Mycoplasma bovis in Nordic European Countries: Emergence and Dominance of a New Clone
Abstract
1. Introduction
2. Results
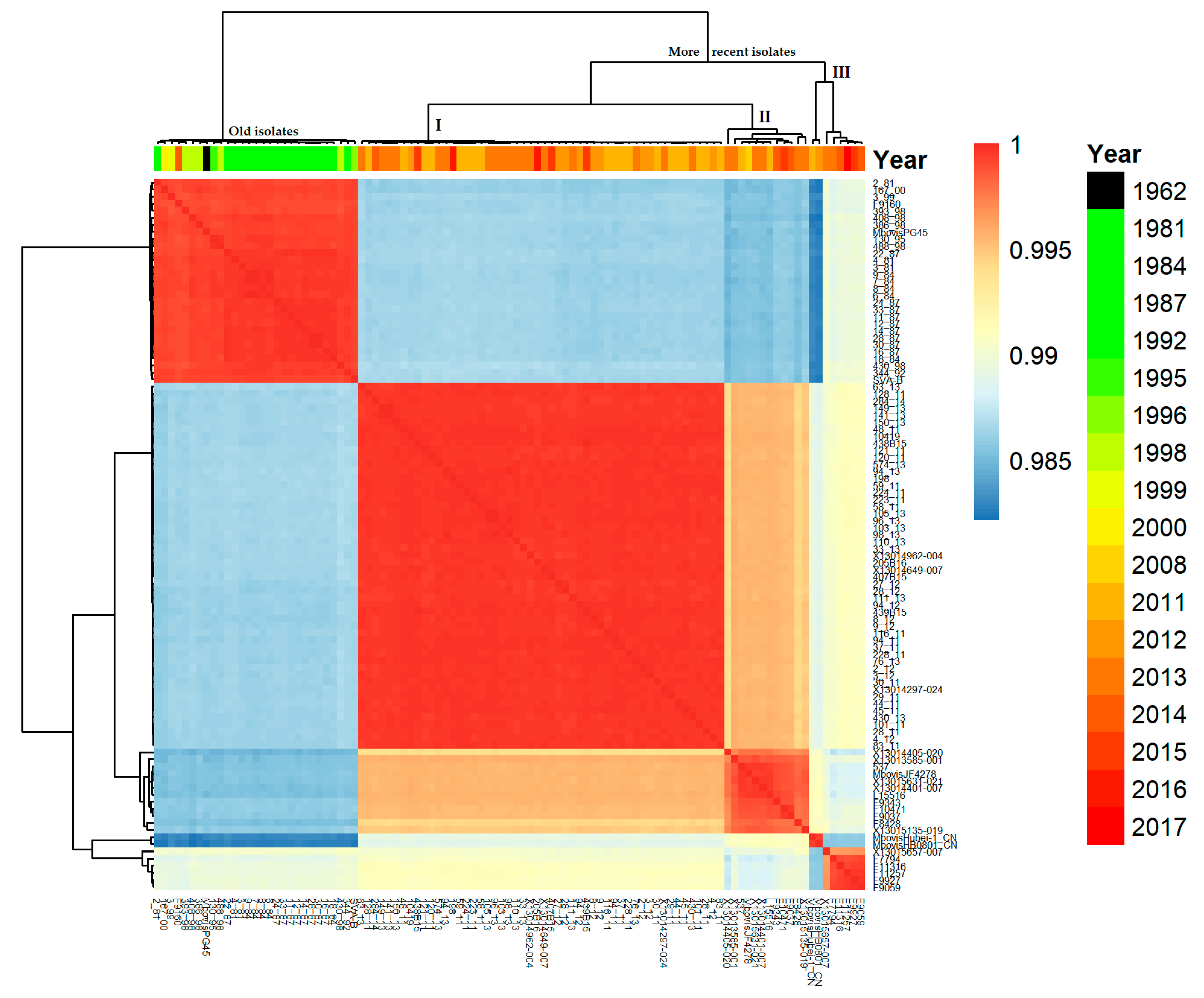
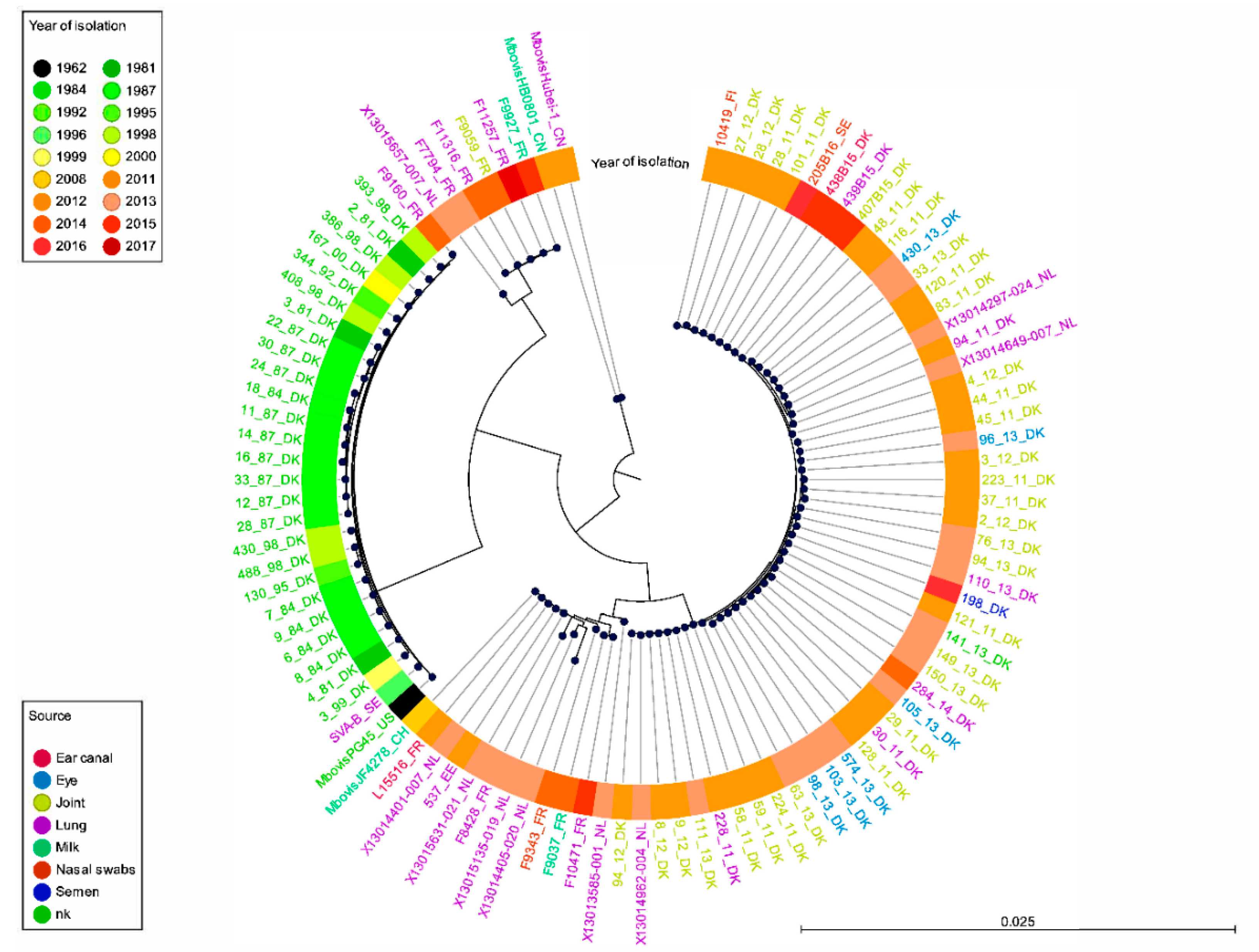
2.1. Genetic Structure of Our M. bovis Population
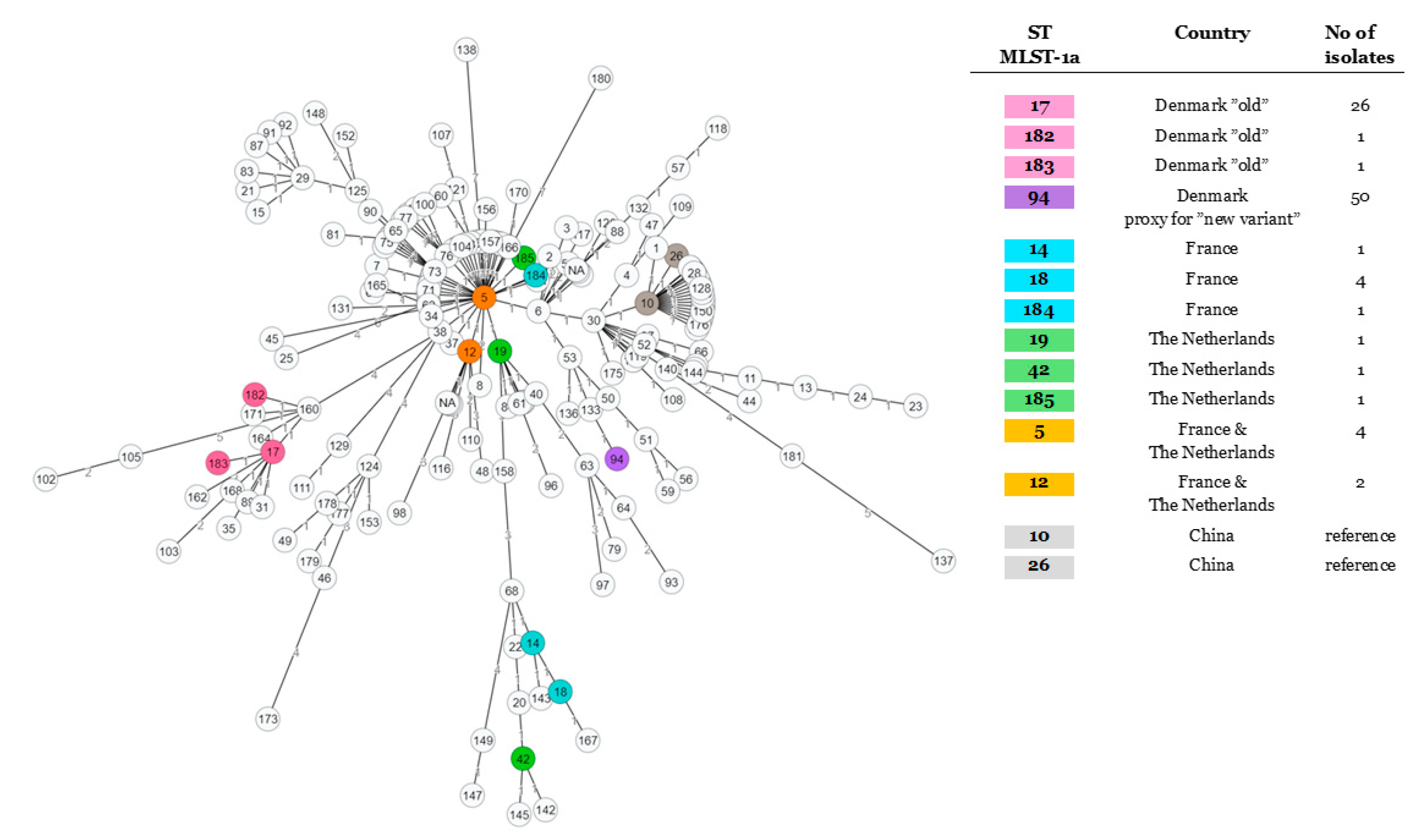
2.2. Studying of the M. bovis Population Structure Using Different Gene-Based Typing Schemes and an Enlarged Worldwide Set of Isolates
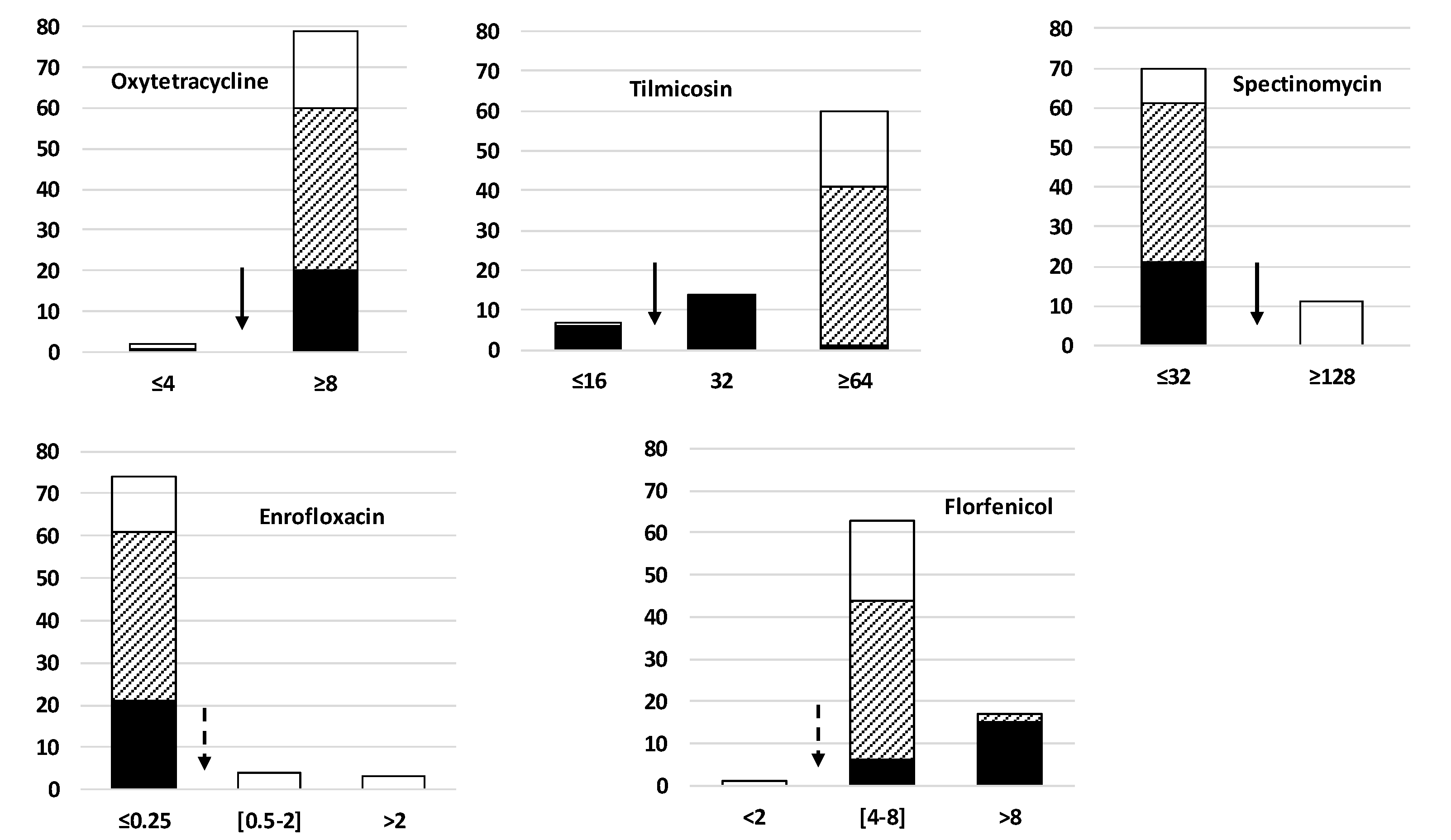
2.3. Diversity of Antimicrobial Resistance Phenotypes in the Study Population
3. Discussion
4. Materials and Methods
4.1. Isolates, Culture, and DNA Extraction
4.2. Sequencing and in Silico Analyses
4.3. Data Availability
4.4. Minimum Inhibitory Concentration (MIC) Assays
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Calcutt, M.J.; Lysnyansky, I.; Sachse, K.; Fox, L.K.; Nicholas, R.A.J.; Ayling, R.D. Gap analysis of Mycoplasma bovis disease, diagnosis and control: An aid to identify future development requirements. Transbound. Emerg. Dis. 2018, 65 (Suppl. 1), 91–109. [Google Scholar] [CrossRef] [PubMed]
- Hale, H.H.; Helmboldt, C.F.; Plastridge, W.N.; Stula, E.F. Bovine mastitis caused by a Mycoplasma species. Cornell Vet. 1962, 52, 582–591. [Google Scholar] [PubMed]
- Nicholas, R.A. Bovine mycoplasmosis: Silent and deadly. Vet. Rec. 2011, 168, 459–462. [Google Scholar] [CrossRef]
- Maunsell, F.P.; Woolums, A.R.; Francoz, D.; Rosenbusch, R.F.; Step, D.L.; Wilson, D.J.; Janzen, E.D. Mycoplasma bovis infections in cattle. J. Vet. Intern. Med. 2011, 25, 772–783. [Google Scholar] [CrossRef]
