The Destructive Fungal Pathogen Botrytis cinerea—Insights from Genes Studied with Mutant Analysis
Abstract
1. Introduction
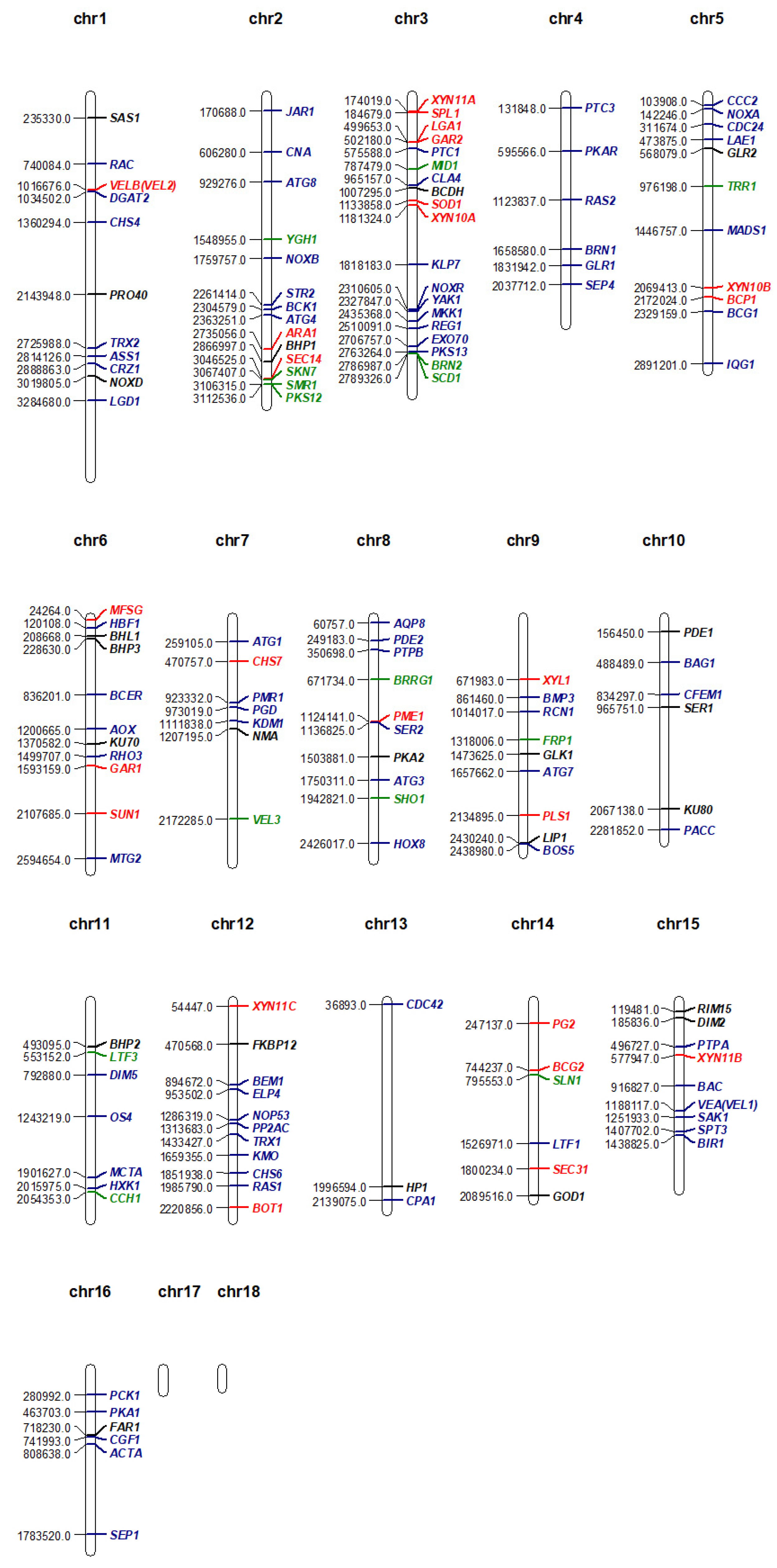
2. The features of B. cinerea Genome
2.1. Genomic Sequences
2.2. Transcriptomic and Secretomic Analysis
3. Molecular Dissection of B. cinerea Biology
3.1. Hyphal Growth and Virulence
| Gene Code (New) | Fungal Strains | Mutant Name | Gene Full Name | Mutant Type | Mutant Phenotypes | Host Species | Other Functions of Encoded Protein | Reference | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hyphal Growth | Sclerotial Formation | Oxalate Production | Virulence | Compound Appressoria Formation (Penetration) | Conidiation/Sporulation | Induce Host HR/Resistance | Secretion Signal | ||||||||
| Bcin01g00550 | B05.10 strain | sas1 | secretion-related Rab/GTPase gene | deletion | + | + | NA | + | NA | + | NA | No | Protein secretion, sporulation | [46] | |
| Bcin01g02000 | B05.10 strain | rac | Small GTPases | deletion | + | + | NA | + | NA | + | NA | No | Polar growth, reproduction | [47] | |
| Bcin01g02730 | B05.10 strain | vel2 | velvet-like gene | deletion | + | + | + | + | NA | + | NA | No | Light response, acidification | [48] | |
| Bcin01g02730 | 38B1 strain | velB | velvet-like gene | deletion | + | + | NA | + | - | + | NA | No | Negative role in asexual development and melanin biosynthesis | [49] | |
| Bcin01g02790 | Chickpea isolate from fields of Govind Ballabh Pant University | dgat2 | diacylglycerol O-acyl transferase 2 | T-DNA, deletion | - | + | + | + | NA | + | NA | No | Penetration and consequently virulence | [50] | |
| Bcin01g03790 | Bd90 strain | chs4 | chitin synthases | deletion | - | - | NA | - | NA | - | NA | No | [51] | ||
| Bcin01g06080 | 38B1 strain | pro40 | scaffold protein | deletion | NA | NA | + | NA | NA | NA | NA | No | [52] | ||
| Bcin01g07770 | B05.10 strain | trx2 | thioredoxin | deletion | - | NA | NA | - | - | - | NA | No | Resist to oxidative stress; strx1/trx2 double mutant has retarded growth as trr1 | [53] | |
| Bcin01g08050 | B05.10 strain | ass1 | argininosuccinate synthase | deletion | + | NA | NA | + | NA | NA | NA | No | Production of L-arginine | [54] | |
| Bcin01g08230 | B05.10 strain | crz1 | calcineurin-Responsive Zinc Finger Transcription Factor | deletion | + | + | NA | + | NA | + | NA | No | Acts downstream of calcineurin but not the only target of calcineurin | [55] | |
| Bcin01g08690 | B05.10 strain | noxD | component of the NADPH oxidase complex | deletion | NA | + | NA | + | - | + | NA | No | Interact with NOXA | [56] | |
| Bcin01g09450 | B05.10 strain | lgd1 | galactonate dehydratase gene | deletion | - | NA | NA | + | NA | NA | defence-related genes were not induced | No | Arabidopsis thaliana and Nicotiana benthamiana, not Solanum lycopersicum | D-galacturonic acid catabolism | [57] |
| Bcin02g00190 | B05.10 strain | jar1 | Histone 3 Lysine 4 (H3K4) demethylation | deletion | - | + | NA | + | + | + | NA | No | Oxidative and low-oxygen stress adaptation | [58] | |
| Bcin02g01540 | B05.10 strain | cnA | catalytic subunit of calcineurin | deletion | + | + | NA | + | NA | + | NA | No | [59] | ||
| Bcin02g02570 | B05.10 strain | atg8 | autophagy-related gene | deletion | + | + | NA | + | NA | + | NA | No | Interact with ATG4, lipid droplet metabolism | [60] | |
| Bcin02g04360 | B05.10 strain | ygh1 | alpha/beta hydrolases | deletion (heterokaryotic) | + | + | NA | - | NA | + | NA | No | Required for the formation of the key intermediate T4HN | [61] | |
| Bcin02g04930 | B05.10 strain | noxB | NADPH oxidases | deletion | - | + | - | + | + | - | NA | No | Penetration | [62] | |
| Bcin02g06470 | B05.10 strain | str2 | cystathionine γ-synthase | deletion | + | + | NA | + | NA | + | NA | No | Response to various stresses | [63] | |
| Bcin02g06590 | 38B1 strain | bck1 | MAPK cascade | deletion | + | NA | - | + | + | + | NA | No | Melanin biosynthesis | [52] | |
| Bcin02g06770 | B05.10 strain | atg4 | cysteine protease | deletion | + | + | NA | + | + | + | NA | No | Autophagy | [64] | |
| Bcin02g07700 | IK2018/B05.10 | ara1 | α-1,5-L-endo-arabinanase | deletion | - | NA | NA | + | NA | NA | NA | Yes | Arabidopsis thaliana | Host dependent, secondary lesion formation during infection | [65] |
| Bcin02g07970 | B05.10 strain | bhp1 | hydrophobin encoding gene | deletion | - | - | NA | - | - | - | NA | Yes | Development of apothecia | [66,67] | |
| Bcin02g08570 | B05.10 strain | sec14 | protein secretion related gene | deletion | - | NA | NA | + | NA | - | NA | No | Protein secretion | [68] | |
| Bcin02g08650 | 38B1 strain | skn7 | response regulator in the high-osmolarity glycerol pathway | deletion | - | + | NA | - | NA | + | NA | No | Regulation of vegetative differentiation and in the response to various stresses | [69] | |
| Bcin02g08760 | B05.10 strain | smr1 | sclerotial melanogenesis-regulatory gene | deletion | NA | + | NA | NA | NA | NA | NA | No | Sclerotial melanogenesis | [70] | |
| Bcin02g08770 | B05.10 strain | pks12 | polyketide synthase | deletion | - | + | NA | - | NA | - | NA | No | [61] | ||
| Bcin03g00500 | B05.10 strain | spl1 | cerato-platanin family protein | deletion | - | NA | NA | + | NA | NA | + | Yes | a variety of hosts | HR and PR gene induction, BAK1 required | [71] |
| Bcin03g01490 | B05.10 strain | lga1 | keto-3-deoxy-L-galactonate aldolase gene | deletion | - | NA | NA | + | NA | NA | defence-related genes were not induced | No | Arabidopsis thaliana and Nicotiana benthamiana, not Solanum lycopersicum | D-galacturonic acid catabolism | [57] |
| Bcin03g01500 | B05.10 strain | gar2 | galacturonate reductase genes | deletion | - | NA | NA | + | NA | NA | defence-related genes were not induced | No | Arabidopsis thaliana and Nicotiana benthamiana, not Solanum lycopersicum | D-galacturonic acid catabolism | [57] |
| Bcin03g01720 | 38B1 strain | ptc1 | Type 2C Ser/Thr phosphatases | deletion | + | + | NA | + | NA | + | NA | No | Melanin biosynthesis, regulation of multiple stress tolerance and virulence | [72] | |
| Bcin03g02380 | B05.10 strain | mid1 | calcium channel protein | deletion | + | - | NA | - | - | - | NA | No | Vegetative growth under conditions of low extracellular calcium | [41] | |
| Bcin03g02930 | B05.10 strain | cla4 | Rac effectors | deletion | + | NA | NA | + | NA | + | NA | No | Cell cycle regulating processes downstream of RAC | [73] | |
| Bcin03g03060 | B05.10 strain | bcdh | UDP-glucose-4,6-dehydratase | deletion | - | - | NA | - | NA | - | NA | No | Production of rhamnose-containing glycan | [74] | |
| Bcin03g03390 | B05.10 strain | sod1 | Cu-Zn-superoxide dismutase | deletion | NA | NA | NA | + | NA | NA | NA | No | Phaseolus vulgaris | [75] | |
| Bcin03g05410 | collected from chickpea feld of Govind Ballabh Pant University | klp7 | kinesin | T-DNA | + | NA | + | + | + | + | NA | No | Reduced activities of polygalacturonase (PG) and pectin methyl esterases (PME) | [76] | |
| Bcin03g06840 | B05.10 strain | noxR | regulatory subunit of the Nox complex | deletion | - | + | - | + | + | - | NA | No | Activation of both NOX enzymes | [62] | |
| Bcin03g06910 | B05.10 strain | yak1 | dual-specificity tyrosine phosphorylation-regulated protein kinase | deletion | - | + | NA | + | + | + | NA | No | Adaptation to oxidative stress and triadimefon | [77] | |
| Bcin03g07190 | 38B1 strain | mkk1 | MAPK kinase | deletion | + | NA | + | + | + | + | NA | No | Melanin biosynthesis, negatively regulates oxalic acid biosynthesis | [52] | |
| Bcin03g07420 | B05.10 strain | reg1 | ortholog of the F. oxysporum transcriptional regulator FoSge1 | deletion | - | - | NA | + | - | + | NA | No | Phaseolus vulgaris | Toxin production | [78] |
| Bcin03g07900 | B05.10 strain | exo70 | exocyst subunit gene | deletion | + | + | NA | + | NA | + | NA | No | [79] | ||
| Bcin03g08050 | B05.10 strain | pks13 | polyketide synthase | deletion | + | + | + | + | NA | + | NA | No | Melanin synthesis, mutant shows enhanced growth rate and virulence, white sclerotia | [80] | |
| Bcin03g08100 | B05.10 strain | brn2 | tetrahydroxynaphthalene (THN) reductases | deletion | - | + | NA | - | NA | + | NA | No | [61] | ||
| Bcin03g08110 | B05.10 strain | scd1 | scytalone dehydratases | deletion | - | + | NA | - | NA | + | NA | No | [61] | ||
