Structural and Functional Analysis of BBA03, Borrelia burgdorferi Competitive Advantage Promoting Outer Surface Lipoprotein
Abstract
1. Introduction
2. Results
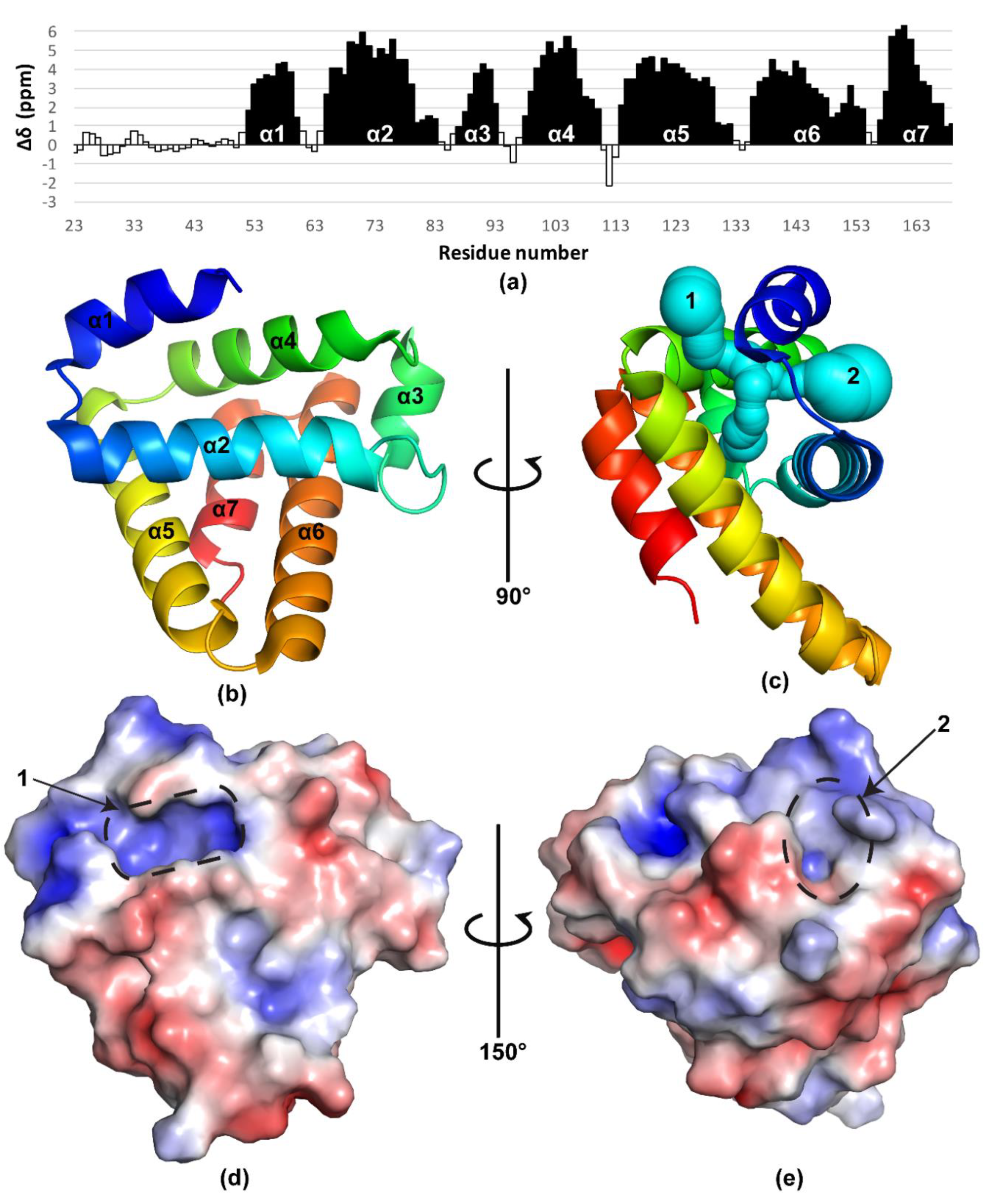
2.1. BBA03 Structure Determination
2.2. Structure Analysis
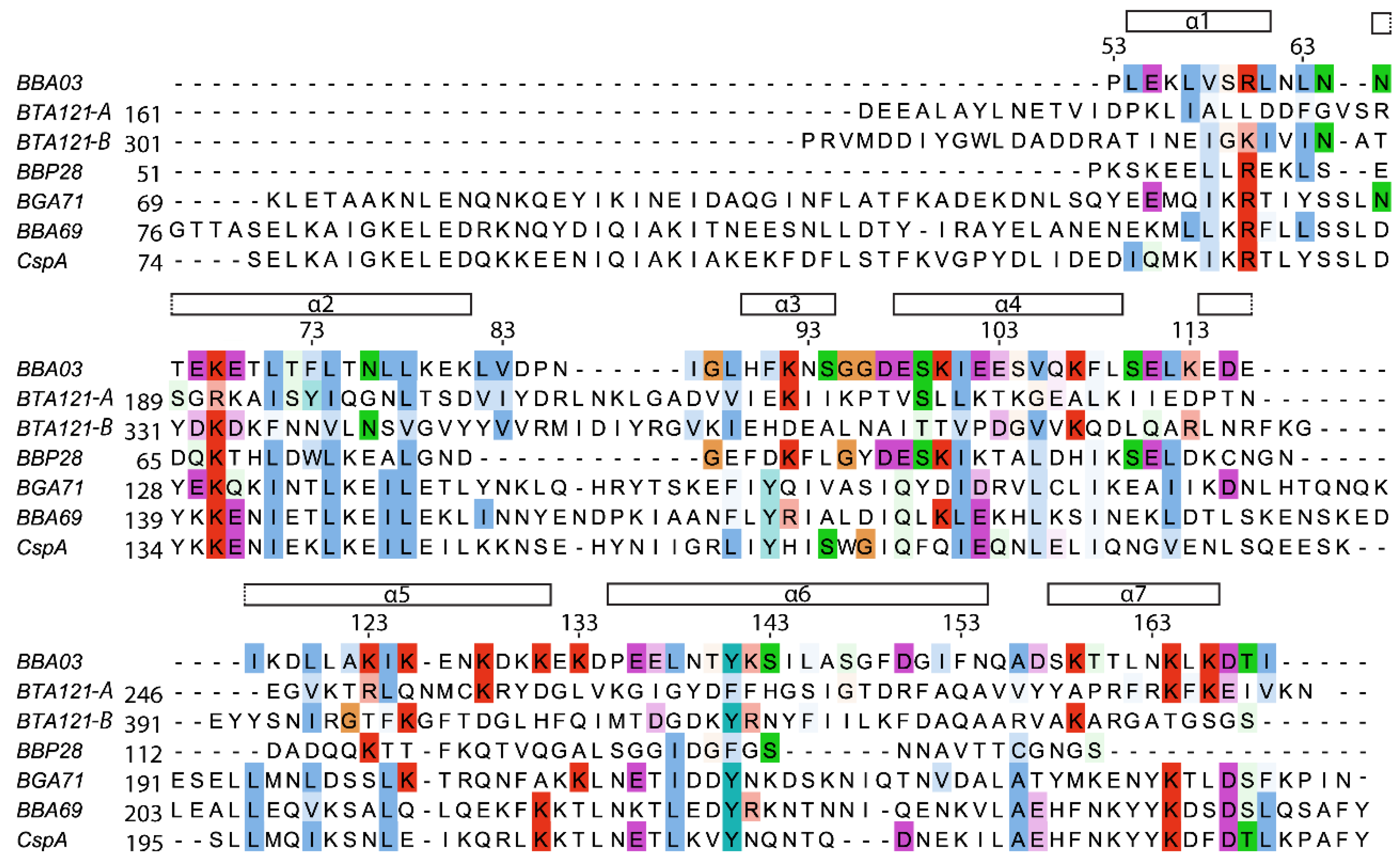
2.3. Structure Comparison
2.4. Sequence Comparison
3. Discussion
4. Materials and Methods
4.1. Protein Sample Preparation
4.2. NMR Spectroscopy
4.3. NMR Relaxation Data
4.4. NMR Structure Determination
4.5. Molecular Docking
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Mead, P.S. Epidemiology of Lyme Disease. Infect. Dis. Clin. N. Am. 2015, 29, 187–210. [Google Scholar] [CrossRef]
- Dunham-Ems, S.M.; Caimano, M.J.; Pal, U.; Wolgemuth, C.W.; Eggers, C.H.; Balic, A.; Radolf, J.D. Live imaging reveals a biphasic mode of dissemination of Borrelia burgdorferi within ticks. J. Clin. Investig. 2009, 119, 3652–3665. [Google Scholar] [CrossRef]
- Müllegger, R.R.; Glatz, M.J.A.J.O.C.D. Skin Manifestations of Lyme Borreliosis. Am. J. Clin. Dermatol. 2008, 9, 355–368. [Google Scholar] [CrossRef]
- Patton, S.K.; Phillips, B. CE: Lyme Disease: Diagnosis, Treatment, and Prevention. AJN Am. J. Nurs. 2018, 118, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Casjens, S.R.; Mongodin, E.F.; Qiu, W.-G.; Luft, B.J.; Schutzer, S.E.; Gilcrease, E.B.; Huang, W.M.; Vujadinovic, M.; Aron, J.K.; Vargas, L.C.; et al. Genome stability of Lyme disease spirochetes: Comparative genomics of Borrelia burgdorferi plasmids. PLoS ONE 2012, 7, e33280. [Google Scholar] [CrossRef] [PubMed]
- Tilly, K.; Bestor, A.; Rosa, P.A. Functional Equivalence of OspA and OspB, but Not OspC, in Tick Colonization by Borrelia burgdorferi. Infect Immun. 2016, 84, 1565–1573. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Xu, Q.; McShan, K.; Liang, F.T. Both decorin-binding proteins A and B are critical for the overall virulence of Borrelia burgdorferi. Infect. Immun. 2008, 76, 1239–1246. [Google Scholar] [CrossRef] [PubMed]
