 28 (7.3) | |
| EHV-2 only | 3 (0.8) | ||
| EHV-5 only | 14 (3.7) | ||
| Double detection | EHV-1, EHV-2 | 5 (1.3) | |
| EHV-1, EHV-5 | 16 (4.2) | ||
| EHV-2, EHV-5 | 13 (3.4) | ||
| Triple detection | EHV-1, EHV-2, EHV-5 | 3 (0.8) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seo, M.-G.; Ouh, I.-O.; Lee, S.K.; Lee, J.-S.; Kwon, O.-D.; Kwak, D. Molecular Detection and Genetic Characteristics of Equine Herpesvirus in Korea. Pathogens 2020, 9, 110. https://doi.org/10.3390/pathogens9020110
Seo M-G, Ouh I-O, Lee SK, Lee J-S, Kwon O-D, Kwak D. Molecular Detection and Genetic Characteristics of Equine Herpesvirus in Korea. Pathogens. 2020; 9(2):110. https://doi.org/10.3390/pathogens9020110
Chicago/Turabian StyleSeo, Min-Goo, In-Ohk Ouh, Sang Kyu Lee, Jong-Seok Lee, Oh-Deog Kwon, and Dongmi Kwak. 2020. "Molecular Detection and Genetic Characteristics of Equine Herpesvirus in Korea" Pathogens 9, no. 2: 110. https://doi.org/10.3390/pathogens9020110
APA StyleSeo, M.-G., Ouh, I.-O., Lee, S. K., Lee, J.-S., Kwon, O.-D., & Kwak, D. (2020). Molecular Detection and Genetic Characteristics of Equine Herpesvirus in Korea. Pathogens, 9(2), 110. https://doi.org/10.3390/pathogens9020110