| Bcin04g00340 | 38B1 strain | ptc3 | Type 2C Ser/Thr phosphatases | deletion | + | + | NA | + | NA | + | NA | No | Melanin biosynthesis, regulation of multiple stress tolerance and virulence | [72] | |
| Bcin04g01630 | B05.10 strain | pkaR | regulatory regulatory subunit of cAMP-dependent protein kinase | deletion | + | - | - | + | NA | - | NA | No | [81] | ||
| Bcin04g03140 | B05.10 strain | ras2 | fungal-specific Ras GTPase | deletion | + | - | - | + | NA | - | NA | No | Conidial germination | [81] | |
| Bcin04g04800 | B05.10 strain | brn1 | tetrahydroxynaphthalene (THN) reductases | deletion | + | + | + | + | NA | + | NA | No | Melanin synthesis, mutant shows enhanced growth rate and virulence, orange sclerotia | [80] | |
| Bcin04g05300 | B05.10 strain | glr1 | GSH reductase | deletion | - | NA | NA | + | + | NA | NA | No | Conidia germination | [53] | |
| Bcin04g05920 | B05.10 strain | sep4 | septin gene | deletion | - | + | NA | + | + | + | NA | No | Melanin and chitin accumulation in hyphal tips | [82] | |
| Bcin05g00240 | HYOGO11 | ccc2 | copper-transporting ATPase | deletion | - | + | NA | + | + | + | NA | No | Melanization | [83] | |
| Bcin05g00350 | B05.10 strain | noxA | NADPH oxidases | deletion | - | + | - | + | - | - | NA | No | Colonize the host tissue | [62] | |
| Bcin05g00760 | B05.10 strain | cdc24 | GEF (guanine nucleotide exchange factor) | deletion (heterokaryotic) | + | NA | NA | + | + | + | NA | No | [84] | ||
| Bcin05g01210 | B05.10 strain | lae1 | putative interaction partner of BcVEL1 | deletion | - | + | + | + | NA | + | NA | No | Putative interaction partner of VEL1 | [53] | |
| Bcin05g01430 | B05.10 strain | glr2 | GSH reductase | deletion | - | NA | NA | - | - | NA | NA | No | [53] | ||
| Bcin05g02680 | B05.10 strain | trr1 | thioredoxin reductase | deletion | + | NA | NA | + | - | - | NA | No | Resist to oxidative stress | [53] | |
| Bcin05g04030 | B05.10 strain | mads1 | MADS-box transcription factor | deletion | + | + | NA | + | NA | + | NA | No | Regulates the expression of light-responsive genes | [68] | |
| Bcin05g06320 | T4 strain | bcp1 | Cyclophilin A | deletion | - | NA | NA | + | NA | - | NA | No | [85] | ||
| Bcin05g06770 | B05.10 strain | bcg1 | Gαi subunits (I) | deletion | + | NA | NA | + | - | NA | NA | No | Protease secretion | [44] | |
| Bcin05g08290 | B05.10 strain | iqg1 | fungal homolog of the RasGAP scaffold protein IQGAP | deletion | _ | + | NA | + | + | + | NA | No | Resistance against oxidative and membrane stress | [86] | |
| Bcin06g00026 | B05.10 strain | mfsG | Major Facilitator Superfamily transporter | deletion | NA | NA | NA | + | NA | NA | NA | No | Increases tolerance to glucosinolates | [87] | |
| Bcin06g00240 | B05.10 strain | hbf1 | hyphal branching-related factor 1 | T-DNA, deletion | - | + | NA | + | + | - | NA | No | Hyphal branching | [88] | |
| Bcin06g00450 | B05.10 strain | bhl1 | Botrytis hydrophobin-like gene | deletion | - | - | NA | - | - | - | NA | Yes | [67] | ||
| Bcin06g00510 | B05.10 strain | bhp3 | hydrophobin encoding gene | deletion | - | - | NA | - | - | - | NA | Yes | Development of apothecia | [66,67] | |
| Bcin06g02380 | B05.10 strain | bcer | UDP-4-keto-6-deoxyglucose-3,5-epimerase/-4-reductas | deletion | + | + | NA | + | NA | + | NA | No | Production of rhamnose-containing glycan | [74] | |
| Bcin06g03440 | B05.10 strain | aox | alternative oxidase | deletion | - | + | NA | + | NA | + | NA | No | Adaptation to environmental stress | [89] | |
| Bcin06g03990 | B05.10 strain | ku70 | inhibitor of NHEJ | deletion | - | NA | NA | - | NA | - | NA | No | Ku deficiencies improved HR efficiency | [90] | |
| Bcin06g04390 | B05.10 strain | rho3 | small GTPases of the Rho family | deletion | + | + | NA | + | + | + | NA | No | [91] | ||
| Bcin06g04660 | B05.10 strain | gar1 | galacturonate reductase genes | deletion | - | NA | NA | + | NA | NA | defence-related genes were not induced | No | Arabidopsis thaliana and Nicotiana benthamiana, not Solanum lycopersicum | D-galacturonic acid catabolism | [57] |
| Bcin06g06040 | B05.10 strain | sun1 | Group-I SUN family of proteins | deletion | - | + | NA | + | NA | + | NA | Yes | Production of reproductive structures and adhesion to plant surface | [92] | |
| Bcin06g07300 | B05.10 strain | mtg2 | Obg protein | deletion | + | + | NA | + | NA | + | NA | No | Asexual development, environmental stress response | [30] | |
| Bcin07g00720 | B05.10 strain | atg1 | autophagy-related gene | deletion | + | + | NA | + | + | + | NA | No | Lipid metabolism | [93] | |
| Bcin07g01300 | Bd90 strain | chs7 | chitin synthases | deletion | - | - | NA | + | NA | - | NA | No | Phaseolus vulgaris, ecotype Col-0 of Arabidopsis thaliana | Virulence depends on host plants | [51] |
| Bcin07g02480 | B05.10 strain | pmr1 | P-type Ca2+/Mn2+-ATPase | deletion | + | + | NA | + | NA | + | NA | No | Solanum lycopersicum leaves and fruit and Malus domestica fruit | Biofilm formation | [94] |
| Bcin07g02610 | B05.10 strain | pgd | 6-phosphogluconate dehydrogenase | deletion | + | NA | NA | + | NA | + | NA | No | Influenced by NOX | [95] | |
| Bcin07g03050 | B05.10 strain | kdm1 | histone 3 lysine 36 (H3K36)-specific demethylas | T-DNA, deletion | + | + | NA | + | NA | + | NA | No | Stress responses and photomorphogenesis | [96] | |
| Bcin07g03340 | B05.10 strain | nma | high-temperature requirement (HtrA) family of serine proteases | deletion | - | NA | NA | - | NA | - | NA | No | Pro-apoptotic activity | [97] | |
| Bcin07g05880 | B05.10 strain | vel3 | velvet-like gene | deletion | - | - | - | - | NA | + | NA | No | Light response, acidification | [48] | |
| Bcin08g00120 | B05.10 strain | aqp8 | aquaporin 8 | deletion | + | + | NA | + | + | + | NA | No | Pigment metabolism | [98] | |
| Bcin08g00550 | B05.10 strain | pde2 | phosphodiesterase | deletion | + | + | NA | + | NA | + | NA | No | Camp signaling pathway | [99] | |
| Bcin08g00850 | 38B1 strain | ptpB | putative protein tyrosine phosphatase (PTP) gene | deletion | + | + | NA | + | NA | + | NA | No | Negative role in melanin biosynthesis; PTPA and PTPB have opposite functions in conidiation | [100] | |
| Bcin08g01740 | 38B1 strain | brrg1 | putative response regulator protein | deletion | - | NA | NA | - | NA | + | NA | No | Sensitivity to fungicides and osmotic stress | [101] | |
| Bcin08g02970 | Bd90 strain | pme1 | pectin methylesterase | deletion | - | - | NA | + | NA | - | NA | Yes | [102] | ||
| Bcin08g02990 | B05.10 strain | ser2 | subtilisin-like protease 2 | deletion | + | + | NA | + | + | + | NA | Yes | [103] | ||
| Bcin08g03910 | B05.10 strain | pka2 | catalytic subunit of cAMP-dependent protein kinase | deletion | - | - | - | - | NA | - | NA | No | [81] | ||
| Bcin08g04530 | B05.10 strain | atg3 | ubiquitin-like (UBL) protein-activating enzymes | deletion | + | + | NA | + | NA | + | NA | No | Autophagy | [104] | |
| Bcin08g05150 | B05.10 strain | sho1 | biosensors of HOG pathway | deletion | + | + | NA | - | NA | + | NA | No | Redundant for sln1 mutant | [105] | |
| Bcin08g06620 | B05.10/T4strain | hox8 | homeobox transcription factor encoding gene | deletion | + | - | NA | + | + | + | NA | No | [106] | ||
| Bcin09g01800 | B05.10 strain | xyl1 | Xylanase | deletion | - | NA | NA | + | NA | NA | + | Yes | Trigger PTI | [107] | |
| Bcin09g02390 | B05.10 strain | bmp3 | cell wall integrity MAPK | deletion | + | + | NA | + | + | + | NA | No | Melanin biosynthesis | [108] | |
| Bcin09g02820 | B05.10 strain | rcn1 | calcipressin | deletion | + | - | NA | + | NA | - | NA | No | Positive modulator of CNA | [59] | |
| Bcin09g03710 | B05.10 strain | frp1 | FRP1 F-box gene | deletion | - | + | NA | - | NA | - | NA | No | Sexual reproduction | [109] | |
| Bcin09g04170 | B05.10 strain | glk1 | glucokinase | deletion | - | NA | NA | - | NA | - | NA | No | [110] | ||
| Bcin09g04730 | B05.10 strain | atg7 | ubiquitin-like (UBL) protein-activating enzymes | deletion | + | + | NA | + | NA | + | NA | No | Autophagy | [104] | |
| Bcin09g06130 | B05.10 strain | pls1 | tetraspanin | deletion | - | - | - | + | + | - | NA | No | Penetration; sexual development | [111] | |