- Vähänikkilä, N.; Pohjanvirta, T.; Haapala, V.; Simojoki, H.; Soveri, T.; Browning, G.F.; Pelkonen, S.; Wawegama, N.K.; Autio, T. Characterisation of the course of Mycoplasma bovis infection in naturally infected dairy herds. Vet. Microbiol. 2019, 231, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Perez-Casal, J.; Prysliak, T.; Maina, T.; Suleman, M.; Jimbo, S. Status of the development of a vaccine against Mycoplasma bovis. Vaccine 2017, 35, 2902–2907. [Google Scholar] [CrossRef] [PubMed]
- Gautier-Bouchardon, A.V. Antimicrobial Resistance in Mycoplasma spp. Microbiol. Spectr. 2018, 6, 425–446. [Google Scholar]
- Owen, J.R.; Noyes, N.; Young, A.E.; Prince, D.J.; Blanchard, P.C.; Lehenbauer, T.W.; Aly, S.S.; Davis, J.H.; O’Rourke, S.M.; Abdo, Z.; et al. Whole-Genome Sequencing and Concordance Between Antimicrobial Susceptibility Genotypes and Phenotypes of Bacterial Isolates Associated with Bovine Respiratory Disease. G3 Genes Genomes Genet. 2017, 7, 3059–3071. [Google Scholar] [CrossRef]
- Parker, A.M.; Shukla, A.; House, J.K.; Hazelton, M.S.; Bosward, K.L.; Kokotovic, B.; Sheehy, P.A. Genetic characterization of Australian Mycoplasma bovis isolates through whole genome sequencing analysis. Vet. Microbiol. 2016, 196, 118–125. [Google Scholar] [CrossRef]
- Sulyok, K.M.; Kreizinger, Z.; Wehmann, E.; Lysnyansky, I.; Banyai, K.; Marton, S.; Jerzsele, A.; Ronai, Z.; Turcsanyi, I.; Makrai, L.; et al. Mutations Associated with Decreased Susceptibility to Seven Antimicrobial Families in Field and Laboratory-Derived Mycoplasma bovis Strains. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef]
- Yair, Y.; Borovok, I.; Mikula, I.; Falk, R.; Fox, L.K.; Gophna, U.; Lysnyansky, I. Genomics-based epidemiology of bovine Mycoplasma bovis strains in Israel. BMC Genomics 2020, 21, 70. [Google Scholar] [CrossRef] [PubMed]