- Bestor, A.; Rego, R.O.M.; Tilly, K.; Rosa, P.A. Competitive advantage of Borrelia burgdorferi with outer surface protein BBA03 during tick-mediated infection of the mammalian host. Infect. Immun. 2012, 80, 3501–3511. [Google Scholar] [CrossRef]
- Patton, T.G.; Dietrich, G.; Dolan, M.C.; Piesman, J.; Carroll, J.A.; Gilmore, R.D., Jr. Functional analysis of the Borrelia burgdorferi bba64 gene product in murine infection via tick infestation. PLoS ONE 2011, 6, e19536. [Google Scholar] [CrossRef]
- Kumar, M.; Kaur, S.; Kariu, T.; Yang, X.; Bossis, I.; Anderson, J.F.; Pal, U. Borrelia burgdorferi BBA52 is a potential target for transmission blocking Lyme disease vaccine. Vaccine 2011, 29, 9012–9019. [Google Scholar] [CrossRef]
- Sehnal, D.; Pravda, L.; Svobodová Vařeková, R.; Koča, J.; Toušek, D.; Berka, K.; Bazgier, V.; Navrátilová, V.; Otyepka, M. MOLEonline: A web-based tool for analyzing channels, tunnels and pores (2018 update). Nucleic Acids Res. 2018, 46, W368–W373. [Google Scholar] [CrossRef]
- Chen, C.; Tian, W.; Lei, X.; Liang, J.; Zhao, J. CASTp 3.0: Computed atlas of surface topography of proteins. Nucleic Acids Res. 2018, 46, W363–W367. [Google Scholar] [CrossRef]
- Baker, N.A.; Sept, D.; Joseph, S.; Holst, M.J.; McCammon, J.A. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA 2001, 98, 10037–10041. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.J.; Headd, J.J.; Moriarty, N.W.; Prisant, M.G.; Videau, L.L.; Deis, L.N.; Verma, V.; Keedy, D.A.; Hintze, B.J.; Chen, V.B.; et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. A Publ. Protein Soc. 2018, 27, 293–315. [Google Scholar] [CrossRef]
- Holm, L. Using Dali for Protein Structure Comparison. Methods Mol. Biol. (Clifton. N. J.) 2020, 2112, 29–42. [Google Scholar] [CrossRef]
- Luo, Z.; Kelleher, A.J.; Darwiche, R.; Hudspeth, E.M.; Shittu, O.K.; Krishnavajhala, A.; Schneiter, R.; Lopez, J.E.; Asojo, O.A. Crystal Structure of Borrelia turicatae protein, BTA121, a differentially regulated gene in the tick-mammalian transmission cycle of relapsing fever spirochetes. Sci. Rep. 2017, 7, 15310. [Google Scholar] [CrossRef]
- Fridmanis, J.; Otikovs, M.; Brangulis, K.; Tārs, K.; Jaudzems, K. Solution NMR structure of Borrelia burgdorferi outer surface lipoprotein BBP28, a member of the mlp protein family. Proteins: Struct. Funct. Bioinform. 2020. [Google Scholar] [CrossRef]