| Bcin09g06880 | B05.10 strain | lip1 | lipase gene | deletion | NA | NA | NA | - | NA | NA | NA | Yes | Catabolite repression | [112] | |
| Bcin09g06900 | 38B1 strain | bos5 | mitogen-activated protein kinase kinase | deletion | + | NA | NA | + | - | + | NA | No | Adaptation to iprodione and ionic stress | [113] | |
| Bcin10g00450 | B05.10 strain | pde1 | phosphodiesterase | deletion | - | - | NA | - | NA | - | NA | No | Camp signaling pathway, enhance PDE2 function | [99] | |
| Bcin10g01250 | B05.10 strain | bag1 | Bcl-2 associated athanogene | deletion | + | + | NA | + | + | + | NA | No | Hyphal melanization, response to multiple abiotic stresses and UPR pathway | [114] | |
| Bcin10g02180 | B05.10 strain | cfem1 | CFEM protein with putative GPIanchored site | deletion | - | - | NA | + | NA | + | NA | Yes | Stress tolerance | [115] | |
| Bcin10g02530 | B05.10 strain | ser1 | subtilisin-like protease 1 | deletion | - | - | NA | - | - | - | NA | Yes | [103] | ||
| Bcin10g05490 | B05.10 strain | ku80 | inhibitor of NHEJ | deletion | - | NA | NA | - | NA | - | NA | No | Ku deficiencies improved HR efficiency | [90] | |
| Bcin10g05950 | B05.10 strain | pacC | PacC transcription factor | deletion | + | + | + | + | NA | + | + | No | Production of reactive oxygen species; enzyme secretion | [116] | |
| Bcin11g01450 | B05.10 strain | bhp2 | hydrophobin encoding gene | deletion | - | - | NA | - | - | - | NA | Yes | Development of apothecia | [66,67] | |
| Bcin11g01720 | B05.10 strain | ltf3 | putative C2H2 transcription factor | deletion | - | NA | NA | - | NA | + | NA | No | [78] | ||
| Bcin11g02360 | B05.10 strain | dim5 | Histone H3 Lysine 9 Methyltransferase | deletion | + | + | NA | + | NA | + | NA | No | [117] | ||
| Bcin11g03560 | 38B1 strain | os4 | mitogen-activated protein kinase kinase kinase gene | deletion | + | NA | NA | + | - | + | NA | No | Adaption to hyperosmotic and oxidative stresses | [118] | |
| Bcin11g05350 | B05.10 strain | mctA | putative monocarboxylate transporter | deletion | - | + | NA | + | NA | + | NA | No | Pyruvate uptake | [119] | |
| Bcin11g05700 | B05.10 strain | hxk1 | hexokinase | deletion | + | NA | NA | + | NA | + | NA | No | fruit | Sugar metabolism | [110] |
| Bcin11g05810 | B05.10 strain | cch1 | calcium channel protein | deletion | + | - | NA | - | - | - | NA | No | Vegetative growth under conditions of low extracellular calcium | [41] | |
| Bcin12g01360 | B05.10 strain | fkbp12 | FK506-binding protein | deletion | - | NA | NA | - | NA | - | NA | No | Sulfur repression of the synthesis of a secreted serine protease | [120] | |
| Bcin12g02530 | B05.10 strain | bem1 | scaffold protein | deletion | - | NA | NA | + | + | + | NA | No | Part of a polarity complex involving the GEF CDC24 | [84] | |
| Bcin12g02750 | B05.10 strain | elp4 | elongator complex protein | deletion | + | + | NA | + | NA | NA | NA | No | Mycelia differentiation, melanization, various environmental stress response | [121] | |
| Bcin12g03770 | B05.10 strain | nop53 | pre-rRNA processing factor | deletion | + | + | NA | + | + | + | NA | No | Oxidative and osmotic stress adaptation | [122] | |
| Bcin12g03880 | B05.10 strain | pp2ac | a catalytic subunit of a PP2A serine/threonine protein phosphatase | T-DNA, RNAi | + | + | NA | + | NA | - | NA | No | Resistance to H2O2 | [123] | |
| Bcin12g04280 | B05.10 strain | trx1 | thioredoxin | deletion | - | NA | NA | + | - | - | NA | No | Resist to oxdative stress; bstrx1bctrx2 double mutant has retarded growth as bctrr1 | [53] | |
| Bcin12g04900 | BC22 strain | kmo | kynurenine 3-monooxygenase (KMO) | T-DNA | + | + | + | + | NA | + | NA | No | Cell wall degrading enzymes activity | [124] | |
| Bcin12g05360 | Bd90 strain | chs6 | chitin synthases | deletion (heterokaryotic ) | + | - | NA | + | NA | + | NA | No | Sexual cycle | [51] | |
| Bcin12g05760 | B05.10 strain | ras1 | Small GTPases | deletion | + | + | NA | + | NA | + | NA | No | Polar growth, reproduction | [47] | |
| Bcin12g06380 | T4 strain | bot1 | P450 monooxygenase | deletion | NA | NA | NA | + | NA | NA | NA | No | Strain-specific virulence factor | [125] | |
| Bcin13g00090 | B05.10 strain | cdc42 | small GTPase | deletion | + | + | NA | + | + | + | NA | No | Conidial germination and nuclear distribution | [126] | |
| Bcin13g05340 | B05.10 strain | hp1 | heterochromatin protein 1 | deletion | - | - | NA | - | NA | - | NA | No | [117] | ||
| Bcin13g05610 | B05.10 strain | cpa1 | capping protein (CP) subunit | deletion | + | + | NA | + | + | + | NA | No | Conidial germination; interact with CPB1 | [127] | |
| Bcin14g00610 | B05.10 strain | pg2 | endopolygalacturonase enzyme | deletion | NA | NA | NA | + | NA | NA | NA | Yes | Solanum lycopersicum and Vicia faba | [128] | |
| Bcin14g01730 | B05.10 strain | bcg2 | group II of Gα subunits | deletion | - | NA | NA | + | - | NA | NA | No | [44] | ||
| Bcin14g01870 | B05.10 strain | sln1 | biosensors of high-osmolarity glycerol(HOG) pathway | deletion | + | + | NA | - | NA | + | NA | No | Redundant for shn1 mutant | [105] | |
| Bcin14g03930 | B05.10 strain | ltf1 | light-responsive transcription factor 1 | deletion | + | NA | NA | + | + | + | NA | No | ROS homoeostasis, light-dependent differentiation | [129] | |
| Bcin14g04650 | B05.10 strain | sec31 | protein secretion related gene | deletion | - | NA | NA | + | NA | - | NA | No | Protein secretion | [68] | |
| Bcin14g05500 | B05.10 strain | god1 | putative glucose oxidase gene | deletion | NA | NA | NA | - | NA | NA | NA | Yes | Phaseolus vulgaris | [75] | |
| Bcin15g00280 | 38B1 strain | rim15 | Per-Arnt-Sim (PAS) kinase | deletion | + | NA | + | - | NA | NA | NA | No | [52] | ||
| Bcin15g00450 | B05.10 strain | dim2 | DNA methyltransferase | deletion | - | - | NA | - | NA | - | NA | No | [117] | ||
| Bcin15g01330 | 38B1 strain | ptpA | putative protein tyrosine phosphatase (PTP) gene | deletion | + | + | NA | + | NA | + | NA | No | Negative role in melanin biosynthesis; bcptpa and bcptpb have opposite functions in conidiation | [100] | |
| Bcin15g02590 | B05.10 strain | bac | adenylate cyclase | deletion | + | NA | NA | + | NA | + | NA | No | [130] | ||
| Bcin15g03390 | 38B1 strain | veA | velvet-like gene | deletion | - | + | NA | + | - | + | NA | No | Negative role in asexual development and melanin biosynthesis | [49] | |
| Bcin15g03390 | B05.10 strain | vel1 | VELVET Gene | deletion | NA | + | + | + | NA | + | NA | No | [131] | ||
| Bcin15g03580 | B05.10 strain | sak1 | Hog-type stress-activated MAPK | deletion | NA | + | NA | + | NA | + | NA | No | Early stages of infection; regulation of secondary metabolism | [132] | |
| Bcin15g04040 | B05.10 strain | spt3 | SPT3 subunit of a Spt-Ada-Gcn5-acetyltransferase | T-DNA, deletion | + | + | NA | + | NA | + | NA | No | Resistance to H2O2 | [123] | |
| Bcin15g04140 | B05.10 strain | bir1 | baculovirus IAP (inhibitor of apoptosis protein) repeat | partial knockout | + | NA | NA | + | NA | + | NA | No | Anti-apoptotic activity | [97] | |
| Bcin16g00630 | B05.10 strain | pck1 | phosphoenolpyruvatecarboxykinase gene | T-DNA, deletion | - | + | NA | + | + | + | NA | No | [133] | ||
| Bcin16g01130 | B05.10 strain | pka1 | catalytic subunit of cAMP-dependent protein kinase | deletion | + | - | - | + | NA | - | NA | No | [81] | ||
| Bcin16g01780 | B05.10 strain | far1 | scaffold protein | deletion | - | NA | NA | - | - | - | NA | No | No obvious phenotypes | [84] | |
| Bcin16g01820 | B05.10 strain | cgf1 | conidial germination-associated factor 1 | deletion | - | - | NA | + | + | + | NA | No | ROS production, osmotic and oxidative stress adaptation | [134] | |
| Bcin16g02020 | B05.10 strain | actA | actin protein | deletion | + | NA | NA | + | NA | + | NA | No | Hyphae structure | [135] | |
| Bcin16g04910 | B05.10 strain | sep1 | formin | T-DNA | + | NA | NA | + | + | + | NA | No | Septum formation and polarized growth | [84] | |
| Bcin03g03480 | B05.10 strain | xyn10A | xylanases of family GH10 | RNAi | - | - | NA | + | NA | - | NA | Yes | [136] | ||
| Bcin05g06020 | xyn10B | xylanases of family GH10 | Yes | ||||||||||||
| Bcin03g00480 | xyn11A | xylanases of family GH11 | Yes | ||||||||||||
| Bcin15g01600 | xyn11B | xylanases of family GH11 | Yes | ||||||||||||
