Genetic Diversity of Cryptosporidium in Bactrian Camels (Camelus bactrianus) in Xinjiang, Northwestern China
Abstract
1. Introduction
2. Results
2.1. Occurrence of Cryptosporidium
2.2. Cryptosporidium Species and Subtypes
3. Discussion
4. Materials and Methods
4.1. Ethics Approval
4.2. Sample Collection
4.3. DNA Extraction and PCR Amplification
4.4. Sequencing and Phylogenetic Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Xiao, L.; Feng, Y. Molecular epidemiologic tools for waterborne pathogens Cryptosporidium spp. and Giardia duodenalis. Food Waterborne Parasitol. 2017, 8–9, 14–32. [Google Scholar] [CrossRef] [PubMed]
- Efstratiou, A.; Ongerth, J.E.; Karanis, P. Waterborne transmission of protozoan parasites: Review of worldwide outbreaks—An update 2011–2016. Water Res. 2017, 114, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.J.; Li, J.Q.; Chen, Y.C.; Zhang, L.X.; Xiao, L.H. Widespread occurrence of Cryptosporidium infections in patients with HIV/AIDS: Epidemiology, clinical feature, diagnosis, and therapy. Acta Trop. 2018, 187, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Shaik, J.S.; Grigg, M.E. Genomics and molecular epidemiology of Cryptosporidium species. Acta Trop. 2018, 184, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Liang, N.; Wu, Y.; Sun, M.; Chang, Y.; Lin, X.; Yu, L.; Hu, S.; Zhang, X.; Zheng, S.; Cui, Z.; et al. Molecular epidemiology of Cryptosporidium spp. in dairy cattle in Guangdong Province, South China. Parasitology 2019, 146, 28–32. [Google Scholar] [CrossRef]
- Ryan, U.; Fayer, R.; Xiao, L. Cryptosporidium species in humans and animals: Current understanding and research needs. Parasitology 2014, 141, 1667–1685. [Google Scholar] [CrossRef]
- Feng, Y.; Ryan, U.M.; Xiao, L. Genetic diversity and population structure of Cryptosporidium. Trends Parasitol. 2018, 34, 997–1011. [Google Scholar] [CrossRef]
- Sazmand, A.; Joachim, A. Parasitic diseases of camels in Iran (1931–2017)—A literature review. Parasite 2017, 24, 21. [Google Scholar] [CrossRef]
- Sazmand, A.; Rasooli, A.; Nouri, M.; Hamidinejat, H.; Hekmatimoghaddam, S. Prevalence of Cryptosporidium spp. in Camels and involved people in Yazd Province, Iran. Iran. J. Parasitol. 2012, 7, 80–84. [Google Scholar]