- Brangulis, K.; Akopjana, I.; Petrovskis, I.; Kazaks, A.; Tars, K. Structural analysis of the outer surface proteins from Borrelia burgdorferi paralogous gene family 54 that are thought to be the key players in the pathogenesis of Lyme disease. J. Struct. Biol. 2020, 210, 107490. [Google Scholar] [CrossRef]
- Hammerschmidt, C.; Klevenhaus, Y.; Koenigs, A.; Hallström, T.; Fingerle, V.; Skerka, C.; Pos, K.M.; Zipfel, P.F.; Wallich, R.; Kraiczy, P. BGA66 and BGA71 facilitate complement resistance of Borrelia bavariensis by inhibiting assembly of the membrane attack complex. Mol. Microbiol. 2016, 99, 407–424. [Google Scholar] [CrossRef]
- Koenigs, A.; Hammerschmidt, C.; Jutras, B.L.; Pogoryelov, D.; Barthel, D.; Skerka, C.; Kugelstadt, D.; Wallich, R.; Stevenson, B.; Zipfel, P.F.; et al. BBA70 of Borrelia burgdorferi is a novel plasminogen-binding protein. J. Biol. Chem. 2013, 288, 25229–25243. [Google Scholar] [CrossRef]
- Brangulis, K.; Akopjana, I.; Petrovskis, I.; Kazaks, A.; Kraiczy, P.; Tars, K. Crystal structure of the membrane attack complex assembly inhibitor BGA71 from the Lyme disease agent Borrelia bavariensis. Sci. Rep. 2018, 8, 11286. [Google Scholar] [CrossRef] [PubMed]
- Brangulis, K.; Akopjana, I.; Petrovskis, I.; Kazaks, A.; Tars, K. Crystal structure of Borrelia burgdorferi outer surface protein BBA69 in comparison to the paralogous protein CspA. Ticks Tick Borne Dis. 2019, 10, 1135–1141. [Google Scholar] [CrossRef]
- Brangulis, K.; Tars, K.; Petrovskis, I.; Kazaks, A.; Ranka, R.; Baumanis, V. Structure of an outer surface lipoprotein BBA64 from the Lyme disease agent Borrelia burgdorferi which is critical to ensure infection after a tick bite. Acta Crystallogr. Sect. D Biol. Crystallogr. 2013, 69, 1099–1107. [Google Scholar] [CrossRef] [PubMed]
- Haupt, K.; Kraiczy, P.; Wallich, R.; Brade, V.; Skerka, C.; Zipfel, P.F. Binding of human factor H-related protein 1 to serum-resistant Borrelia burgdorferi is mediated by borrelial complement regulator-acquiring surface proteins. J. Infect. Dis. 2007, 196, 124–133. [Google Scholar] [CrossRef] [PubMed]