| Bcin12g00090 | xyn11C | xylanases of family GH11 | Yes | ||||||||||||
3.2. Sclerotia Development
3.3. Signaling Events Leading to Conidiation
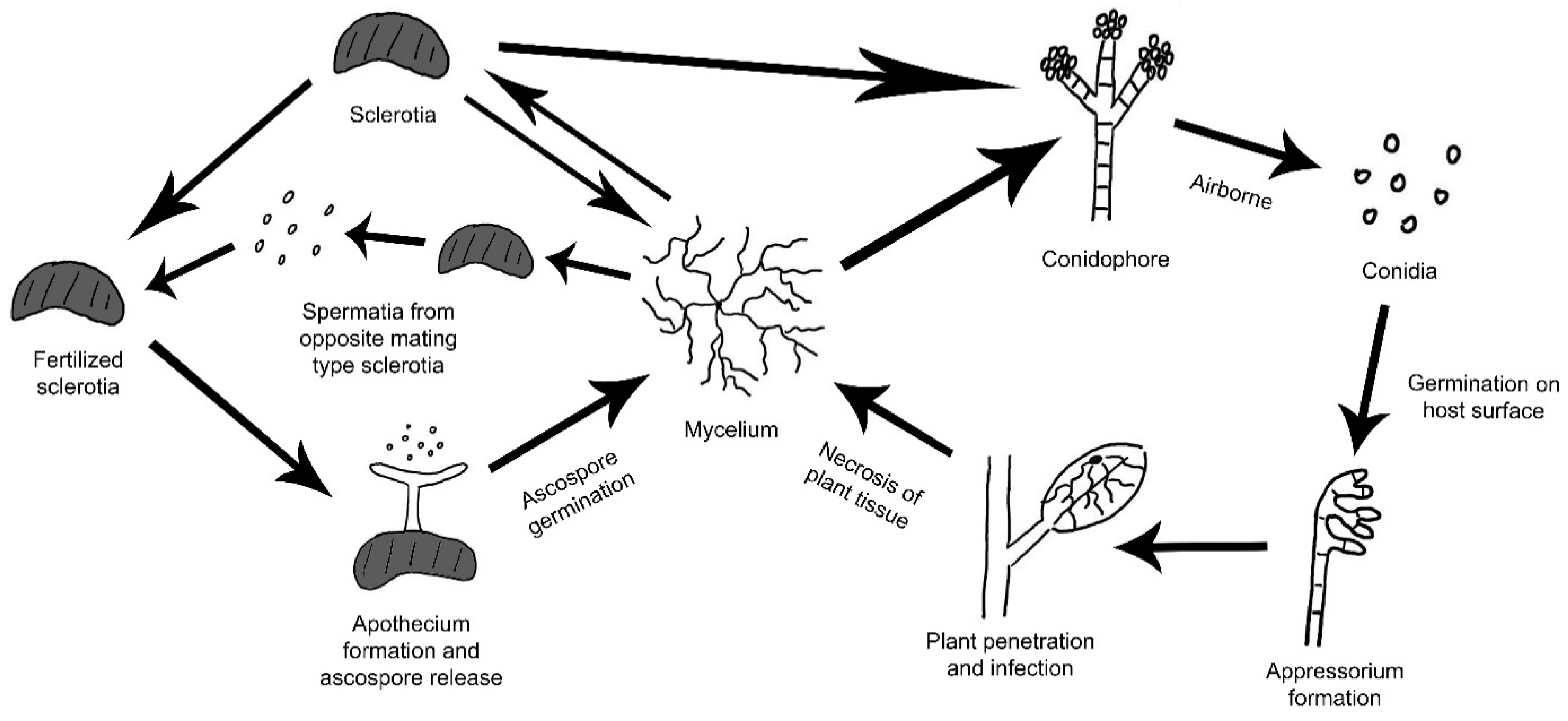
3.4. Infection and Pathogenicity Mechanisms
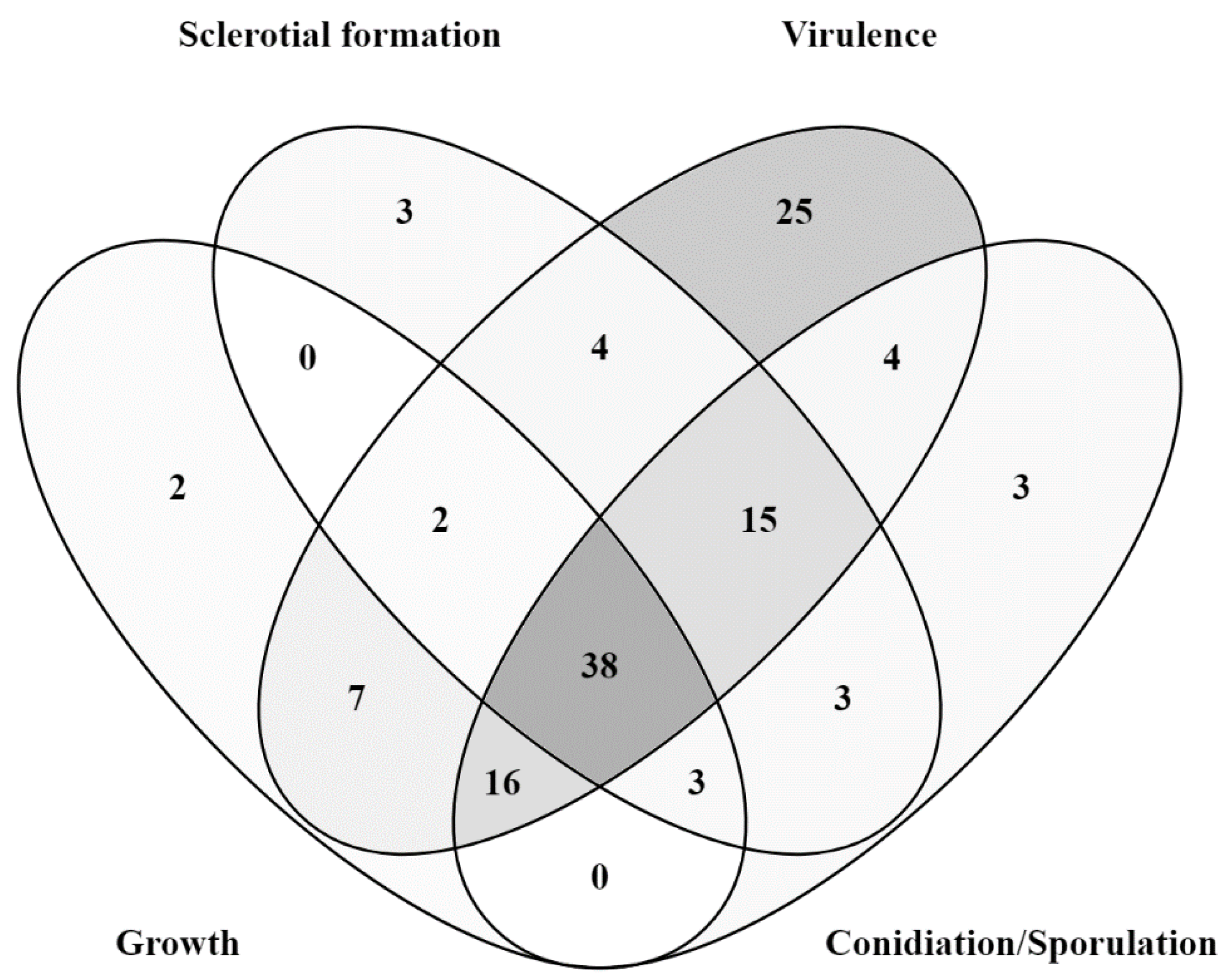
3.5. Generalist Genes–Mutants with Defects in Multiple Aspects of B. Cinerea Biology, Including Growth, Virulence and Sclerotia or Conidia Development
3.6. Melanization and Its Effects on Development and Pathogenicity
4. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Persoon, C.H. Dispositio methodica fungorum. Neues Mag. Bot. 1794, 1, 81–128. Available online: http://www.mycobank.org/BioloMICS.aspx?TableKey=14682616000000061&Rec=16179&Fields=All (accessed on 7 May 2020).
- Mirzaei, S.; Goltapeh, E.M.; Shams-Bakhsh, M.; Safaie, N. Identification of Botrytis spp. on plants grown in Iran. J. Phytopathol. 2007, 156, 21–28. [Google Scholar] [CrossRef]
- Gregory, P. Studies on Sclerotinia and Botrytis. Trans. Br. Mycol. Soc. 1949, 32, 1–IN3. [Google Scholar] [CrossRef]
- Whetzel, H.H. A Synopsis of the genera and species of the Sclerotiniaceae, a family of stromatic inoperculate discomycetes. Mycologia 1945, 37, 648. [Google Scholar] [CrossRef]
- Zhong, S.; Zhang, J.; Zhang, G.-Z. Botrytis polyphyllae: A new botrytis species causing gray mold on Paris polyphylla. Plant Dis. 2019, 103, 1721–1727. [Google Scholar] [CrossRef] [PubMed]
- Grant-Downton, R.T.; Terhem, R.B.; Kapralov, M.V.; Mehdi, S.; Rodriguez-Enriquez, M.J.; Gurr, S.J.; Van Kan, J.A.L.; Dewey, F.M. A novel Botrytis species is associated with a newly emergent foliar disease in cultivated hemerocallis. PLoS ONE 2014, 9, e89272. [Google Scholar] [CrossRef] [PubMed]
- Staats, M.; Van Baarlen, P.; Van Kan, J.A.L. Molecular phylogeny of the plant pathogenic genus Botrytis and the evolution of host specificity. Mol. Biol. Evol. 2004, 22, 333–346. [Google Scholar] [CrossRef]
- Williamson, B.; Tudzynski, B.; Tudzynski, P.; Van Kan, J.A.L. Botrytis cinerea: The cause of grey mould disease. Mol. Plant Pathol. 2007, 8, 561–580. [Google Scholar] [CrossRef]
- Amselem, J.; Cuomo, C.A.; Van Kan, J.A.L.; Viaud, M.; Benito, E.P.; Couloux, A.; Coutinho, P.M.; De Vries, R.P.; Dyer, P.S.; Fillinger, S.; et al. Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet. 2011, 7, e1002230. [Google Scholar] [CrossRef] [PubMed]
- Jarvis, W.R. The infection of strawberry and raspberry fruits by Botrytis cinerea Fr. Ann. Appl. Biol. 1962, 50, 569–575. [Google Scholar] [CrossRef]
- Viret, O.; Keller, M.; Jaudzems, V.G.; Cole, F.M. Botrytis cinerea infection of grape flowers: Light and electron microscopical studies of infection sites. Phytopathology 2004, 94, 850–857. [Google Scholar] [CrossRef]
- McNicol, R.J.; Williamson, B.; Dolan, A. Infection of red raspberry styles and carpels by Botrytis cinerea and its possible role in post-harvest grey mould. Ann. Appl. Biol. 1985, 106, 49–53. [Google Scholar] [CrossRef]
- Neri, F.; Cappellin, L.; Spadoni, A.; Cameldi, I.; Alarcon, A.A.; Aprea, E.; Romano, A.; Gasperi, F.; Biasioli, F. Role of strawberry volatile organic compounds in the development of Botrytis cinerea infection. Plant Pathol. 2014, 64, 709–717. [Google Scholar] [CrossRef]
- Prusky, D.; Lichter, A. Activation of quiescent infections by postharvest pathogens during transition from the biotrophic to the necrotrophic stage. FEMS Microbiol. Lett. 2007, 268, 1–8. [Google Scholar] [CrossRef]
- Gentile, A.C. Carbohydrate metabolism and oxalic acid synthesis by Botrytis cinerea. Plant Physiol. 1954, 29, 257–261. [Google Scholar] [CrossRef]
- Sasanuma, I.; Suzuki, T. Effect of calcium on cell-wall degrading enzymes of Botrytis cinerea. Biosci. Biotechnol. Biochem. 2016, 80, 1730–1736. [Google Scholar] [CrossRef]
- Xiao, C.L. Postharvest fruit rots in d’Anjou pears caused by Botrytis cinerea, Potebniamyces pyri, and Sphaeropsis pyriputrescens. Plant Heal. Prog. 2006, 7, 40. [Google Scholar] [CrossRef]
- Araújo, A.E.; Maffia, L.A.; Mizubuti, E.S.G.; Alfenas, A.C.; De Capdeville, G.; Grossi, J.A.S. Survival of Botrytis cinerea as mycelium in rose crop debris and as sclerotia in soil. Fitopatol. Bras. 2005, 30, 516–521. [Google Scholar] [CrossRef][Green Version]
- Backhouse, D.; Willetts, H.J. A histochemical study of sclerotia of Botrytis cinerea and Botrytis fabae. Can. J. Microbiol. 1984, 30, 171–178. [Google Scholar] [CrossRef]
- Rodríguez-García, C.; Medina, V.; Alonso, A.; Ayllón, M.A. Mycoviruses of Botrytis cinerea isolates from different hosts. Ann. Appl. Biol. 2013, 164, 46–61. [Google Scholar] [CrossRef]
- Weiberg, A.; Wang, M.; Lin, F.-M.; Zhao, H.; Zhang, Z.; Kaloshian, I.; Huang, H.-D.; Jin, H. Fungal small RNAs suppress plant immunity by hijacking Host RNA interference pathways. Science 2013, 342, 118–123. [Google Scholar] [CrossRef]
- Qushim, B.; Wu, F.; Guan, Z.; Peres, N. The economic impact of botrytis fruit rot on strawberry production in Florida. In Proceedings of the 2018 Annual Meeting, Jacksonville, FL, USA, 2–6 February 2018. [Google Scholar]
- Dean, R.; Van Kan, J.A.L.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef]
- Nishimoto, R. Global trends in the crop protection industry. J. Pestic. Sci. 2019, 44, 141–147. [Google Scholar] [CrossRef]
- Droby, S.; Wisniewski, M.; Macarisin, D.; Wilson, C. Twenty years of postharvest biocontrol research: Is it time for a new paradigm? Postharvest Biol. Technol. 2009, 52, 137–145. [Google Scholar] [CrossRef]
- Katan, T. Resistance to 3,5-dichlorophenyl-N-cyclic imide (’dicarboximide’) fungicides in the grey mould pathogen Botrytis cinerea on protected crops. Plant Pathol. 1982, 31, 133–141. [Google Scholar] [CrossRef]
- Gubler, W.D. Control of Botrytis bunch rot of grape with canopy management. Plant Dis. 1987, 71, 599. [Google Scholar] [CrossRef]
- Naegele, R.P. Evaluation of host resistance to Botrytis bunch rot in Vitis spp. and its correlation with Botrytis leaf spot. HortScience 2018, 53, 204–207. [Google Scholar] [CrossRef]
- Kan, J.A.L.V.; Stassen, J.H.M.; Mosbach, A.; Van Der Lee, T.A.J.; Faino, L.; Farmer, A.D.; Papasotiriou, D.G.; Zhou, S.; Seidl, M.F.; Cottam, E.; et al. A gapless genome sequence of the fungus Botrytis cinerea. Mol. Plant Pathol. 2016, 18, 75–89. [Google Scholar] [CrossRef] [PubMed]
- Shao, W.; Zhang, Y.; Wang, J.; Lv, C.; Chen, C. BcMtg2 is required for multiple stress tolerance, vegetative development and virulence in Botrytis cinerea. Sci. Rep. 2016, 6, 28673. [Google Scholar] [CrossRef] [PubMed]
- Staats, M.; Van Kan, J.A.L. Genome update of Botrytis cinerea Strains B05.10 and T4. Eukaryot. Cell 2012, 11, 1413–1414. [Google Scholar] [CrossRef] [PubMed]
- Porquier, A.; Moraga, J.; Morgant, G.; Dalmais, B.; Simon, A.; Sghyer, H.; Collado, I.G.; Viaud, M. Botcinic acid biosynthesis in Botrytis cinerea relies on a subtelomeric gene cluster surrounded by relics of transposons and is regulated by the Zn2Cys6 transcription factor BcBoa13. Curr. Genet. 2019, 65, 965–980. [Google Scholar] [CrossRef]
- Bokor, A.A.; Van Kan, J.A.; Poulter, R.T. Sexual mating of Botrytis cinerea illustrates PRP8 intein HEG activity. Fungal Genet. Biol. 2010, 47, 392–398. [Google Scholar] [CrossRef]
- Blanco-Ulate, B.; Morales-Cruz, A.; Amrine, K.C.H.; Labavitch, J.M.; Powell, A.L.T.; Cantu, D. Genome-wide transcriptional profiling of Botrytis cinerea genes targeting plant cell walls during infections of different hosts. Front. Plant Sci. 2014, 5, 435. [Google Scholar] [CrossRef] [PubMed]
- Atwell, S.; Corwin, J.A.; Soltis, N.E.; Subedy, A.; Denby, K.; Kliebenstein, D.J. Whole genome resequencing of Botrytis cinerea isolates identifies high levels of standing diversity. Front. Microbiol. 2015, 6, 996. [Google Scholar] [CrossRef]
- Lamb, B.C.; Mandaokar, S.; Bahsoun, B.; Grishkan, I.; Nevo, E. Differences in spontaneous mutation frequencies as a function of environmental stress in soil fungi at “Evolution Canyon,” Israel. Proc. Natl. Acad. Sci. USA 2008, 105, 5792–5796. [Google Scholar] [CrossRef]
- Leroch, M.; Kleber, A.; Silva, E.; Coenen, T.; Koppenhöfer, D.; Shmaryahu, A.; Valenzuela, P.D.T.; Hahn, M. Transcriptome profiling of Botrytis cinerea conidial germination reveals upregulation of infection-related genes during the prepenetration stage. Eukaryot. Cell 2013, 12, 614–626. [Google Scholar] [CrossRef]
