Molecular Detection and Characterization of Borrelia garinii (Spirochaetales: Borreliaceae) in Ixodes nipponensis (Ixodida: Ixodidae) Parasitizing a Dog in Korea
Abstract
1. Introduction
2. Results
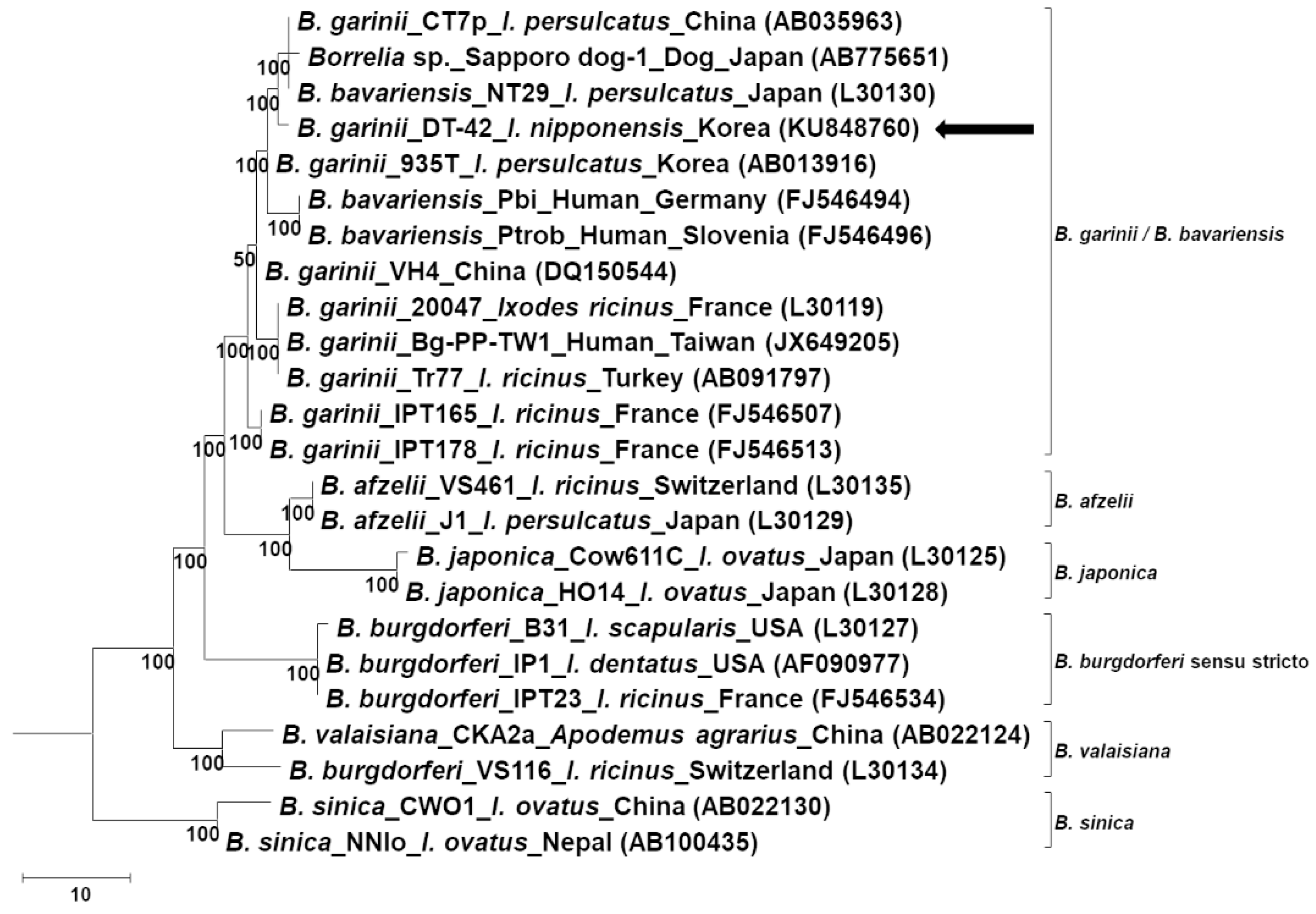
2.1. Molecular Identification of Borrelia Spp.
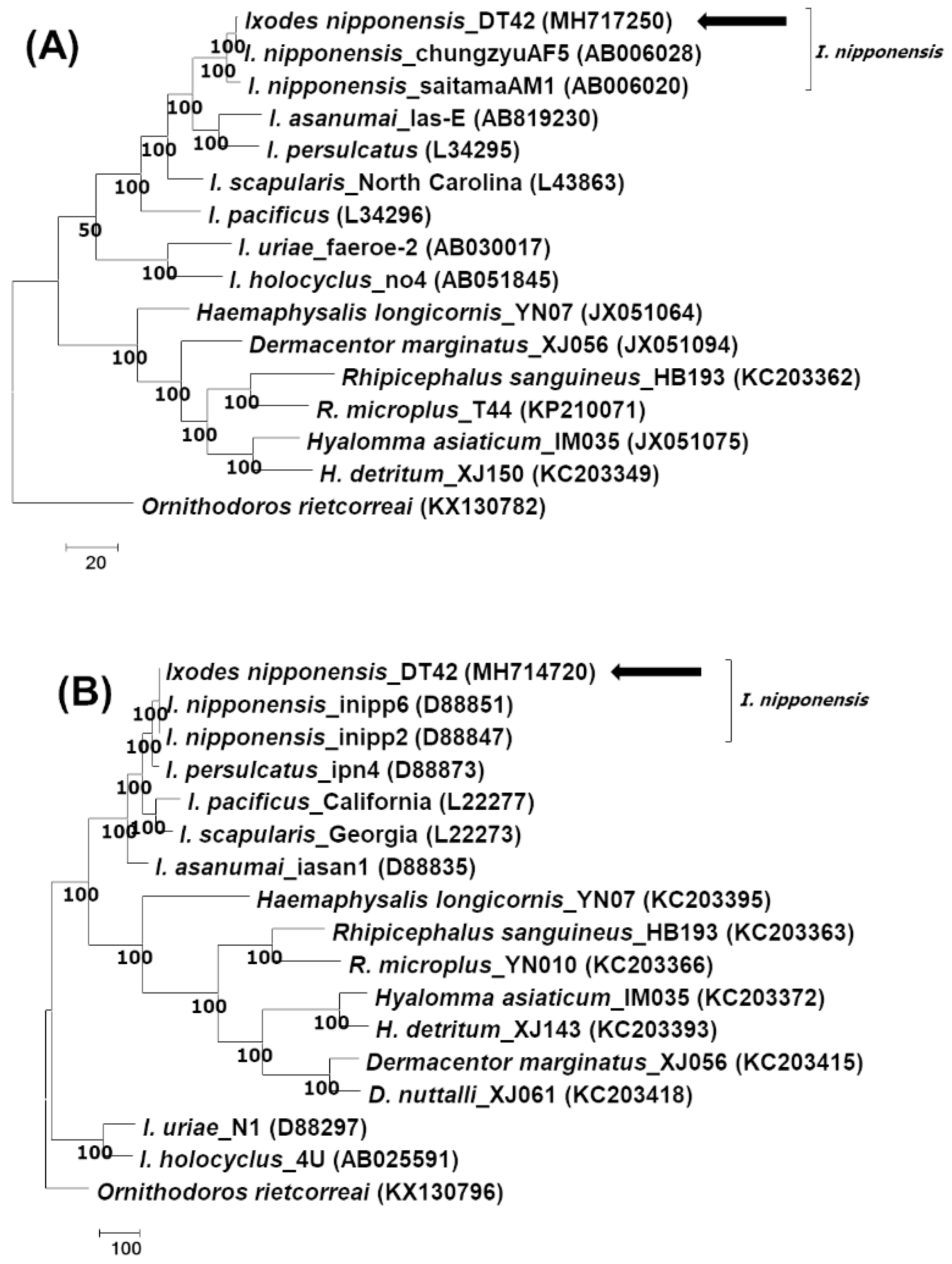
2.2. Identification of Tick Species
2.3. Molecular Characterization and Phylogenetic Analysis of B. garinii
3. Discussion
4. Materials and Methods
4.1. Tick Collection and Species Identification
4.2. DNA Extraction, PCR, and Sequencing
4.3. Phylogenetic Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Stanek, G.; Reiter, M. The expanding Lyme Borrelia complex-clinical significance of genomic species? Clin. Microbiol. Infect. 2011, 17, 487–493. [Google Scholar] [CrossRef] [PubMed]