- Hallström, T.; Siegel, C.; Mörgelin, M.; Kraiczy, P.; Skerka, C.; Zipfel, P.F. CspA from Borrelia burgdorferi inhibits the terminal complement pathway. mBio 2013, 4, e00481-13. [Google Scholar] [CrossRef] [PubMed]
- Brangulis, K.; Akopjana, I.; Petrovskis, I.; Kazaks, A.; Zelencova, D.; Jekabsons, A.; Jaudzems, K.; Tars, K. BBE31 from the Lyme disease agent Borrelia burgdorferi, known to play an important role in successful colonization of the mammalian host, shows the ability to bind glutathione. Biochim. Et Biophys. Acta. Gen. Subj. 2020, 1864, 129499. [Google Scholar] [CrossRef] [PubMed]
- Gilmore, R.D., Jr.; Howison, R.R.; Schmit, V.L.; Carroll, J.A. Borrelia burgdorferi expression of the bba64, bba65, bba66, and bba73 genes in tissues during persistent infection in mice. Microb. Pathog. 2008, 45, 355–360. [Google Scholar] [CrossRef]
- Brangulis, K.; Petrovskis, I.; Kazaks, A.; Tars, K.; Ranka, R. Crystal structure of the infectious phenotype-associated outer surface protein BBA66 from the Lyme disease agent Borrelia burgdorferi. Ticks Tick-Borne Dis. 2014, 5, 63–68. [Google Scholar] [CrossRef]
- Patton, T.G.; Brandt, K.S.; Nolder, C.; Clifton, D.R.; Carroll, J.A.; Gilmore, R.D. Borrelia burgdorferi bba66 gene inactivation results in attenuated mouse infection by tick transmission. Infect. Immun. 2013, 81, 2488–2498. [Google Scholar] [CrossRef]
- Brangulis, K.; Petrovskis, I.; Kazaks, A.; Baumanis, V.; Tars, K. Structural characterization of the Borrelia burgdorferi outer surface protein BBA73 implicates dimerization as a functional mechanism. Biochem. Biophys. Res. Commun. 2013, 434, 848–853. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed]
- Casjens, S.; Palmer, N.; van Vugt, R.; Huang, W.M.; Stevenson, B.; Rosa, P.; Lathigra, R.; Sutton, G.; Peterson, J.; Dodson, R.J.; et al. A bacterial genome in flux: The twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 2000, 35, 490–516. [Google Scholar] [CrossRef] [PubMed]
- Brisson, D.; Drecktrah, D.; Eggers, C.H.; Samuels, D.S. Genetics of Borrelia burgdorferi. Annu. Rev. Genet. 2012, 46, 515–536. [Google Scholar] [CrossRef] [PubMed]