- Zhang, W.; Corwin, J.A.; Copeland, D.; Feusier, J.; Eshbaugh, R.; Cook, D.E.; Atwell, S.; Kliebenstein, D.J. Plant-necrotroph co-transcriptome networks illuminate a metabolic battlefield. eLife 2019, 8. [Google Scholar] [CrossRef]
- Choi, J.; Park, J.; Kim, D.; Jung, K.; Kang, S.; Lee, Y.-H. Fungal secretome database: Integrated platform for annotation of fungal secretomes. BMC Genom. 2010, 11, 105. [Google Scholar] [CrossRef]
- González-Fernández, R.; Valero-Galván, J.; Gómez-Gálvez, F.J.; Jorrín-Novo, J.V. Unraveling the in vitro secretome of the phytopathogen Botrytis cinerea to understand the interaction with its hosts. Front. Plant Sci. 2015, 6, 839. [Google Scholar] [CrossRef] [PubMed]
- Harren, K.; Tudzynski, B. Cch1 and Mid1 are functionally required for vegetative growth under low-calcium conditions in the phytopathogenic ascomycete Botrytis cinerea. Eukaryot. Cell 2013, 12, 712–724. [Google Scholar] [CrossRef]
- Nguyen, Q.B.; Kadotani, N.; Kasahara, S.; Tosa, Y.; Mayama, S.; Nakayashiki, H. Systematic functional analysis of calcium-signalling proteins in the genome of the rice-blast fungus, Magnaporthe oryzae, using a high-throughput RNA-silencing system. Mol. Microbiol. 2008, 68, 1348–1365. [Google Scholar] [CrossRef]
- Bormann, J.; Tudzynski, P. Deletion of Mid1, a putative stretch-activated calcium channel in Claviceps purpurea, affects vegetative growth, cell wall synthesis and virulence. Microbiology 2009, 155, 3922–3933. [Google Scholar] [CrossRef][Green Version]
- Gronover, C.S.; Kasulke, D.; Tudzynski, P.; Tudzynski, B. The role of G protein alpha subunits in the infection process of the gray mold fungus Botrytis cinerea. Mol. Plant-Microbe Interact. 2001, 14, 1293–1302. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Dean, R.A. G Protein α subunit genes control growth, development, and pathogenicity of Magnaporthe grisea. Mol. Plant-Microbe Interact. 1997, 10, 1075–1086. [Google Scholar] [CrossRef]
- Zhang, Z.; Qin, G.; Li, B.; Tian, S. Knocking out Bcsas1 in Botrytis cinerea impacts growth, development, and secretion of extracellular proteins, which decreases virulence. Mol. Plant-Microbe Interact. 2014, 27, 590–600. [Google Scholar] [CrossRef]
- Dub, A.M.; Kokkelink, L.; Tudzynski, B.; Tudzynski, P.; Sharon, A. Involvement of Botrytis cinerea small GTPases BcRAS1 and BcRAC in differentiation, virulence, and the cell cycle. Eukaryot. Cell 2013, 12, 1609–1618. [Google Scholar] [CrossRef]
- Müller, N.; Leroch, M.; Schumacher, J.; Zimmer, D.; Könnel, A.; Klug, K.; Leisen, T.; Scheuring, D.; Sommer, F.K.; Mühlhaus, T.; et al. Investigations on VELVET regulatory mutants confirm the role of host tissue acidification and secretion of proteins in the pathogenesis of Botrytis cinerea. New Phytol. 2018, 219, 1062–1074. [Google Scholar] [CrossRef]
- Yang, Q.; Chen, Y.; Ma, Z. Involvement of BcVeA and BcVelB in regulating conidiation, pigmentation and virulence in Botrytis cinerea. Fungal Genet. Biol. 2013, 50, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Sharma, E.; Tayal, P.; Anand, G.; Mathur, P.; Kapoor, R. Functional analysis of diacylglycerol O-acyl transferase 2 gene to decipher its role in virulence of Botrytis cinerea. Curr. Genet. 2017, 64, 443–457. [Google Scholar] [CrossRef]
- Morcx, S.; Kunz, C.; Choquer, M.; Assié, S.; Blondet, E.; Simond-Côte, E.; Gajek, K.; Chapeland-Leclerc, F.; Expert, D.; Soulie, M. Disruption of Bcchs4, Bcchs6 or Bcchs7 chitin synthase genes in Botrytis cinerea and the essential role of class VI chitin synthase (Bcchs6). Fungal Genet. Biol. 2013, 52, 1–8. [Google Scholar] [CrossRef]
- Yin, Y.; Wu, S.; Chui, C.; Ma, T.; Jiang, H.; Hahn, M.; Ma, Z. The MAPK kinase BcMkk1 suppresses oxalic acid biosynthesis via impeding phosphorylation of BcRim15 by BcSch9 in Botrytis cinerea. PLoS Pathog. 2018, 14, e1007285. [Google Scholar] [CrossRef]
- Viefhues, A.; Heller, J.; Temme, N.; Tudzynski, P. Redox systems in Botrytis cinerea: Impact on development and virulence. Mol. Plant-Microbe Interact. 2014, 27, 858–874. [Google Scholar] [CrossRef] [PubMed]
- Patel, R.M.; Van Kan, J.A.L.; Bailey, A.M.; Foster, G. Inadvertent gene silencing of argininosuccinate synthase (bcass1) in Botrytis cinerea by the pLOB1 vector system. Mol. Plant Pathol. 2010, 11, 613–624. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, J.; De Larrinoa, I.F.; Tudzynski, B. Calcineurin-responsive zinc finger transcription factor CRZ1 of Botrytis cinerea is required for growth, development, and full virulence on bean plants. Eukaryot. Cell 2008, 7, 584–601. [Google Scholar] [CrossRef] [PubMed]
- Siegmund, U.; Marschall, R.; Tudzynski, P. BcNoxD, a putative ER protein, is a new component of the NADPH oxidase complex in Botrytis cinerea. Mol. Microbiol. 2014, 95, 988–1005. [Google Scholar] [CrossRef]
- Zhang, L.; Van Kan, J.A.L. Botrytis cinereamutants deficient ind-galacturonic acid catabolism have a perturbed virulence on Nicotiana benthamiana and Arabidopsis, but not on tomato. Mol. Plant Pathol. 2012, 14, 19–29. [Google Scholar] [CrossRef]
- Hou, J.; Feng, H.; Chang, H.; Liu, Y.; Li, G.; Yang, S.; Sun, C.; Zhang, M.; Yuan, Y.; Sun, J.; et al. The H3K4 demethylase Jar1 orchestrates ROS production and expression of pathogenesis-related genes to facilitate Botrytis cinerea virulence. New Phytol. 2019, 225, 930–947. [Google Scholar] [CrossRef]
- Harren, K.; Schumacher, J.; Tudzynski, B. The Ca2+/calcineurin-dependent signaling pathway in the gray mold Botrytis cinerea: The role of calcipress in in modulating calcineurin activity. PLoS ONE 2012, 7, e41761. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.; Liu, N.; Sang, C.; Shi, D.; Zhou, M.; Chen, C.-J.; Qin, Q.; Chen, W. The autophagy gene BcATG8 regulates the vegetative differentiation and pathogenicity of Botrytis cinerea. Appl. Environ. Microbiol. 2018, 84, e02455-17. [Google Scholar] [CrossRef]
- Schumacher, J. DHN melanin biosynthesis in the plant pathogenic fungus Botrytis cinerea is based on two developmentally regulated key enzyme (PKS)-encoding genes. Mol. Microbiol. 2015, 99, 729–748. [Google Scholar] [CrossRef]
- Segmüller, N.; Kokkelink, L.; Giesbert, S.; Odinius, D.; Van Kan, J.; Tudzynski, P. NADPH oxidases are involved in differentiation and pathogenicity in Botrytis cinerea. Mol. Plant-Microbe Interact. 2008, 21, 808–819. [Google Scholar] [CrossRef]
- Shao, W.; Yang, Y.; Zhang, Y.; Lv, C.; Ren, W.; Chen, C. Involvement of BcStr2 in methionine biosynthesis, vegetative differentiation, multiple stress tolerance and virulence in Botrytis cinerea. Mol. Plant Pathol. 2015, 17, 438–447. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Ren, W.; Li, F.; Chen, C.-J.; Ma, Z. Involvement of the cysteine protease BcAtg4 in development and virulence of Botrytis cinerea. Curr. Genet. 2018, 65, 293–300. [Google Scholar] [CrossRef]
- Nafisi, M.; Stranne, M.; Zhang, L.; Van Kan, J.A.L.; Sakuragi, Y. The endo-arabinanase BcAra1 is a novel host-specific virulence factor of the necrotic fungal phytopathogen Botrytis cinerea. Mol. Plant-Microbe Interact. 2014, 27, 781–792. [Google Scholar] [CrossRef] [PubMed]
- Terhem, R.B.; Van Kan, J.A.L. Functional analysis of hydrophobin genes in sexual development of Botrytis cinerea. Fungal Genet. Biol. 2014, 71, 42–51. [Google Scholar] [CrossRef]
- Mosbach, A.; Leroch, M.; Mendgen, K.; Hahn, M. Lack of evidence for a role of hydrophobins in conferring surface hydrophobicity to conidia and hyphae of Botrytis cinerea. BMC Microbiol. 2011, 11, 10. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, H.; Qin, G.; He, C.; Li, B.; Tian, S. The MADS-Box transcription factor Bcmads1 is required for growth, sclerotia production and pathogenicity of Botrytis cinerea. Sci. Rep. 2016, 6, 33901. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Yin, D.; Yin, Y.; Cao, Y.; Ma, Z. The response regulator BcSkn7 is required for vegetative differentiation and adaptation to oxidative and osmotic stresses in Botrytis cinerea. Mol. Plant Pathol. 2014, 16, 276–287. [Google Scholar] [CrossRef]
- Zhou, Y.; Yang, L.; Wu, M.; Chen, W.; Li, G.; Zhang, J. A single-nucleotide deletion in the transcription factor gene bcsmr1 causes sclerotial-melanogenesis deficiency in Botrytis cinerea. Front. Microbiol. 2017, 8, 2492. [Google Scholar] [CrossRef]