- Takano, A.; Nakao, M.; Masuzawa, T.; Takada, N.; Yano, Y.; Ishiguro, F.; Fujita, H.; Ito, T.; Ma, X.; Oikawa, Y.; et al. Multilocus sequence typing implicates rodents as the main reservoir host of human-pathogenic Borrelia garinii in Japan. J. Clin. Microbiol. 2011, 49, 2035–2039. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Burgdorfer, W.; Barbour, A.G.; Hayes, S.F.; Benach, J.L.; Grunwaldt, E.; Davis, J.P. Lyme disease-a tick-borne spirochetosis? Science 1982, 216, 1317–1319. [Google Scholar] [CrossRef] [PubMed]
- Park, K.H.; Lee, S.H.; Won, W.J.; Jang, W.J.; Chang, W.H. Isolation of Borrelia burgdorferi, the causative agent of Lyme disease, from Ixodes ticks in Korea. J. Korean Soc. Microbiol. 1992, 27, 307–312. [Google Scholar]
- Shih, C.M.; Chang, H.M.; Chen, S.L.; Chao, L.L. Genospecies identification and characterization of Lyme disease spirochetes of genospecies Borrelia burgdorferi sensu lato isolated from rodents in Taiwan. J. Clin. Microbiol. 1998, 36, 3127–3132. [Google Scholar] [PubMed]
- De Michelis, S.; Sewell, H.S.; Collares-Pereira, M.; Santos-Reis, M.; Schouls, L.M.; Benes, V.; Holmes, E.C.; Kurtenbach, K. Genetic diversity of Borrelia burgdorferi sensu lato in ticks from mainland Portugal. J. Clin. Microbiol. 2000, 38, 2128–2133. [Google Scholar]
- Goossens, H.A.; van den Bogaard, A.E.; Nohlmans, M.K. Dogs as sentinels for human Lyme borreliosis in The Netherlands. J. Clin. Microbiol. 2001, 39, 844–848. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Lyme Disease Data Tables: Historical Data. 2018. Available online: https://www.cdc.gov/lyme/stats/tables.html (accessed on 23 July 2018).
- Lee, M.G.; Chung, K.Y.; Choi, Y.S.; Cho, S.N. Lyme disease. Korean J. Dermatol. 1993, 31, 601–605. (In Korean) [Google Scholar] [CrossRef][Green Version]
- KCDC. Infectious Diseases Surveillance Yearbook 2018; Korea Centers for Disease Control and Prevention: Chungbuk, Korea, 2019.