- Tilly, K.; Rosa, P.A.; Stewart, P.E. Biology of infection with Borrelia burgdorferi. Infect Dis. Clin. N. Am. 2008, 22, 217–234. [Google Scholar] [CrossRef] [PubMed]
- Bestor, A.; Stewart, P.E.; Jewett, M.W.; Sarkar, A.; Tilly, K.; Rosa, P.A. Use of the Cre-lox recombination system to investigate the lp54 gene requirement in the infectious cycle of Borrelia burgdorferi. Infect. Immun. 2010, 78, 2397–2407. [Google Scholar] [CrossRef]
- Maruskova, M.; Seshu, J. Deletion of BBA64, BBA65, and BBA66 Loci Does Not Alter the Infectivity of Borrelia burgdorferi in the Murine Model of Lyme Disease. Infect. Immun. 2008, 76, 5274. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; He, M.; He, J.J.; Yang, X.F. Role of the Surface Lipoprotein BBA07 in the Enzootic Cycle of Borrelia burgdorferi. Infect. Immun. 2010, 78, 2910. [Google Scholar] [CrossRef]
- Lin, Y.-P.; Frye, A.M.; Nowak, T.A.; Kraiczy, P. New Insights Into CRASP-Mediated Complement Evasion in the Lyme Disease Enzootic Cycle. Front Cell Infect Microbiol 2020, 10. [Google Scholar] [CrossRef]
- Kerstholt, M.; Netea, M.G.; Joosten, L.A.B. Borrelia burgdorferi hijacks cellular metabolism of immune cells: Consequences for host defense. Ticks Tick-Borne Dis. 2020, 11, 101386. [Google Scholar] [CrossRef]
- Fraser, C.M.; Casjens, S.; Huang, W.M.; Sutton, G.G.; Clayton, R.; Lathigra, R.; White, O.; Ketchum, K.A.; Dodson, R.; Hickey, E.K.; et al. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 1997, 390, 580–586. [Google Scholar] [CrossRef]
- Toledo, A.; Huang, Z.; Coleman, J.L.; London, E.; Benach, J.L. Lipid rafts can form in the inner and outer membranes of Borrelia burgdorferi and have different properties and associated proteins. Mol. Microbiol. 2018, 108, 63–76. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Toledo, A.M.; Benach, J.L.; London, E. Ordered Membrane Domain-Forming Properties of the Lipids of Borrelia burgdorferi. Biophys. J. 2016, 111, 2666–2675. [Google Scholar] [CrossRef]