- Baker, S.; Thomson, N.; Weill, F.X.; Holt, K.E. Genomic insights into the emergence and spread of antimicrobial-resistant bacterial pathogens. Science 2018, 360, 733–738. [Google Scholar] [CrossRef]
- Register, K.B.; Bayles, D.O.; Ma, H.; Windeyer, M.C.; Perez-Casal, J.; Bras, A.L.; Suleman, M.; Woodbury, M.; Jelinski, M.D.; Alt, D.P. Complete Genome Sequences of 16 Mycoplasma bovis Isolates from Canadian Bison and Cattle. Microbiol. Resour. Announc. 2020, 9. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zheng, H.; Liu, Y.; Jiang, Y.; Xin, J.; Chen, W.; Song, Z. The complete genome sequence of Mycoplasma bovis strain Hubei-1. PLoS ONE 2011, 6, e20999. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Guo, A.; Cui, P.; Chen, Y.; Mustafa, R.; Ba, X.; Hu, C.; Bai, Z.; Chen, X.; Shi, L.; et al. Comparative Geno-Plasticity Analysis of Mycoplasma bovis HB0801 (Chinese Isolate). PLoS ONE 2012, 7, e38239. [Google Scholar] [CrossRef]
- Wise, K.S.; Calcutt, M.J.; Foecking, M.F.; Roske, K.; Madupu, R.; Methe, B.A. Complete genome sequence of Mycoplasma bovis type strain PG45 (ATCC 25523). Infect. Immun. 2011, 79, 982–983. [Google Scholar] [CrossRef] [PubMed]
- Josi, C.; Burki, S.; Vidal, S.; Dordet-Frisoni, E.; Citti, C.; Falquet, L.; Pilo, P. Large-Scale Analysis of the Mycoplasma bovis Genome Identified Non-essential, Adhesion- and Virulence-Related Genes. Front. Microbiol. 2019, 10, 2085. [Google Scholar] [CrossRef]
- Feenstra, A.; Bisgaard Madsen, E.; Friis, N.F.; Meyling, A.; Ahrens, P. A field study of Mycoplasma bovis infection in cattle. Zent. Vet. B 1991, 38, 195–202. [Google Scholar] [CrossRef]