- Lee, S.H.; Yun, S.H.; Choi, E.; Park, Y.S.; Lee, S.E.; Cho, G.J.; Kwon, O.D.; Kwak, D. Serological detection of Borrelia burgdorferi among horses in Korea. Korean J. Parasitol. 2016, 54, 97–101. [Google Scholar] [CrossRef]
- Suh, G.H.; Ahn, K.S.; Ahn, J.H.; Kim, H.J.; Leutenegger, C.; Shin, S.S. Serological and molecular prevalence of canine vector-borne diseases (CVBDs) in Korea. Parasites Vectors 2017, 10, 146. [Google Scholar] [CrossRef]
- Park, S.H.; Hwang, K.J.; Chu, H.; Park, M.Y. Serological detection of Lyme borreliosis agents in patients from Korea, 2005–2009. Osong Public Health Res. Perspect. 2011, 2, 29–33. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kang, J.G.; Chae, J.B.; Cho, Y.K.; Jo, Y.S.; Shin, N.S.; Lee, H.; Choi, K.S.; Yu, D.H.; Park, J.; Park, B.K. Molecular detection of Anaplasma, Bartonella, and Borrelia theileri in raccoon dogs (Nyctereutes procyonoides) in Korea. Am. J. Trop. Med. Hyg. 2018, 98, 1061–1068. [Google Scholar] [CrossRef] [PubMed]
- Smith, F.D.; Ballantyne, R.; Morgan, E.R.; Wall, R. Estimating Lyme disease risk using pet dogs as sentinels. Comp. Immunol. Microbiol. Infect. Dis. 2012, 35, 163–167. [Google Scholar] [CrossRef] [PubMed]
- Irwin, P.J.; Robertson, I.D.; Westman, M.E.; Perkins, M.; Straubinger, R.K. Searching for Lyme borreliosis in Australia: Results of a canine sentinel study. Parasites Vectors 2017, 10, 114. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, S.; Helps, C.; Tasker, S.; Newbury, H.; Wall, R. Prevalence and distribution of Borrelia and Babesia species in ticks feeding on dogs in the UK. Med. Vet. Entomol. 2018, 32, 14–22. [Google Scholar] [CrossRef]
- Duncan, A.W.; Correa, M.T.; Levine, J.F.; Breitschwerdt, E.B. The dog as a sentinel for human infection: Prevalence of Borrelia burgdorferi C6 antibodies in dogs from southeastern and mid-Atlantic states. Vector Borne Zoonotic Dis. 2004, 4, 221–229. [Google Scholar] [CrossRef]
- Song, T.Y.; Yang, E.M.; Kim, C.J. A pediatric case of severe fever with thrombocytopenia syndrome in Korea. J. Korean Med. Sci. 2017, 32, 704–707. [Google Scholar] [CrossRef]
- Kim, U.J.; Kim, D.M.; Kim, S.E.; Kang, S.J.; Jang, H.C.; Park, K.H.; Jung, S.I. Case report: Detection of the identical virus in a patient presenting with severe fever with thrombocytopenia syndrome encephalopathy and the tick that bit her. BMC Infect. Dis. 2018, 18, 181. [Google Scholar] [CrossRef]
- Lee, S.H.; Kim, H.J.; Byun, J.W.; Lee, M.J.; Kim, N.H.; Kim, D.H.; Kang, H.E.; Nam, H.M. Molecular detection and phylogenetic analysis of severe fever with thrombocytopenia syndrome virus in shelter dogs and cats in the Republic of Korea. Ticks Tick Borne Dis. 2017, 8, 626–630. [Google Scholar] [CrossRef]
- Lee, S.H.; Mossaad, E.; Ibrahim, A.M.; Ismail, A.A.; Moumouni, P.F.A.; Liu, M.; Ringo, A.E.; Gao, Y.; Guo, H.; Li, J.; et al. Detection and molecular characterization of tick-borne pathogens infecting sheep and goats in Blue Nile and West Kordofan states in Sudan. Ticks Tick Borne Dis. 2018, 9, 598–604. [Google Scholar] [CrossRef]