- Brown, C.R.; Dennis, E.A. Borrelia burgdorferi infection induces lipid mediator production during Lyme arthritis. Biochimie 2017, 141, 86–90. [Google Scholar] [CrossRef]
- Dümmler, A.; Lawrence, A.-M.; de Marco, A. Simplified screening for the detection of soluble fusion constructs expressed in E. coli using a modular set of vectors. Microb. Cell Factories 2005, 4, 34. [Google Scholar] [CrossRef] [PubMed]
- D’Auvergne, E.J.; Gooley, P.R. Optimisation of NMR dynamic models II. A new methodology for the dual optimisation of the model-free parameters and the Brownian rotational diffusion tensor. J. Biomol. NMR 2008, 40, 121–133. [Google Scholar] [CrossRef]
- Kay, L.E.; Torchia, D.A.; Bax, A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: Application to staphylococcal nuclease. Am. Chem. Soc. 1989, 28, 8972–8979. [Google Scholar]
- Keller, R. The Computer Aided Resonance Assignment Tutorial, 1st ed.; CANTINA Verlag: Goldau, Switzerland, 2004. [Google Scholar]
- Serrano, P.; Pedrini, B.; Mohanty, B.; Geralt, M.; Herrmann, T.; Wüthrich, K. The J-UNIO protocol for automated protein structure determination by NMR in solution. J. Biomol. NMR 2012, 53, 341–354. [Google Scholar] [CrossRef]
- Herrmann, T.; Güntert, P.; Wüthrich, K. Protein NMR Structure Determination with Automated NOE Assignment Using the New Software CANDID and the Torsion Angle Dynamics Algorithm DYANA. J. Mol. Biol. 2002, 319, 209–227. [Google Scholar] [CrossRef]
- Herrmann, T.; Güntert, P.; Wüthrich, K. Protein NMR structure determination with automated NOE-identification in the NOESY spectra using the new software ATNOS. J. Biomol. NMR 2002, 24, 171–189. [Google Scholar] [CrossRef]
- Güntert, P.; Mumenthaler, C.; Wüthrich, K. Torsion angle dynamics for NMR structure calculation with the new program Dyana11Edited by P. E. Wright. J. Mol. Biol. 1997, 273, 283–298. [Google Scholar] [CrossRef]
- Brunger, A.T. CNS (Crystallography and NMR System). In Encyclopedia of Biophysics; Roberts, G.C.K., Ed.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 326–327. [Google Scholar]
- Schrödinger Release 2020-2: Maestro, Schrödinger, New York. 2020. Available online: https://www.schrodinger.com/ (accessed on 9 October 2020).