- Zhang, Q.; Zhang, Z.; Ai, S.; Wang, X.; Zhang, R.; Duan, Z. Cryptosporidium spp., Enterocytozoon bieneusi, and Giardia duodenalis from animal sources in the Qinghai-Tibetan Plateau Area (QTPA) in China. Comp. Immunol. Microbiol. Infect. Dis. 2019, 67, 101346. [Google Scholar] [CrossRef]
- Baroudi, D.; Zhang, H.; Amer, S.; Khelef, D.; Roellig, D.M.; Wang, Y.; Feng, Y.; Xiao, L. Divergent Cryptosporidium parvum subtype and Enterocytozoon bieneusi genotypes in dromedary camels in Algeria. Parasitol. Res. 2018, 117, 905–910. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Wang, X.; Zhou, C.; Li, P.; Xu, Q.; Zhao, C.; Liu, W.; Xu, W. Investigation on Cryptosporidium infections in wild animals in zoo in Anhui Province. J. Zoo Wildl. Med. 2016, 47, 846–854. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhou, X.; Zhong, Z.; Deng, J.; Chen, W.; Cao, S.; Fu, H.; Zuo, Z.; Hu, Y.; Peng, G. Multilocus genotype and subtype analysis of Cryptosporidium andersoni derived from a Bactrian camel (Camelus bactrianus) in China. Parasitol. Res. 2014, 113, 2129–2136. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Zhang, L.; Ning, C.; Feng, Y.; Jian, F.; Xiao, L.; Lu, B.; Ai, W.; Dong, H. Multilocus phylogenetic analysis of Cryptosporidium andersoni (Apicomplexa) isolated from a bactrian camel (Camelus bactrianus) in China. Parasitol. Res. 2008, 102, 915–920. [Google Scholar] [CrossRef]
- Zahedi, A.; Lee, G.K.C.; Greay, T.L.; Walsh, A.L.; Blignaut, D.J.C.; Ryan, U.M. First report of Cryptosporidium parvum in a dromedary camel calf from Western Australia. Acta Parasitol. 2018, 63, 422–427. [Google Scholar] [CrossRef]
- Xiao, L.; Escalante, L.; Yang, C.; Sulaiman, I.; Escalante, A.A.; Montali, R.J.; Fayer, R.; Lal, A.A. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl. Environ. Microbiol. 1999, 65, 1578–1583. [Google Scholar] [CrossRef]
- Kvác, M.; Sak, B.; Kvetonová, D.; Ditrich, O.; Hofmannová, L.; Modrý, D.; Vítovec, J.; Xiao, L. Infectivity, pathogenicity, and genetic characteristics of mammalian gastric Cryptosporidium spp. in domestic ruminants. Vet. Parasitol. 2008, 153, 363–367. [Google Scholar] [CrossRef]
- Morgan, U.M.; Xiao, L.; Monis, P.; Sulaiman, I.; Pavlasek, I.; Blagburn, B.; Olson, M.; Upton, S.J.; Khramtsov, N.V.; Lal, A. Molecular and phylogenetic analysis of Cryptosporidium muris from various hosts. Parasitology 2000, 120, 457–464. [Google Scholar] [CrossRef]