- Pal, U.; Li, X.; Wang, T.; Montgomery, R.R.; Ramamoorthi, N.; Bao, F.; Yang, X.; Pypaert, M.; Pradhan, D.; Kantor, F.S. TROSPA, an Ixodes scapularis receptor for Borrelia burgdorferi. Cell 2004, 119, 457–468. [Google Scholar] [CrossRef] [PubMed]
- Kee, S.; Hwang, K.J.; Oh, H.B.; Kim, M.B.; Shim, J.C.; Ree, H.I.; Park, K.S. Isolation and identification of Borrelia burgdorferi in Korea. J. Korean Soc. Microbiol. 1994, 29, 301–310. [Google Scholar]
- Park, K.H.; Chang, W.H.; Schwan, T.G. Identification and characterization of lyme disease spirochetes, Borrelia burgdorferi sensu lato, isolated in Korea. J. Clin. Microbiol. 1993, 31, 1831–1837. [Google Scholar] [PubMed]
- Park, K.H.; Choi, Y.J.; Kim, J.; Park, H.J.; Song, D.; Jang, W.J. Reclassification of Borrelia spp. isolated in South Korea using multilocus sequence typing. Jpn. J. Infect. Dis. 2018, 71, 350–353. [Google Scholar] [CrossRef] [PubMed]
- Margos, G.; Vollmer, S.A.; Cornet, M.; Garnier, M.; Fingerle, V.; Wilske, B.; Bormane, A.; Vitorino, L.; Collares-Pereira, M.; Drancourt, M.; et al. A new Borrelia species defined by multilocus sequence analysis of housekeeping genes. Appl. Environ. Microbiol. 2009, 75, 5410–5416. [Google Scholar] [CrossRef]
- Zygner, W.; Jaros, S.; Wędrychowicz, H. Prevalence of Babesia canis, Borrelia afzelii, and Anaplasma phagocytophilum infection in hard ticks removed from dogs in Warsaw (central Poland). Vet. Parasitol. 2008, 153, 139–142. [Google Scholar] [CrossRef]
- Földvári, G.; Márialigeti, M.; Solymosi, N.; Lukács, Z.; Majoros, G.; Kósa, J.P.; Farkas, R. Hard ticks infesting dogs in Hungary and their infection with Babesia and Borrelia species. Parasitol. Res. 2007, 101, 25–34. [Google Scholar] [CrossRef]
- Wang, J.; Kelly, P.; Zhang, J.; Shi, Z.; Song, C.; Zheng, X.; Zhang, Y.; Hao, Y.; Dong, H.; El-Mahallawy, H.S.; et al. Detection of Dirofilaria immitis antigen and antibodies against Anaplasma phagocytophilum, Borrelia burgdorferi and Ehrlichia canis in dogs from ten provinces of China. Acta Parasitol. 2018, 63, 412–415. [Google Scholar] [CrossRef]
- Chao, L.L.; Liu, L.L.; Ho, T.Y.; Shih, C.M. First detection and molecular identification of Borrelia garinii spirochete from Ixodes ovatus tick ectoparasitized on stray cat in Taiwan. PLoS ONE 2014, 9, e110599. [Google Scholar] [CrossRef]
- Fomenko, N.V.; Livanova, N.N.; Chernousova, N.Y. Diversity of Borrelia burgdorferi sensu lato in natural foci of Novosibirsk region. Int. J. Med. Microbiol. 2008, 298, 139–148. [Google Scholar] [CrossRef]
- Inokuma, H.; Maetani, S.; Fujitsuka, J.; Takano, A.; Sato, K.; Fukui, T.; Masuzawa, T.; Kawabata, H. Astasia and pyrexia related to Borrelia garinii infection in two dogs in Hokkaido, Japan. J. Vet. Med. Sci. 2013, 75, 975–978. [Google Scholar] [CrossRef] [PubMed]