- Arede, M.; Nielsen, P.K.; Ahmed, S.S.; Halasa, T.; Nielsen, L.R.; Toft, N. A space-time analysis of Mycoplasma bovis: Bulk tank milk antibody screening results from all Danish dairy herds in 2013–2014. Acta Vet. Scand. 2016, 58, 16. [Google Scholar] [CrossRef]
- Kusiluka, L.J.; Kokotovic, B.; Ojeniyi, B.; Friis, N.F.; Ahrens, P. Genetic variations among Mycoplasma bovis strains isolated from Danish cattle. FEMS Microbiol. Lett. 2000, 192, 113–138. [Google Scholar] [CrossRef][Green Version]
- Kusiluka, L.J.; Ojeniyi, B.; Friis, N.F. Increasing prevalence of Mycoplasma bovis in Danish cattle. Acta Vet. Scand. 2000, 41, 139–146. [Google Scholar]
- Petersen, M.B. Mycoplasma bovis in Dairy Cattle: Clinical Epidemiology and Antibody Measurements for Decision Making. Ph.D. Thesis, Department of Veterinary and Animal Sciences, University of Copenhagen, Copenhagen, Denmark, 2018; p. 160. [Google Scholar]
- Gulliksen, S.M.; Jor, E.; Lie, K.I.; Loken, T.; Akerstedt, J.; Osteras, O. Respiratory infections in Norwegian dairy calves. J. Dairy Sci. 2009, 92, 5139–5146. [Google Scholar] [CrossRef] [PubMed]
- Becker, C.A.; Thibault, F.M.; Arcangioli, M.A.; Tardy, F. Loss of diversity within Mycoplasma bovis isolates collected in France from bovines with respiratory diseases over the last 35 years. Infect. Genet. Evol. 2015, 33, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Poumarat, F.; Martel, J.L. In vitro antibiotic sensitivity of French strains of Mycoplasma bovis. Ann. Rech. Vet. 1989, 20, 145–152. [Google Scholar] [PubMed]
- Register, K.B.; Thole, L.; Rosenbush, R.F.; Minion, F.C. Multilocus sequence typing of Mycoplasma bovis reveals host-specific genotypes in cattle versus bison. Vet. Microbiol. 2015, 175, 92–98. [Google Scholar] [CrossRef]