- Frías, M.; González, C.; Brito, N. BcSpl1, a cerato-platanin family protein, contributes to Botrytis cinerea virulence and elicits the hypersensitive response in the host. New Phytol. 2011, 192, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.Q.; Jiang, J.; Mayr, C.; Hahn, M.; Ma, Z. Involvement of two type 2C protein phosphatases BcPtc1 and BcPtc3 in the regulation of multiple stress tolerance and virulence of Botrytis cinerea. Environ. Microbiol. 2013, 15, 2696–2711. [Google Scholar] [CrossRef] [PubMed]
- Minz-Dub, A.; Sharon, A. The Botrytis cinerea PAK kinase BcCla4 mediates morphogenesis, growth and cell cycle regulating processes downstream of BcRac. Mol. Microbiol. 2017, 104, 487–498. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Salas, O.; Bowler, K.; Oren-Young, L.; Bar-Peled, M.; Sharon, A. Genetic alteration of UDP-rhamnose metabolism in Botrytis cinerealeads to the accumulation of UDP-KDG that adversely affects development and pathogenicity. Mol. Plant Pathol. 2016, 18, 263–275. [Google Scholar] [CrossRef]
- Rolke, Y.; Liu, S.; Quidde, T.; Williamson, B.; Schouten, A.; Weltring, K.-M.; Siewers, V.; Tenberge, K.B.; Tudzynski, B.; Tudzynski, P. Functional analysis of H2O2-generating systems in Botrytis cinerea: The major Cu-Zn-superoxide dismutase (BCSOD1) contributes to virulence on French bean, whereas a glucose oxidase (BCGOD1) is dispensable. Mol. Plant Pathol. 2004, 5, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Tayal, P.; Raj, S.; Sharma, E.; Kumar, M.; Dayaman, V.; Verma, N.; Jogawat, A.; Dua, M.; Kapoor, R.; Johri, A.K. A Botrytis cinerea KLP-7 kinesin acts as a virulence determinant during plant infection. Sci. Rep. 2017, 7, 10664. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Zhang, J.; Hu, J.; Wang, X.; Lv, B.; Liang, W. Involvement of BcYak1 in the regulation of vegetative differentiation and adaptation to oxidative stress of Botrytis cinerea. Front. Microbiol. 2018, 9, 281. [Google Scholar] [CrossRef]
- Brandhoff, B.; Simon, A.; Dornieden, A.; Schumacher, J. Regulation of conidiation in Botrytis cinerea involves the light-responsive transcriptional regulators BcLTF3 and BcREG1. Curr. Genet. 2017, 63, 931–949. [Google Scholar] [CrossRef]
- Guan, W.; Feng, J.; Wang, R.; Ma, Z.; Wang, W.-X.; Wang, K.; Zhu, T. Functional analysis of the exocyst subunit BcExo70 in Botrytis cinerea. Curr. Genet. 2019, 66, 85–95. [Google Scholar] [CrossRef]
- Zhang, C.; He, Y.; Zhu, P.; Chen, L.; Wang, Y.; Ni, B.; Xu, L. Loss of bcbrn1 and bcpks13 in Botrytis cinerea not only blocks melanization but also increases vegetative growth and virulence. Mol. Plant-Microbe Interact. 2015, 28, 1091–1101. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, J.; Kokkelink, L.; Huesmann, C.; Jimenez-Teja, D.; Collado, I.G.; Barakat, R.; Tudzynski, P.; Tudzynski, B. The cAMP-dependent signaling pathway and its role in conidial germination, growth, and virulence of the gray mold Botrytis cinerea. Mol. Plant-Microbe Interact. 2008, 21, 1443–1459. [Google Scholar] [CrossRef]
- Feng, H.; Li, G.; Du, S.; Yang, S.; Li, X.-Q.; De Figueiredo, P.; Qin, Q.-M. The septin protein Sep4 facilitates host infection by plant fungal pathogens via mediating initiation of infection structure formation. Environ. Microbiol. 2017, 19, 1730–1749. [Google Scholar] [CrossRef] [PubMed]
- Saitoh, Y.; Izumitsu, K.; Morita, A.; Tanaka, C. A copper-transporting ATPase BcCCC2 is necessary for pathogenicity of Botrytis cinerea. Mol. Genet. Genom. 2010, 284, 33–43. [Google Scholar] [CrossRef]
- Giesbert, S.; Siegmund, U.; Schumacher, J.; Kokkelink, L.; Tudzynski, P. Functional analysis of BcBem1 and its interaction partners in Botrytis cinerea: Impact on differentiation and virulence. PLoS ONE 2014, 9, e95172. [Google Scholar] [CrossRef] [PubMed]
- Viaud, M.; Brunet-Simon, A.; Brygoo, Y.; Pradier, J.-M.; Levis, C. Cyclophilin A and calcineurin functions investigated by gene inactivation, cyclosporin A inhibition and cDNA arrays approaches in the phytopathogenic fungus Botrytis cinerea. Mol. Microbiol. 2003, 50, 1451–1465. [Google Scholar] [CrossRef] [PubMed]
- Marschall, R.; Tudzynski, P. BcIqg1, a fungal IQGAP homolog, interacts with NADPH oxidase, MAP kinase and calcium signaling proteins and regulates virulence and development in Botrytis cinerea. Mol. Microbiol. 2016, 101, 281–298. [Google Scholar] [CrossRef]
- Vela-Corcía, D.; Srivastava, D.A.; Dafa-Berger, A.; Rotem, N.; Barda, O.; Levy, M. MFS transporter from Botrytis cinerea provides tolerance to glucosinolate-breakdown products and is required for pathogenicity. Nat. Commun. 2019, 10, 1–11. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, J.; Li, G.; Zhang, M.; Zhang, Y.; Wang, Y.; Hou, J.; Yang, S.; Sun, J.; Qin, Q. A novel Botrytis cinerea- specific gene BcHBF1 enhances virulence of the grey mould fungus via promoting host penetration and invasive hyphal development. Mol. Plant Pathol. 2019, 20, 731–747. [Google Scholar] [CrossRef]
- Lin, Z.; Wu, J.; Jamieson, P.A.; Zhang, C.Q. Alternative oxidase is involved in the pathogenicity, development, and oxygen stress response of Botrytis cinerea. Phytopathology 2019, 109, 1679–1688. [Google Scholar] [CrossRef]
- Choquer, M.; Robin, G.; Le PãªCheur, P.; Giraud, C.; Levis, C.; Viaud, M. Ku70orKu80deficiencies in the fungus Botrytis cinerea facilitate targeting of genes that are hard to knock out in a wild-type context. FEMS Microbiol. Lett. 2008, 289, 225–232. [Google Scholar] [CrossRef]
- An, B.; Li, B.; Qin, G.; Tian, S. Function of small GTPase Rho3 in regulating growth, conidiation and virulence of Botrytis cinerea. Fungal Genet. Biol. 2015, 75, 46–55. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Hernández, A.; González, M.; González, C.; Van Kan, J.A.L.; Brito, N. BcSUN1, a B. cinerea SUN-Family Protein, Is Involved in Virulence. Front. Microbiol. 2017, 8, 35. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.; Zhang, Z.; Shao, W.; Yang, Y.; Zhou, M.; Chen, C.-J. The autophagy-related geneBcATG1is involved in fungal development and pathogenesis in Botrytis cinerea. Mol. Plant Pathol. 2016, 18, 238–248. [Google Scholar] [CrossRef] [PubMed]
- Plaza, V.; Lagües, Y.; Carvajal, M.; Pérez-García, L.A.; Mora-Montes, H.M.; Canessa, P.; Larrondo, L.F.; Castillo, L. bcpmr1 encodes a P-type Ca2+/Mn2+-ATPase mediating cell-wall integrity and virulence in the phytopathogen Botrytis cinerea. Fungal Genet. Biol. 2015, 76, 36–46. [Google Scholar] [CrossRef]
- Li, H.; Zhang, Z.; He, C.; Qin, G.; Tian, S. Comparative proteomics reveals the potential targets of BcNoxR, a putative regulatory subunit of NADPH oxidase of Botrytis cinerea. Mol. Plant-Microbe Interact. 2016, 29, 990–1003. [Google Scholar] [CrossRef]
- Schumacher, J.; Studt, L.; Tudzynski, P. The putative H3K36 demethylase BcKDM1 affects virulence, stress responses and photomorphogenesis in Botrytis cinerea. Fungal Genet. Biol. 2019, 123, 14–24. [Google Scholar] [CrossRef]
- Shlezinger, N.; Doron, A.; Sharon, A. Apoptosis-like programmed cell death in the grey mould fungus Botrytis cinerea: Genes and their role in pathogenicity. Biochem. Soc. Trans. 2011, 39, 1493–1498. [Google Scholar] [CrossRef]
- An, B.; Li, B.; Li, H.; Zhang, Z.; Qin, G.; Tian, S. Aquaporin8 regulates cellular development and reactive oxygen species production, a critical component of virulence in Botrytis cinerea. New Phytol. 2015, 209, 1668–1680. [Google Scholar] [CrossRef]
- Harren, K.; Brandhoff, B.; Knödler, M.; Tudzynski, B. The high-affinity phosphodiesterase BcPde2 has impact on growth, differentiation and virulence of the phytopathogenic ascomycete Botrytis cinerea. PLoS ONE 2013, 8, e78525. [Google Scholar] [CrossRef]
- Yang, Q.; Yu, F.; Yin, Y.; Ma, Z. Involvement of protein tyrosine phosphatases BcPtpA and BcPtpB in regulation of vegetative development, virulence and multi-stress tolerance in Botrytis cinerea. PLoS ONE 2013, 8, e61307. [Google Scholar] [CrossRef][Green Version]
- Yan, L.; Yang, Q.; Jiang, J.; Michailides, T.J.; Ma, Z. Involvement of a putative response regulator Brrg-1 in the regulation of sporulation, sensitivity to fungicides, and osmotic stress in Botrytis cinerea. Appl. Microbiol. Biotechnol. 2010, 90, 215–226. [Google Scholar] [CrossRef]