- Smith, R.P., Jr.; Muzaffar, S.B.; Lavers, J.; Lacombe, E.H.; Cahill, B.K.; Lubelczyk, C.B.; Kinsler, A.; Mathers, A.J.; Rand, P.W. Borrelia garinii in seabird ticks (Ixodes uriae), Atlantic Coast, North America. Emerg. Infect. Dis. 2006, 12, 1909–1912. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.; Irwin, P.J.; Lee, S.; Oh, M.; Ahn, K.; Myung, B.; Shin, S. Comparison of selected canine vector-borne diseases between urban animal shelter and rural hunting dogs in Korea. Parasites Vectors 2010, 3, 32. [Google Scholar] [CrossRef] [PubMed]
- Jung, B.Y.; Gebeyehu, E.B.; Seo, M.G.; Byun, J.W.; Kim, H.Y.; Kwak, D. Prevalence of vector-borne diseases in shelter dogs in Korea. Vet. Rec. 2012, 171, 249. [Google Scholar] [CrossRef]
- Kim, B.J.; Kim, H.; Won, S.; Kim, H.C.; Chong, S.T.; Klein, T.A.; Kim, K.G.; Seo, H.Y.; Chae, J.S. Ticks collected from wild and domestic animals and natural habitats in the Republic of Korea. Korean J. Parasitol. 2014, 52, 281–285. [Google Scholar] [CrossRef]
- Choe, H.C.; Fudge, M.; Sames, W.J.; Robbins, R.G.; Lee, I.Y.; Chevalier, N.A.; Chilcoat, C.D.; Lee, S.H. Tick surveillance of dogs in the Republic of Korea. Syst. Appl. Acarol. 2011, 16, 215–222. [Google Scholar] [CrossRef]
- Sun, Y.; Xu, R. Ability of Ixodes persulcatus, Haemaphysalis concinna and Dermacentor silvarum ticks to acquire and transstadially transmit Borrelia garinii. Exp. Appl. Acarol. 2003, 31, 151. [Google Scholar] [CrossRef]
- VanBik, D.; Lee, S.H.; Seo, M.G.; Jeon, B.R.; Goo, Y.K.; Park, S.J.; Rhee, M.H.; Kwon, O.D.; Kim, T.H.; Geraldino, P.J.L.; et al. Borrelia species detected in ticks feeding on wild Korean water deer (Hydropotes inermis) using molecular and genotypic analyses. J. Med. Entomol. 2017, 54, 1397–1402. [Google Scholar] [CrossRef]
- Lee, K.; Takano, A.; Taylor, K.; Sashika, M.; Shimozuru, M.; Konnai, S.; Kawabata, H.; Tsubota, T. A relapsing fever group Borrelia sp. similar to Borrelia lonestari found among wild sika deer (Cervus nippon yesoensis) and Haemaphysalis spp. ticks in Hokkaido, Japan. Ticks Tick Borne Dis. 2014, 5, 841–847. [Google Scholar] [CrossRef]
- Figlerowicz, M.; Urbanowicz, A.; Lewandowski, D.; Jodynis-Liebert, J.; Sadowski, C. Functional insights into recombinant TROSPA protein from Ixodes ricinus. PLoS ONE 2013, 8, e76848. [Google Scholar] [CrossRef]
- Schwartz, J.J.; Gazumyan, A.; Schwartz, I. rRNA gene organization in the Lyme disease spirochete, Borrelia burgdorferi. J. Bacteriol. 1992, 174, 3757–3765. [Google Scholar] [CrossRef]
- Yang, X.F.; Pal, U.; Alani, S.M.; Fikrig, E.; Norgard, M.V. Essential role for OspA/B in the life cycle of the Lyme disease spirochete. J. Exp. Med. 2004, 199, 641–648. [Google Scholar] [CrossRef]
- Comstedt, P.; Hanner, M.; Schüler, W.; Meinke, A.; Schlegl, R.; Lundberg, U. Characterization and optimization of a novel vaccine for protection against Lyme borreliosis. Vaccine 2015, 33, 5982–5988. [Google Scholar] [CrossRef]