- Rosales, R.S.; Churchward, C.P.; Schnee, C.; Sachse, K.; Lysnyansky, I.; Catania, S.; Iob, L.; Ayling, R.D.; Nicholas, R.A. Global multilocus sequence typing analysis of Mycoplasma bovis isolates reveals two main population clusters. J. Clin. Microbiol. 2015, 53, 789–794. [Google Scholar] [CrossRef]
- Register, K.B.; Lysnyansky, I.; Jelinski, M.D.; Boatwright, W.D.; Waldner, M.; Bayles, D.O.; Pilo, P.; Alt, D.P. Comparison of Two Multilocus Sequence Typing Schemes for Mycoplasma bovis and Revision of the PubMLST Reference Method. J. Clin. Microbiol. 2020, 58. [Google Scholar] [CrossRef]
- Shakya, M.; Ahmed, S.A.; Davenport, K.W.; Flynn, M.C.; Lo, C.C.; Chain, P.S.G. Standardized phylogenetic and molecular evolutionary analysis applied to species across the microbial tree of life. Sci. Rep. 2020, 10, 1723. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals, 4th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2015. [Google Scholar]
- Khalil, D.; Becker, C.A.; Tardy, F. Alterations in the Quinolone Resistance-Determining Regions and Fluoroquinolone Resistance in Clinical Isolates and Laboratory-Derived Mutants of Mycoplasma bovis: Not. All Genotypes May Be Equal. Appl. Environ. Microbiol. 2016, 82, 1060–1068. [Google Scholar] [CrossRef]
- Khalil, D.; Becker, C.A.M.; Tardy, F. Monitoring the Decrease in Susceptibility to Ribosomal RNAs Targeting Antimicrobials and Its Molecular Basis in Clinical Mycoplasma bovis Isolates over Time. Microb. Drug. Resist. 2017, 23, 799–811. [Google Scholar] [CrossRef]
- Haapala, V.; Pohjanvirta, T.; Vähänikkilä, N.; Halkilahti, J.; Simonen, H.; Pelkonen, S.; Soveri, T.; Simojoki, H.; Autio, T. Semen as a source of Mycoplasma bovis mastitis in dairy herds. Vet. Microbiol. 2018, 216, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Heuvelink, A.; Reugebrink, C.; Mars, J. Antimicrobial susceptibility of Mycoplasma bovis isolates from veal calves and dairy cattle in the Netherlands. Vet. Microbiol. 2016, 189, 1–7. [Google Scholar] [CrossRef]