- Valette-Collet, O.; Cimerman, A.; Reignault, P.; Levis, C.; Boccara, M. Disruption of Botrytis cinerea pectin methylesterase gene Bcpme1 reduces virulence on several host plants. Mol. Plant-Microbe Interact. 2003, 16, 360–367. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Xie, J.; Fu, Y.; Jiāng, D.; Chen, T.; Cheng, J. The subtilisin-like protease Bcser2 Affects the sclerotial formation, conidiation and virulence of Botrytis cinerea. Int. J. Mol. Sci. 2020, 21, 603. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.; Sang, C.; Shi, D.; Song, X.; Zhou, M.; Chen, C.-J. Ubiquitin-like activating enzymes BcAtg3 and BcAtg7 participate in development and pathogenesis of Botrytis cinerea. Curr. Genet. 2018, 64, 919–930. [Google Scholar] [CrossRef]
- Ren, W.; Liu, N.; Yang, Y.; Yang, Q.; Chen, C.-J.; Gao, Q. The sensor proteins BcSho1 and BcSln1 are involved in, though not essential to, vegetative differentiation, pathogenicity and osmotic stress tolerance in Botrytis cinerea. Front. Microbiol. 2019, 10, 328. [Google Scholar] [CrossRef]
- Antal, Z.; Rascle, C.; Cimerman, A.; Viaud, M.; Billon-Grand, G.; Choquer, M.; Bruel, C. The homeobox BcHOX8 gene in Botrytis cinerea regulates vegetative growth and morphology. PLoS ONE 2012, 7, e48134. [Google Scholar] [CrossRef]
- Yang, Y.; Yang, X.; Dong, Y.; Qiu, D. The Botrytis cinerea Xylanase BcXyl1 modulates plant immunity. Front. Microbiol. 2018, 9, 2535. [Google Scholar] [CrossRef]
- Rui, O.; Hahn, M. The Slt2-type MAP kinase Bmp3 of Botrytis cinerea is required for normal saprotrophic growth, conidiation, plant surface sensing and host tissue colonization. Mol. Plant Pathol. 2007, 8, 173–184. [Google Scholar] [CrossRef]
- Jonkers, W.; Van Kan, J.A.L.; Tijm, P.; Lee, Y.-W.; Tudzynski, P.; Rep, M.; Michielse, C.B. The FRP1 F-box gene has different functions in sexuality, pathogenicity and metabolism in three fungal pathogens. Mol. Plant Pathol. 2011, 12, 548–563. [Google Scholar] [CrossRef]
- Rui, O.; Hahn, M. The Botrytis cinerea hexokinase, Hxk1, but not the glucokinase, Glk1, is required for normal growth and sugar metabolism, and for pathogenicity on fruits. Microbiol. 2007, 153, 2791–2802. [Google Scholar] [CrossRef]
- Siegmund, U.; Heller, J.; Van Kan, J.A.L.; Tudzynski, P. The NADPH oxidase complexes in Botrytis cinerea: Evidence for a close association with the ER and the Tetraspanin Pls1. PLoS ONE 2013, 8, e55879. [Google Scholar] [CrossRef]
- Reis, H.; Pfiffi, S.; Hahn, M. Molecular and functional characterization of a secreted lipase from Botrytis cinerea. Mol. Plant Pathol. 2005, 6, 257–267. [Google Scholar] [CrossRef]
- Yan, L.; Yang, Q.Q.; Sundin, G.W.; Li, H.; Ma, Z. The mitogen-activated protein kinase kinase BOS5 is involved in regulating vegetative differentiation and virulence in Botrytis cinerea. Fungal Genet. Biol. 2010, 47, 753–760. [Google Scholar] [CrossRef]
- Zhang, H.; Li, Y.; Dickman, M.B.; Wang, Z. Cytoprotective co-chaperone BcBAG1 is a component for fungal development, virulence, and Unfolded Protein Response (UPR) of Botrytis cinerea. Front. Microbiol. 2019, 10, 685. [Google Scholar] [CrossRef]
- Zhu, W.; Wei, W.; Wu, Y.; Zhou, Y.; Peng, F.; Zhang, S.; Chen, P.; Xu, X. BcCFEM1, a CFEM domain-containing protein with putative GPI-anchored site, is involved in pathogenicity, conidial production, and stress tolerance in Botrytis cinerea. Front. Microbiol. 2017, 8, 1807. [Google Scholar] [CrossRef]
- Rascle, C.; Dieryckx, C.; Dupuy, J.-W.; Muszkieta, L.; Souibgui, E.; Droux, M.; Bruel, C.; Girard, V.; Poussereau, N. The pH regulator PacC: A host-dependent virulence factor in Botrytis cinerea. Environ. Microbiol. Rep. 2018, 10, 555–568. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, X.; Zhao, Y.; Cheng, J.; Xie, J.; Fu, Y.; Jiang, D.; Chen, T. Histone H3 lysine 9 methyltransferase DIM5 is required for the development and virulence of Botrytis cinerea. Front. Microbiol. 2016, 7, 1289. [Google Scholar] [CrossRef]
- Yang, Q.; Yan, L.; Gu, Q.; Ma, Z. The mitogen-activated protein kinase kinase kinase BcOs4 is required for vegetative differentiation and pathogenicity in Botrytis cinerea. Appl. Microbiol. Biotechnol. 2012, 96, 481–492. [Google Scholar] [CrossRef]
- Cui, Z.; Gao, N.; Wang, Q.; Ren, Y.; Wang, K.; Zhu, T. BcMctA, a putative monocarboxylate transporter, is required for pathogenicity in Botrytis cinerea. Curr. Genet. 2015, 61, 545–553. [Google Scholar] [CrossRef]
- Meléndez, H.G.; Billon-Grand, G.; Fevre, M.; Mey, G. Role of the Botrytis cinerea FKBP12 ortholog in pathogenic development and in sulfur regulation. Fungal Genet. Biol. 2009, 46, 308–320. [Google Scholar] [CrossRef] [PubMed]
- Shao, W.; Lv, C.; Zhang, Y.; Wang, J.; Chen, C.-J. Involvement of BcElp4 in vegetative development, various environmental stress response and virulence of Botrytis cinerea. Microb. Biotechnol. 2017, 10, 886–895. [Google Scholar] [CrossRef]
- Cao, S.-N.; Yuan, Y.; Qin, Y.; Zhang, M.-Z.; De Figueiredo, P.; Li, G.-H.; Qin, Q.-M. The pre-rRNA processing factor Nop53 regulates fungal development and pathogenesis via mediating production of reactive oxygen species. Environ. Microbiol. 2018, 20, 1531–1549. [Google Scholar] [CrossRef]
- Giesbert, S.; Schumacher, J.; Kupas, V.; Espino, J.; Segmüller, N.; Haeuser-Hahn, I.; Schreier, P.H.; Tudzynski, P. Identification of pathogenesis-associated genes by T-DNA–mediated insertional mutagenesis in Botrytis cinerea: A Type 2A phosphoprotein phosphatase and an SPT3 transcription factor have significant impact on virulence. Mol. Plant-Microbe Interact. 2012, 25, 481–495. [Google Scholar] [CrossRef]
- Zhang, K.; Yuan, X.; Zang, J.; Wang, M.; Zhao, F.; Li, P.; Cao, H.; Han, J.; Xing, J.; Dong, J. The kynurenine 3-monooxygenase encoding gene, BcKMO, is involved in the growth, development, and pathogenicity of Botrytis cinerea. Front. Microbiol. 2018, 9, 1039. [Google Scholar] [CrossRef] [PubMed]
- Siewers, V.; Viaud, M.; Jimenez-Teja, D.; Collado, I.G.; Gronover, C.S.; Pradier, J.-M.; Tudzynski, B.; Tudzynski, P. Functional analysis of the cytochrome P450 monooxygenase gene bcbot1 of Botrytis cinerea indicates that botrydial is a strain-specific virulence factor. Mol. Plant-Microbe Interact. 2005, 18, 602–612. [Google Scholar] [CrossRef]
- Kokkelink, L.; Minz, A.; Al-Masri, M.; Giesbert, S.; Barakat, R.; Sharon, A.; Tudzynski, P. The small GTPase BcCdc42 affects nuclear division, germination and virulence of the gray mold fungus Botrytis cinerea. Fungal Genet. Biol. 2011, 48, 1012–1019. [Google Scholar] [CrossRef]
- González-Rodríguez, V.E.; Garrido, C.; Cantoral, J.M.; Schumacher, J. The F-actin capping protein is required for hyphal growth and full virulence but is dispensable for septum formation in Botrytis cinerea. Fungal Biol. 2016, 120, 1225–1235. [Google Scholar] [CrossRef] [PubMed]
- Kars, I.; Krooshof, G.H.; Wagemakers, L.; Joosten, R.; Benen, J.A.; Van Kan, J.A. Necrotizing activity of five Botrytis cinerea endopolygalacturonases produced in Pichia pastoris. Plant J. 2005, 43, 213–225. [Google Scholar] [CrossRef]
- Schumacher, J.; Simon, A.; Cohrs, K.C.; Viaud, M.; Tudzynski, P. The transcription factor BcLTF1 regulates virulence and light responses in the necrotrophic plant pathogen Botrytis cinerea. PLoS Genet. 2014, 10, e1004040. [Google Scholar] [CrossRef] [PubMed]
- Klimpel, A.; Gronover, C.S.; Williamson, B.; Stewart, J.A.; Tudzynski, B. The adenylate cyclase (BAC) in Botrytis cinerea is required for full pathogenicity. Mol. Plant Pathol. 2002, 3, 439–450. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, J.; Pradier, J.-M.; Simon, A.; Traeger, S.; Moraga, J.; Collado, I.G.; Viaud, M.; Tudzynski, B. Natural variation in the VELVET Gene bcvel1 affects virulence and light-dependent differentiation in Botrytis cinerea. PLoS ONE 2012, 7, e47840. [Google Scholar] [CrossRef]