- El-Alfy, E.S.; Abu-Elwafa, S.; Abbas, I.; Al-Araby, M.; Al-Kappany, Y.; Umeda, K.; Nishikawa, Y. Molecular screening approach to identify protozoan and trichostrongylid parasites infecting one-humped camels (Camelus dromedarius). Acta Trop. 2019, 197, 105060. [Google Scholar] [CrossRef]
- Abdelwahab, A.; Abdelmaogood, S. Identification of Cryptosporidium species infecting camels (Camelus dromedarius) in Egypt. J. Am. Sci. 2011, 7, 609–612. [Google Scholar]
- Sazmand, A.; Joachim, A.; Otranto, D. Zoonotic parasites of dromedary camels: So important, so ignored. Parasites Vectors 2019, 12, 610. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Zimmerman, D.; Deem, L. A review of zoonotic pathogens of Dromedary Camels. Ecohealth 2019, 16, 356–377. [Google Scholar] [CrossRef] [PubMed]
- Leoni, F.; Amar, C.; Nichols, G.; Pedraza-Díaz, S.; McLauchlin, J. Genetic analysis of Cryptosporidium from 2414 humans with diarrhoea in England between 1985 and 2000. J. Med. Microbiol. 2006, 55, 703–707. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Ren, J.; Yuan, Z.; Liu, A.; Zhao, H.; Liu, H.; Chu, L.; Pan, W.; Cao, J.; Lin, Y.; et al. Cryptosporidium andersoni as a novel predominant Cryptosporidium species in outpatients with diarrhea in Jiangsu Province, China. BMC Infect. Dis. 2014, 14, 555. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Wang, R.; Huang, J.; Wang, H.; Zhao, J.; Luo, N.; Li, J.; Zhang, Z.; Zhang, L. Cryptosporidiosis caused by Cryptosporidium parvum subtype IIdA15G1 at a dairy farm in Northwestern China. Parasites Vectors 2014, 7, 529. [Google Scholar] [CrossRef] [PubMed]
- Qi, M.; Wang, H.; Jing, B.; Wang, D.; Wang, R.; Zhang, L. Occurrence and molecular identification of Cryptosporidium spp. in dairy calves in Xinjiang, Northwestern China. Vet. Parasitol. 2015, 212, 404–407. [Google Scholar] [CrossRef]
- Thompson, R.C.A.; Ash, A. Molecular epidemiology of Giardia and Cryptosporidium infections. Infect. Genet. Evol. 2016, 40, 315–323. [Google Scholar] [CrossRef]
- Jian, F.; Liu, A.; Wang, R.; Zhang, S.; Qi, M.; Zhao, W.; Shi, Y.; Wang, J.; Wei, J.; Zhang, L.; et al. Common occurrence of Cryptosporidium hominis in horses and donkeys. Infect. Genet. Evol. 2016, 43, 261–266. [Google Scholar] [CrossRef]