- Petersen, M.B.; Krogh, K.; Nielsen, L.R. Factors associated with variation in bulk tank milk Mycoplasma bovis antibody-ELISA results in dairy herds. J. Dairy Sci. 2016, 99, 3815–3823. [Google Scholar] [CrossRef]
- Burki, S.; Spergser, J.; Bodmer, M.; Pilo, P. A dominant lineage of Mycoplasma bovis is associated with an increased number of severe mastitis cases in cattle. Vet. Microbiol. 2016, 196, 63–66. [Google Scholar] [CrossRef]
- Lysnyansky, I.; Freed, M.; Rosales, R.S.; Mikula, I.; Khateb, N.; Gerchman, I.; Straten, M.v.; Levisohn, S. An overview of Mycoplasma bovis mastitis in Israel (2004–2014). Vet. J. 2016, 207, 180–183. [Google Scholar] [CrossRef]
- Becker, C.A.M.; Ambroset, C.; Huleux, A.; Vialatte, A.; Colin, A.; Tricot, A.; Arcangioli, M.A.; Tardy, F. Monitoring Mycoplasma bovis Diversity and Antimicrobial Susceptibility in Calf Feedlots Undergoing a Respiratory Disease Outbreak. Pathogens 2020, 9, 593. [Google Scholar] [CrossRef]
- Abu-Amero, K.K.; Abu-Groun, E.A.; Halablab, M.A.; Miles, R.J. Kinetics and distribution of alcohol oxidising activity in Acholeplasma and Mycoplasma species. FEMS Microbiol. Lett. 2000, 183, 147–151. [Google Scholar] [CrossRef][Green Version]
- Josi, C.; Burki, S.; Stojiljkovic, A.; Wellnitz, O.; Stoffel, M.H.; Pilo, P. Bovine Epithelial in vitro Infection Models for Mycoplasma bovis. Front. Cell Infect. Microbiol. 2018, 8, 329. [Google Scholar] [CrossRef] [PubMed]