- Roos, K.; Wu, C.; Damm, W.; Reboul, M.; Stevenson, J.M.; Lu, C.; Dahlgren, M.K.; Mondal, S.; Chen, W.; Wang, L.; et al. OPLS3e: Extending Force Field Coverage for Drug-Like Small Molecules. J. Chem. Theory Comput. 2019, 15, 1863–1874. [Google Scholar] [CrossRef] [PubMed]
- Rossi, P.; Swapna, G.V.T.; Huang, Y.J.; Aramini, J.M.; Anklin, C.; Conover, K.; Hamilton, K.; Xiao, R.; Acton, T.B.; Ertekin, A.; et al. A microscale protein NMR sample screening pipeline. J. Biomol. Nmr 2010, 46, 11–22. [Google Scholar] [CrossRef] [PubMed]


| NOE upper distance limits | 2602 |
| Intra-residual (|i − j| = 0) | 767 |
| Sequential (|i − j| = 1) | 626 |
| Medium range (1 < |i − j| ≤ 4) | 704 |
| Long range (|i − j| ≥ 5) | 616 |
| Residual CYANA target function value after CNS (Å2) | 9.75 ± 1.05 |
| Residual NOE violations | |
| Number ≥ 0.1 Å | 29 ± 5 |
| Maximum (Å) | 0.38 ± 0.08 |
| PARALLHDG force field energies (kcal/mol) | |
| Total | −5534 ± 101 |
| Van der Waals | −481 ± 19 |
| Electrostatic | −5961 ± 117 |
| Root-mean-square-deviation (RMSD) from mean coordinates (residues 53–169) (Å) | |
| Backbone | 0.50 ± 0.06 |
| All heavy atoms | 0.90 ± 0.06 |
| Molprobity structure statistics [14] | |
| Ramachandran plot favored regions (%) | 94.5 |
| Ramachandran plot allowed regions (%) | 99.3 |
| Average clash score of all atoms and models | 21.62 ± 2.47 |
| Protein | PDB Code | Z-score | Backbone RMSD, Å | Sequence Identity % | Function |
|---|---|---|---|---|---|
| BTA121 [16] | 5VJ4 | 7.7 | 2.3 | 13 | Binds fatty acids [16] |
| BBP28 [17] | 6QBI | 7.4 | 2.5 | 21 | Unknown |
| BGA71 [21] | 6FL0 | 7.0 | 2.9 | 12 | Binds C7, C8 and C9 terminal complement components [19] |
| BBA69 [22] | 6QO1 | 6.1 | 3.2 | 14 | Unknown |
| CspA [23] | 5A2U | 6.1 | 3.3 | 11 | Binds factor H and FHL-1 [24] Binds C7 and C9 terminal complement components [25] |
| BBE31 [26] | 6FZE | 5.4 | 3.1 | 9 | Binds glutathione [26] |
| BBA64 [23] | 4ALY | 4.3 | 3.4 | 18 | Unknown [9] |
| BBA65 [18] | 4BG5 | 4.3 | 3.4 | 14 | Unknown [27] |
| BBA66 [28] | 2YN7 | 4.3 | 3.6 | 12 | Unknown [29] |
| BBA73 [30] | 4B2F | 4.1 | 3.5 | 13 | Unknown |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fridmanis, J.; Bobrovs, R.; Brangulis, K.; Tārs, K.; Jaudzems, K. Structural and Functional Analysis of BBA03, Borrelia burgdorferi Competitive Advantage Promoting Outer Surface Lipoprotein. Pathogens 2020, 9, 826. https://doi.org/10.3390/pathogens9100826
Fridmanis J, Bobrovs R, Brangulis K, Tārs K, Jaudzems K. Structural and Functional Analysis of BBA03, Borrelia burgdorferi Competitive Advantage Promoting Outer Surface Lipoprotein. Pathogens. 2020; 9(10):826. https://doi.org/10.3390/pathogens9100826
Chicago/Turabian StyleFridmanis, Jēkabs, Raitis Bobrovs, Kalvis Brangulis, Kaspars Tārs, and Kristaps Jaudzems. 2020. "Structural and Functional Analysis of BBA03, Borrelia burgdorferi Competitive Advantage Promoting Outer Surface Lipoprotein" Pathogens 9, no. 10: 826. https://doi.org/10.3390/pathogens9100826
APA StyleFridmanis, J., Bobrovs, R., Brangulis, K., Tārs, K., & Jaudzems, K. (2020). Structural and Functional Analysis of BBA03, Borrelia burgdorferi Competitive Advantage Promoting Outer Surface Lipoprotein. Pathogens, 9(10), 826. https://doi.org/10.3390/pathogens9100826