- Laatamna, A.E.; Wagnerová, P.; Sak, B.; Květoňová, D.; Xiao, L.; Rost, M.; McEvoy, J.; Saadi, A.R.; Aissi, M.; Kváč, M. Microsporidia and Cryptosporidium in horses and donkeys in Algeria: Detection of a novel Cryptosporidium hominis subtype family (Ik) in a horse. Vet. Parasitol. 2015, 208, 135–142. [Google Scholar] [CrossRef]
- Lebbad, M.; Winiecka-Krusnell, J.; Insulander, M.; Beser, J. Molecular characterization and epidemiological investigation of Cryptosporidium hominis IkA18G1 and C. hominis monkey genotype IiA17, two unusual subtypes diagnosed in Swedish patients. Exp. Parasitol. 2018, 188, 50–57. [Google Scholar] [CrossRef]
- Liu, X.; Xie, N.; Li, W.; Zhou, Z.; Zhong, Z.; Shen, L.; Cao, S.; Yu, X.; Hu, Y.; Chen, W.; et al. Emergence of Cryptosporidium hominis monkey genotype II and novel subtype family Ik in the Squirrel Monkey (Saimiri sciureus) in China. PLoS ONE 2015, 10, e0141450. [Google Scholar] [CrossRef]
- Li, N.; Xiao, L.; Alderisio, K.; Elwin, K.; Cebelinski, E.; Chalmers, R.; Santin, M.; Fayer, R.; Kvac, M.; Ryan, U. Subtyping Cryptosporidium ubiquitum,a zoonotic pathogen emerging in humans. Emerg. Infect. Dis. 2014, 20, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Zhang, Z.; Zhang, Y.; Yang, Y.; Zhao, J.; Wang, R.; Jian, F.; Ning, C.; Zhang, W.; Zhang, L. Prevalence and molecular characterization of Cryptosporidium spp. and Giardia duodenalis in deer in Henan and Jilin, China. Parasites Vectors 2018, 11, 239. [Google Scholar] [CrossRef]
- Majeed, Q.A.H.; El-Azazy, O.M.E.; Abdou, N.M.I.; Al-Aal, Z.A.; El-Kabbany, A.I.; Tahrani, L.M.A.; AlAzemi, M.S.; Wang, Y.; Feng, Y.; Xiao, L. Epidemiological observations on cryptosporidiosis and molecular characterization of Cryptosporidium spp. in sheep and goats in Kuwait. Parasitol. Res. 2018, 117, 1631–1636. [Google Scholar] [CrossRef] [PubMed]
- Kváč, M.; Vlnatá, G.; Ježková, J.; Horčičková, M.; Konečný, R.; Hlásková, L.; McEvoy, J.; Sak, B. Cryptosporidium occultus sp. n. (Apicomplexa: Cryptosporidiidae) in rats. Eur. J. Protistol. 2018, 63, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Zhang, Z.; Hu, S.; Zhao, W.; Zhao, J.; Kváč, M.; Guo, Y.; Li, N.; Feng, Y.; Xiao, L. Common occurrence of divergent Cryptosporidium species and Cryptosporidium parvum subtypes in farmed bamboo rats (Rhizomys sinensis). Parasites Vectors 2020, 13, 149. [Google Scholar] [CrossRef]