- Gautier-Bouchardon, A.V.; Ferre, S.; Le Grand, D.; Paoli, A.; Gay, E.; Poumarat, F. Overall decrease in the susceptibility of Mycoplasma bovis to antimicrobials over the past 30 years in France. PLoS ONE 2014, 9, e87672. [Google Scholar] [CrossRef] [PubMed]
- Poumarat, F.; Longchambon, D.; Martel, J.L. Application of dot immunobinding on membrane filtration (MF dot) to the study of relationships within “M. mycoides cluster” and within “glucose and arginine-negative cluster” of ruminant mycoplasmas. Vet. Microbiol. 1992, 32, 375–390. [Google Scholar] [CrossRef]
- Kobisch, M.; Friis, N.F. Swine mycoplasmoses. Rev. Sci. Tech. 1996, 15, 1569–1605. [Google Scholar] [CrossRef]
- Kim, D.; Song, L.; Breitwieser, F.P.; Salzberg, S.L. Centrifuge: Rapid and sensitive classification of metagenomic sequences. Genome Res. 2016, 26, 1721–1729. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef] [PubMed]
- Gardner, S.N.; Slezak, T.; Hall, B.G. kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics 2015, 31, 2877–2878. [Google Scholar] [CrossRef]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef]
- Register, K.B.; Jelinski, M.D.; Waldner, M.; Boatwright, W.D.; Anderson, T.K.; Hunter, D.L.; Hamilton, R.G.; Burrage, P.; Shury, T.; Bildfell, R.; et al. Comparison of multilocus sequence types found among North. American isolates of Mycoplasma bovis from cattle, bison, and deer, 2007–2017. J. Vet. Diagn. Investig. 2019, 31, 899–904. [Google Scholar] [CrossRef]
- Gardner, S.N.; Slezak, T. Simulate_PCR for amplicon prediction and annotation from multiplex, degenerate primers and probes. BMC Bioinform. 2014, 15, 237. [Google Scholar] [CrossRef]