- Sadziene, A.; Thomas, D.D.; Bundoc, V.G.; Holt, S.C.; Barbour, A.G. A flagella-less mutant of Borrelia burgdorferi. Structural, molecular, and in vitro functional characterization. J. Clin. Investig. 1991, 88, 82–92. [Google Scholar] [CrossRef][Green Version]
- Park, H.S.; Lee, J.H.; Jeong, E.J.; Koh, S.E.; Park, T.K.; Jang, W.J.; Park, K.H.; Kim, B.J.; Kook, Y.H.; Lee, S.H. Evaluation of groEL gene analysis for identification of Borrelia burgdorferi sensu lato. J. Clin. Microbiol. 2004, 42, 1270–1273. [Google Scholar] [CrossRef][Green Version]
- Chao, L.L.; Shih, C.M. Molecular analysis of Rhipicephalus sanguineus (Acari: Ixodidae), an incriminated vector tick for Babesia vogeli in Taiwan. Exp. Appl. Acarol. 2016, 70, 469–481. [Google Scholar] [CrossRef]
- Lu, X.; Lin, X.D.; Wang, J.B.; Qin, X.C.; Tian, J.H.; Guo, W.P.; Fan, F.N.; Shao, R.; Xu, J.; Zhang, Y.Z. Molecular survey of hard ticks in endemic areas of tick-borne diseases in China. Ticks Tick Borne Dis. 2013, 4, 288–296. [Google Scholar] [CrossRef]
- Lv, J.; Wu, S.; Zhang, Y.; Chen, Y.; Feng, C.; Yuan, X.; Jia, G.; Deng, J.; Wang, C.; Wang, Q. Assessment of four DNA fragments (COI, 16S rDNA, ITS2, 12S rDNA) for species identification of the Ixodida (Acari: Ixodida). Parasites Vectors 2014, 7, 93. [Google Scholar] [CrossRef]
- Barbieri, A.M.; Venzal, J.M.; Marcili, A.; Almeida, A.P.; González, E.M.; Labruna, M.B. Borrelia burgdorferi sensu lato infecting ticks of the Ixodes ricinus complex in Uruguay: First report for the Southern Hemisphere. Vector Borne Zoonotic Dis. 2013, 13, 147–153. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- The Editors. Editorial. Cladistics 2016, 32, 1. [Google Scholar] [CrossRef]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.-H.; Goo, Y.-K.; Geraldino, P.J.L.; Kwon, O.-D.; Kwak, D. Molecular Detection and Characterization of Borrelia garinii (Spirochaetales: Borreliaceae) in Ixodes nipponensis (Ixodida: Ixodidae) Parasitizing a Dog in Korea. Pathogens 2019, 8, 289. https://doi.org/10.3390/pathogens8040289
Lee S-H, Goo Y-K, Geraldino PJL, Kwon O-D, Kwak D. Molecular Detection and Characterization of Borrelia garinii (Spirochaetales: Borreliaceae) in Ixodes nipponensis (Ixodida: Ixodidae) Parasitizing a Dog in Korea. Pathogens. 2019; 8(4):289. https://doi.org/10.3390/pathogens8040289
Chicago/Turabian StyleLee, Seung-Hun, Youn-Kyoung Goo, Paul John L. Geraldino, Oh-Deog Kwon, and Dongmi Kwak. 2019. "Molecular Detection and Characterization of Borrelia garinii (Spirochaetales: Borreliaceae) in Ixodes nipponensis (Ixodida: Ixodidae) Parasitizing a Dog in Korea" Pathogens 8, no. 4: 289. https://doi.org/10.3390/pathogens8040289
APA StyleLee, S.-H., Goo, Y.-K., Geraldino, P. J. L., Kwon, O.-D., & Kwak, D. (2019). Molecular Detection and Characterization of Borrelia garinii (Spirochaetales: Borreliaceae) in Ixodes nipponensis (Ixodida: Ixodidae) Parasitizing a Dog in Korea. Pathogens, 8(4), 289. https://doi.org/10.3390/pathogens8040289