- Ma, J.; Li, P.; Zhao, X.; Xu, H.; Wu, W.; Wang, Y.; Guo, Y.; Wang, L.; Feng, Y.; Xiao, L. Occurrence and molecular characterization of Cryptosporidium spp. and Enterocytozoon bieneusi in dairy cattle, beef cattle and water buffaloes in China. Vet. Parasitol. 2015, 207, 220–227. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, J.; Li, Z.; Xu, C.; Hou, M.; Qi, M. Molecular identification of Cryptosporidium spp. in alpacas (Vicugna pacos) in China. Int. J. Parasitol. Parasites Wildl. 2020, 12, 181–184. [Google Scholar] [CrossRef]
- Xu, N.; Liu, H.; Jiang, Y.; Yin, J.; Yuan, Z.; Shen, Y.; Cao, J. First report of Cryptosporidium viatorum and Cryptosporidium occultus in humans in China, and of the unique novel C. viatorum subtype XVaA3h. BMC Infect. Dis. 2020, 20, 16. [Google Scholar] [CrossRef]
- Ong, C.S.; Eisler, D.L.; Alikhani, A.; Fung, V.W.; Tomblin, J.; Bowie, W.R.; Isaac-Renton, J.L. Novel Cryptosporidium genotypes in sporadic cryptosporidiosis cases: First report of human infections with a cervine genotype. Emerg. Infect. Dis. 2002, 8, 263–268. [Google Scholar] [CrossRef]
- Robinson, G.; Chalmers, R.M.; Stapleton, C.; Palmer, S.R.; Watkins, J.; Francis, C.; Kay, D. A whole water catchment approach to investigating the origin and distribution of Cryptosporidium species. J. Appl. Microbiol. 2011, 111, 717–730. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Alderisio, K.A.; Xiao, L. Distribution of Cryptosporidium genotypes in storm event water samples from three watersheds in New York. Appl. Environ. Microbiol. 2005, 71, 4446–4454. [Google Scholar] [CrossRef] [PubMed]
- Alves, M.; Xiao, L.; Sulaiman, I.; Lal, A.A.; Matos, O.; Antunes, F. Subgenotype analysis of Cryptosporidium isolates from humans, cattle, and zoo ruminants in Portugal. J. Clin. Microbiol. 2003, 41, 2744–2747. [Google Scholar] [CrossRef]
- Xiao, L.; Morgan, U.M.; Limor, J.; Escalante, A.; Arrowood, M.; Shulaw, W.; Thompson, R.C.; Fayer, R.; Lal, A.A. Genetic diversity within Cryptosporidium parvum and related Cryptosporidium species. Appl. Environ. Microbiol. 1999, 65, 3386–3391. [Google Scholar] [CrossRef] [PubMed]
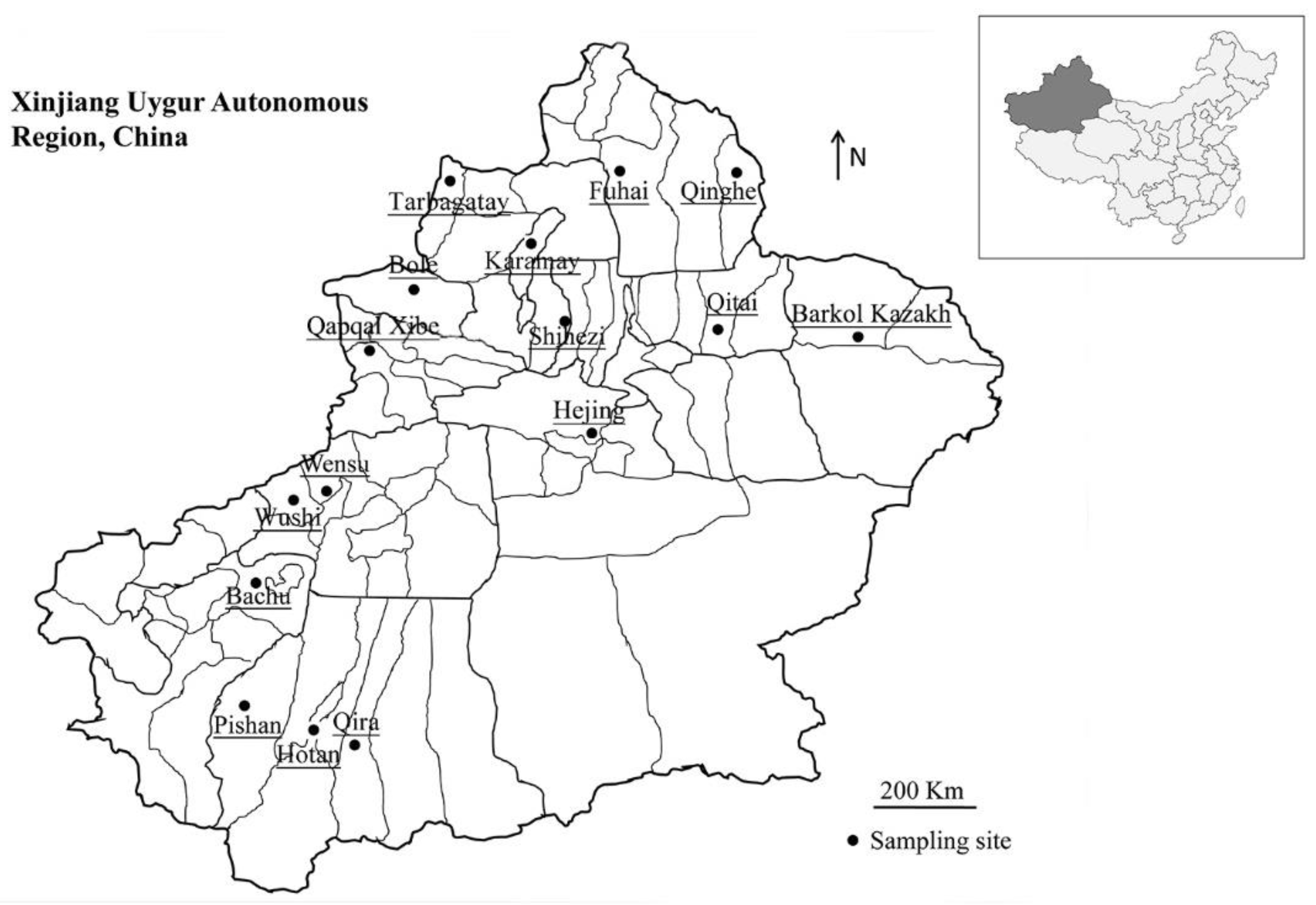