- Heller, J.; Ruhnke, N.; Espino, J.J.; Massaroli, M.; Collado, I.G.; Tudzynski, P. The mitogen-activated protein kinase BcSak1 of Botrytis cinerea is required for pathogenic development and has broad regulatory functions beyond stress response. Mol. Plant-Microbe Interact. 2012, 25, 802–816. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.-K.; Chang, H.-W.; Liu, Y.; Qin, Y.; Ding, Y.-H.; Wang, L.; Zhao, Y.; Zhang, M.-Z.; Cao, S.-N.; Li, L.-T.; et al. The key gluconeogenic gene PCK1 is crucial for virulence of Botrytis cinerea via initiating its conidial germination and host penetration. Environ. Microbiol. 2018, 20, 1794–1814. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Sun, C.; Liu, Y.; Feng, H.; Chang, H.; Cao, S.; Li, G.; Yang, S.; Hou, J.; Zhu-Salzman, K.; et al. Transcriptome analysis and functional validation reveal a novel gene, BcCGF1, that enhances fungal virulence by promoting infection-related development and host penetration. Mol. Plant Pathol. 2020, 21, 834–853. [Google Scholar] [CrossRef]
- Li, H.; Zhang, Z.; Qin, G.; He, C.; Li, B.; Tian, S. Actin is required for cellular development and virulence of Botrytis cinerea via the mediation of secretory proteins. mSystems 2020, 5, 00732-19. [Google Scholar] [CrossRef]
- García, N.; González, M.A.; González, C.; Brito, N. Simultaneous silencing of xylanase genes in Botrytis cinerea. Front. Plant Sci. 2017, 8, 2174. [Google Scholar] [CrossRef]
- Parsley, T.B.; Segers, G.C.; Nuss, D.L.; Dawe, A.L. Analysis of altered G-protein subunit accumulation in Cryphonectria parasitica reveals a third Gα homologue. Curr. Genet. 2003, 43, 24–33. [Google Scholar] [CrossRef]
- Krystofova, S.; Borkovich, K.A. The heterotrimeric G-protein subunits GNG-1 and GNB-1 Form a Gβγ dimer required for normal female fertility, asexual development, and Gα protein levels in Neurospora crassa. Eukaryot. Cell 2005, 4, 365–378. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Jin, Y.; Wang, G. The role of SCF ubiquitin-ligase complex at the beginning of life. Reprod. Biol. Endocrinol. 2019, 17, 1–9. [Google Scholar] [CrossRef]
- Jonkers, W.; Rodrigues, C.D.A.; Rep, M. Impaired colonization and infection of tomato roots by the Δfrp1 mutant of Fusarium oxysporum correlates with reduced CWDE gene expression. Mol. Plant-Microbe Interact. 2009, 22, 507–518. [Google Scholar] [CrossRef]
- Hunter, T. Protein kinases and phosphatases: The Yin and Yang of protein phosphorylation and signaling. Cell 1995, 80, 225–236. [Google Scholar] [CrossRef]
- Yu, Z.; Armant, O.; Fischer, R. Fungi use the SakA (HogA) pathway for phytochrome-dependent light signalling. Nat. Microbiol. 2016, 1, 16019. [Google Scholar] [CrossRef]
- Yen, C.-L.E.; Stone, S.J.; Koliwad, S.; Harris, C.; Farese, R.V.F., Jr. Thematic review series: Glycerolipids.DGAT enzymes and triacylglycerol biosynthesis. J. Lipid Res. 2008, 49, 2283–2301. [Google Scholar] [CrossRef] [PubMed]
- McDonald, A.E. Alternative oxidase: An inter-kingdom perspective on the function and regulation of this broadly distributed ’cyanide-resistant’ terminal oxidase. Funct. Plant Biol. 2008, 35, 535–552. [Google Scholar] [CrossRef] [PubMed]
- Dordas, C. Role of nutrients in controlling plant diseases in sustainable agriculture. A review. Agron. Sustain. Dev. 2008, 28, 33–46. [Google Scholar] [CrossRef]
- Walters, D.R.; Bingham, I. Influence of nutrition on disease development caused by fungal pathogens: Implications for plant disease control. Ann. Appl. Biol. 2007, 151, 307–324. [Google Scholar] [CrossRef]
- Michielse, C.B.; Becker, M.; Heller, J.; Moraga, J.; Collado, I.G.; Tudzynski, P.; Moraga, J. The Botrytis cinerea Reg1 protein, a putative transcriptional regulator, is required for pathogenicity, conidiogenesis, and the production of secondary metabolites. Mol. Plant-Microbe Interact. 2011, 24, 1074–1085. [Google Scholar] [CrossRef]
- Doss, R.P.; Potter, S.W.; Soeldner, A.H.; Christian, J.K.; Fukunaga, L.E. Adhesion of germlings of Botrytis cinerea. Appl. Environ. Microbiol. 1995, 61, 260–265. [Google Scholar] [CrossRef] [PubMed]
- Espino, J.J.; Gutiérrez-Sánchez, G.; Brito, N.; Shah, P.; Orlando, R.; González, C. The Botrytis cinerea early secretome. Proteomics 2010, 10, 3020–3034. [Google Scholar] [CrossRef]
- González, M.; Brito, N.; González, C. Identification of glycoproteins secreted by wild-type Botrytis cinerea and by protein O-mannosyltransferase mutants. BMC Microbiol. 2014, 14, 254. [Google Scholar] [CrossRef]
- Brito, N.; Espino, J.J.; González, C. The endo-β-1,4-xylanase Xyn11A is required for virulence in Botrytis cinerea. Mol. Plant-Microbe Interact. 2006, 19, 25–32. [Google Scholar] [CrossRef]
- Kulkarni, R.D.; Kelkar, H.S.; Dean, R.A. An eight-cysteine-containing CFEM domain unique to a group of fungal membrane proteins. Trends Biochem. Sci. 2003, 28, 118–121. [Google Scholar] [CrossRef]
- Qiu, L.; Wang, J.-J.; Ying, S.; Feng, M.-G. Wee1 and Cdc25 control morphogenesis, virulence and multistress tolerance of Beauveria bassiana by balancing cell cycle-required cyclin-dependent kinase 1 activity. Environ. Microbiol. 2014, 17, 1119–1133. [Google Scholar] [CrossRef]
- Laprade, L.; Boyartchuk, V.L.; Dietrich, W.F.; Winston, F. Spt3 plays opposite roles in filamentous growth in Saccharomyces cerevisiae and Candida albicans and is required for C. albicans virulence. Genetics 2002, 161, 509–519. [Google Scholar]
- Michielse, C.B.; Van Wijk, R.; Reijnen, L.; Cornelissen, B.J.C.; Rep, M. Insight into the molecular requirements for pathogenicity of Fusarium oxysporum f. sp. lycopersici through large-scale insertional mutagenesis. Genome Biol. 2009, 10, R4. [Google Scholar] [CrossRef]
- Donofrio, N.; Oh, Y.; Lundy, R.; Pan, H.; Brown, D.; Jeong, J.; Coughlan, S.; Mitchell, T.; Dean, R. Global gene expression during nitrogen starvation in the rice blast fungus, Magnaporthe grisea. Fungal Genet. Biol. 2006, 43, 605–617. [Google Scholar] [CrossRef] [PubMed]
- Adamo, J.E.; Moskow, J.J.; Gladfelter, A.S.; Viterbo, D.; Lew, D.J.; Brennwald, P.J. Yeast Cdc42 functions at a late step in exocytosis, specifically during polarized growth of the emerging bud. J. Cell Biol. 2001, 155, 581–592. [Google Scholar] [CrossRef] [PubMed]
- Doss, R.P.; Deisenhofer, J.; Von Nidda, H.-A.K.; Soeldner, A.H.; McGuire, R.P. Melanin in the extracellular matrix of germlings of Botrytis cinerea. Phytochemistry 2003, 63, 687–691. [Google Scholar] [CrossRef]
- Liu, W.; Soulié, M.-C.; Perrino, C.; Fillinger, S. The osmosensing signal transduction pathway from Botrytis cinerea regulates cell wall integrity and MAP kinase pathways control melanin biosynthesis with influence of light. Fungal Genet. Biol. 2011, 48, 377–387. [Google Scholar] [CrossRef]
- Zhou, Y.; Li, N.; Yang, J.; Yang, L.; Wu, M.; Chen, W.; Li, G.; Zhang, J. Contrast between orange-and black-colored sclerotial isolates of Botrytis cinerea: Melanogenesis and ecological fitness. Plant. Dis. 2018, 102, 428–436. [Google Scholar] [CrossRef]
- Schumacher, J.; Simon, A.; Cohrs, K.C.; Traeger, S.; Porquier, A.; Dalmais, B.; Viaud, M.; Tudzynski, B. The VELVET complex in the gray mold fungus Botrytis cinerea: Impact of BcLAE1 on differentiation, secondary metabolism, and virulence. Mol. Plant.-Microbe Interact. 2015, 28, 659–674. [Google Scholar] [CrossRef] [PubMed]
- Leisen, T.; Bietz, F.; Werner, J.; Wegner, A.; Schaffrath, U.; Scheuring, D.; Willmund, F.; Mosbach, A.; Scalliet, G.; Hahn, M. CRISPR/Cas with ribonucleoprotein complexes and transiently selected telomere vectors allows highly efficient marker-free and multiple genome editing in Botrytis cinerea. PLoS Pathog. 2020, 16, e1008326. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheung, N.; Tian, L.; Liu, X.; Li, X. The Destructive Fungal Pathogen Botrytis cinerea—Insights from Genes Studied with Mutant Analysis. Pathogens 2020, 9, 923. https://doi.org/10.3390/pathogens9110923
Cheung N, Tian L, Liu X, Li X. The Destructive Fungal Pathogen Botrytis cinerea—Insights from Genes Studied with Mutant Analysis. Pathogens. 2020; 9(11):923. https://doi.org/10.3390/pathogens9110923
Chicago/Turabian StyleCheung, Nicholas, Lei Tian, Xueru Liu, and Xin Li. 2020. "The Destructive Fungal Pathogen Botrytis cinerea—Insights from Genes Studied with Mutant Analysis" Pathogens 9, no. 11: 923. https://doi.org/10.3390/pathogens9110923
APA StyleCheung, N., Tian, L., Liu, X., & Li, X. (2020). The Destructive Fungal Pathogen Botrytis cinerea—Insights from Genes Studied with Mutant Analysis. Pathogens, 9(11), 923. https://doi.org/10.3390/pathogens9110923