- Edgar, R.C. C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2–Approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Marenda, M.S.; Sagne, E.; Poumarat, F.; Citti, C. Suppression subtractive hybridization as a basis to assess Mycoplasma agalactiae and Mycoplasma bovis genomic diversity and species-specific sequences. Microbiology 2005, 151 Pt 2, 475–489. [Google Scholar] [CrossRef]
- Poumarat, F.; Perrin, B.; Longchambon, D. Identification of ruminant mycoplasmas by dot immunobinding on membrane filtration (MF dot). Vet. Microbiol. 1991, 29, 329–338. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tardy, F.; Aspan, A.; Autio, T.; Ridley, A.; Tricot, A.; Colin, A.; Pohjanvirta, T.; Smid, B.; Harders, F.; Lindegaard, M.; et al. Mycoplasma bovis in Nordic European Countries: Emergence and Dominance of a New Clone. Pathogens 2020, 9, 875. https://doi.org/10.3390/pathogens9110875
Tardy F, Aspan A, Autio T, Ridley A, Tricot A, Colin A, Pohjanvirta T, Smid B, Harders F, Lindegaard M, et al. Mycoplasma bovis in Nordic European Countries: Emergence and Dominance of a New Clone. Pathogens. 2020; 9(11):875. https://doi.org/10.3390/pathogens9110875
Chicago/Turabian StyleTardy, Florence, Anna Aspan, Tiina Autio, Anne Ridley, Agnès Tricot, Adélie Colin, Tarja Pohjanvirta, Bregtje Smid, Frank Harders, Mikkel Lindegaard, and et al. 2020. "Mycoplasma bovis in Nordic European Countries: Emergence and Dominance of a New Clone" Pathogens 9, no. 11: 875. https://doi.org/10.3390/pathogens9110875
APA StyleTardy, F., Aspan, A., Autio, T., Ridley, A., Tricot, A., Colin, A., Pohjanvirta, T., Smid, B., Harders, F., Lindegaard, M., Tølbøll Lauritsen, K., Lyhs, U., Wisselink, H. J., & Strube, M. L. (2020). Mycoplasma bovis in Nordic European Countries: Emergence and Dominance of a New Clone. Pathogens, 9(11), 875. https://doi.org/10.3390/pathogens9110875