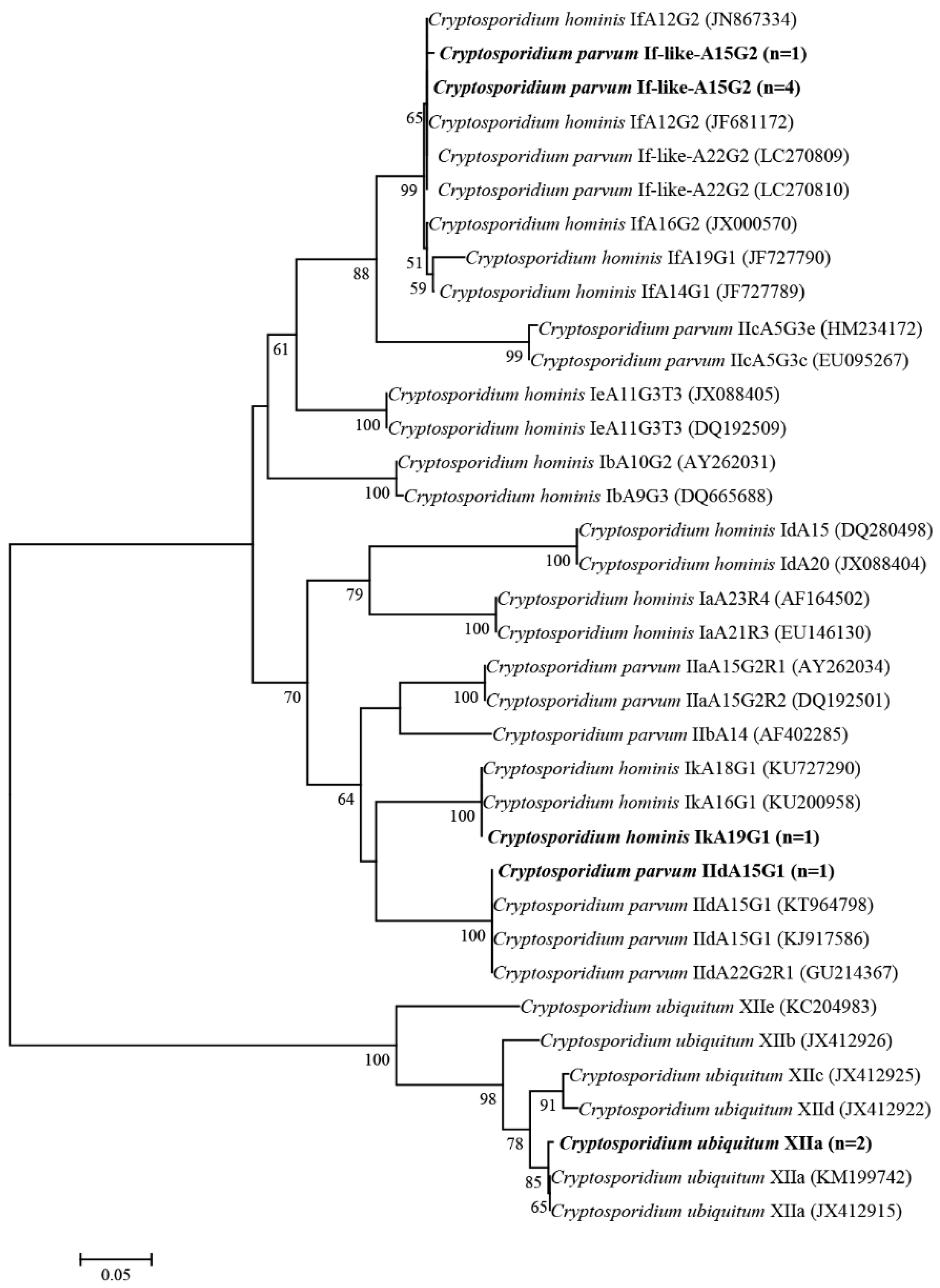
| Collection Sites | N/T (%) [95% Cl] | Cryptosporidium Species/Subtypes (No.) |
|---|---|---|
| Barkol Kazakh | 0/58 (0) | None |
| Qinghe | 0/57 (0) | None |
| Qitai | 2/18 (11.1) [0–27.2] | C. andersoni (1); C. hominis (1)/IkA19G1 (1) |
| Fuhai | 1/26 (3.8) [0–11.8] | C. parvum (1)/If-like-A15G2 (1) |
| Karamay | 3/45 (6.7) [0–14.2] | C. andersoni (3) |
| Shihezi | 5/60 (8.3) [1.1–15.5] | C. parvum (5)/If-like-A15G2 (4), IIdA15G1 (1) |
| Hejing | 5/16 (31.3) [5.7–56.8] | C. andersoni (5) |
| Tarbagatay | 3/16 (18.8) [0–40.2] | C. andersoni (3) |
| Bole | 0/61 (0) | None |
| Qapqal Xibe | 4/12 (33.3) [2–64.6] | C. andersoni (4) |
| Wensu | 1/24 (4.2) [0–12.8] | C. bovis (1) |
| Wushi | 2/10 (20.0) [0–50.2] | C. andersoni (2) |
| Bachu | 0/17 (0) | None |
| Pishan | 3/17 (17.6) [0–37.9] | C. andersoni (1); C. ubiquitum (2)/XIIa (2) |
| Hotan | 0/17 (0) | None |
| Qira | 7/22 (31.8) [10.7–53.0] | C. andersoni (5); C. occultus (2) |
| Total | 36/476 (7.6) [5.2–9.9] | C. andersoni (24); C. bovis (1); C. occultus (2); C. parvum (6)/If-like-A15G2 (5), IIdA15G1 (1); C. hominis (1)/IkA19G1 (1); C. ubiquitum (2)/XIIa (2) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, Y.; Cui, Z.; Zhou, Q.; Jing, B.; Xu, C.; Wang, T.; Qi, M.; Zhang, L. Genetic Diversity of Cryptosporidium in Bactrian Camels (Camelus bactrianus) in Xinjiang, Northwestern China. Pathogens 2020, 9, 946. https://doi.org/10.3390/pathogens9110946
Cao Y, Cui Z, Zhou Q, Jing B, Xu C, Wang T, Qi M, Zhang L. Genetic Diversity of Cryptosporidium in Bactrian Camels (Camelus bactrianus) in Xinjiang, Northwestern China. Pathogens. 2020; 9(11):946. https://doi.org/10.3390/pathogens9110946
Chicago/Turabian StyleCao, Yangwenna, Zhaohui Cui, Qiang Zhou, Bo Jing, Chunyan Xu, Tian Wang, Meng Qi, and Longxian Zhang. 2020. "Genetic Diversity of Cryptosporidium in Bactrian Camels (Camelus bactrianus) in Xinjiang, Northwestern China" Pathogens 9, no. 11: 946. https://doi.org/10.3390/pathogens9110946
APA StyleCao, Y., Cui, Z., Zhou, Q., Jing, B., Xu, C., Wang, T., Qi, M., & Zhang, L. (2020). Genetic Diversity of Cryptosporidium in Bactrian Camels (Camelus bactrianus) in Xinjiang, Northwestern China. Pathogens, 9(11), 946. https://doi.org/10.3390/pathogens9110946






