Spectroscopic Characterization of Bovine, Avian and Johnin Purified Protein Derivative (PPD) with High-Throughput Fourier Transform InfraRed-Based Method
Abstract
1. Introduction
2. Results
2.1. Raw IR Spectra Analysis
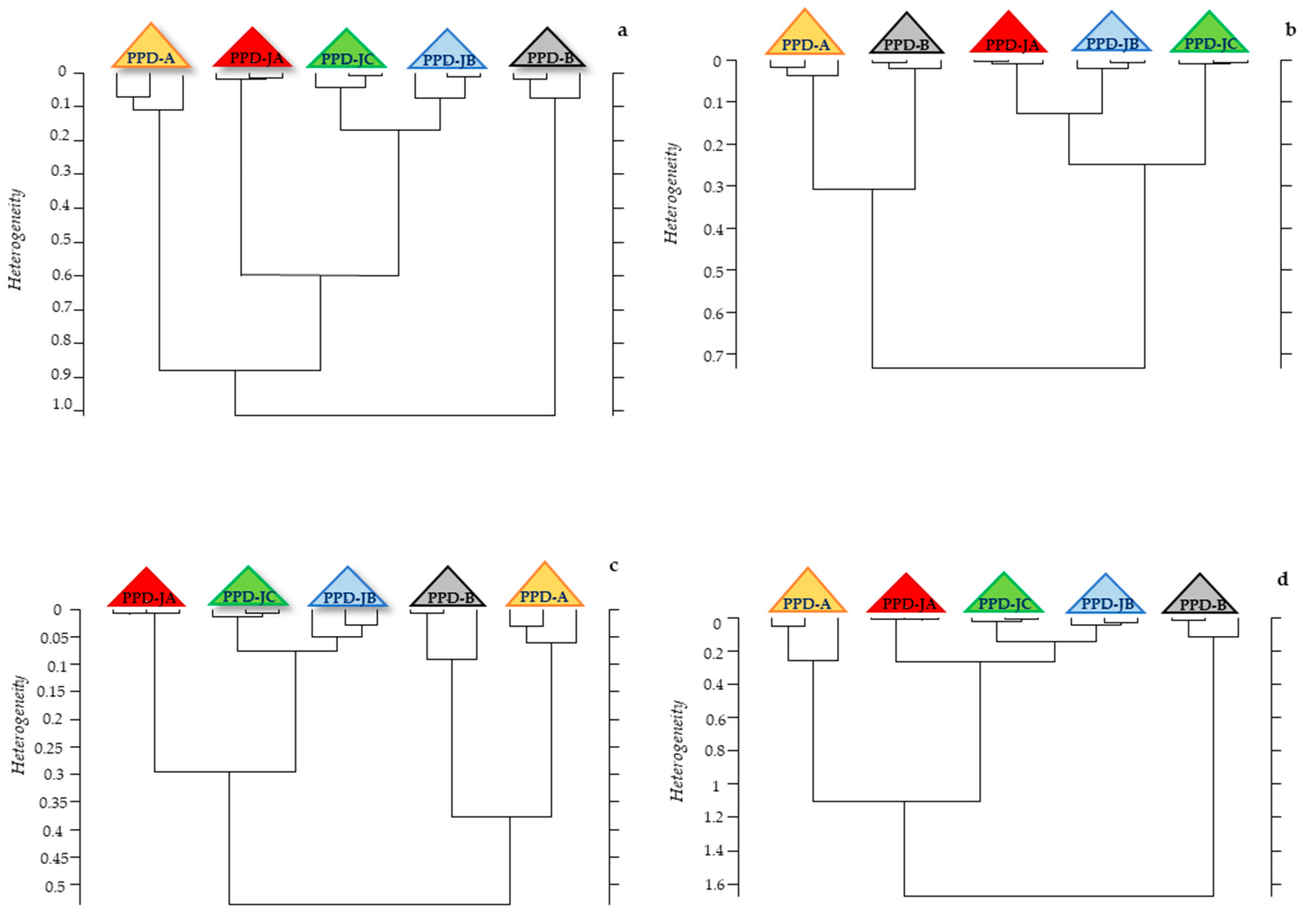
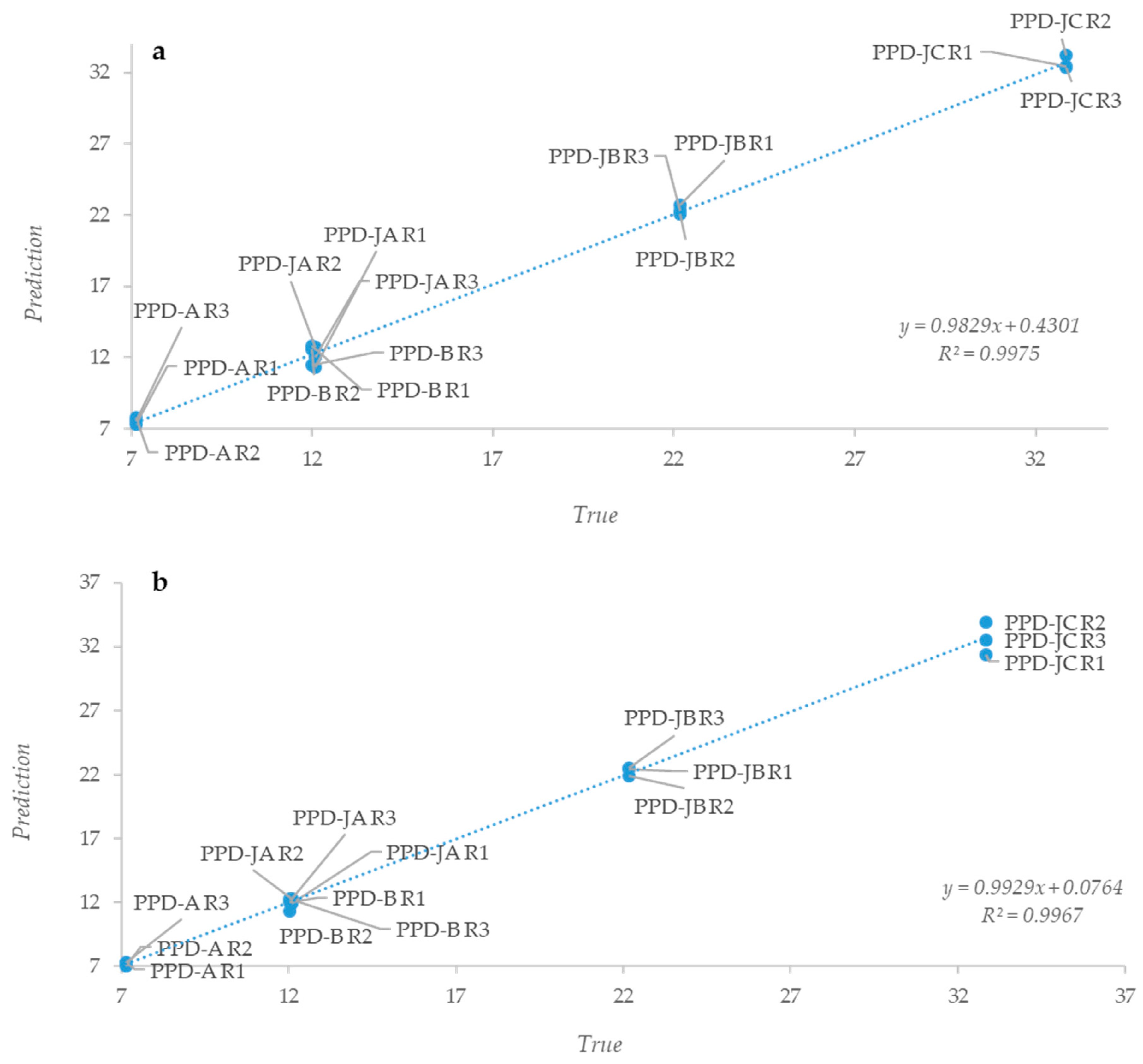
2.2. Statistical Analysis of IR Spectra
3. Discussion
4. Materials and Methods
4.1. PPDs Production
4.2. FTIR-Based Characterization
4.3. Spectra Statistical Analyses
4.4. Data Availability
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tortoli, E. The new mycobacteria: An update. FEMS Immunol. Med. Microbiol. 2006, 48, 159–178. [Google Scholar] [CrossRef] [PubMed]
- Romha, G.; Gebru, G.; Asefa, A.; Mamo, G. Epidemiology of Mycobacterium bovis and Mycobacterium tuberculosis in animals: Transmission dynamics and control challenges of zoonotic TB in Ethiopia. Prev. Vet. Med. 2018, 158, 1–17. [Google Scholar] [CrossRef] [PubMed]
- De la Rua-Domenech, R. Human Mycobacterium bovis infection in the United Kingdom: Incidence, risks, control measures and review of the zoonotic aspects of bovine tuberculosis. Tuberculosis 2006, 86, 77–109. [Google Scholar] [CrossRef] [PubMed]
- Kubica, T.; Rusch-Gerdes, S.; Niemann, S. Mycobacterium bovis subsp. caprae caused one-third of human M. bovis-associated tuberculosis cases reported in Germany between 1999 and 2001. J. Clin. Microbiol. 2003, 41, 3070–3077. [Google Scholar]
- Moravkova, M.; Hlozek, P.; Beran, V.; Pavlik, I.; Preziuso, S.; Cuteri, V.; Bartos, M. Strategy for the detection and differentiation of Mycobacterium avium species in isolates and heavily infected tissues. Res. Vet. Sci. 2008, 85, 257–264. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Latent Tuberculosis Infection: Updated and Consolidated Guidelines for Programmatic Management; World Health Organization: Geneva, Switzerland, 2018. [Google Scholar]
- Auguste, P.; Tsertsvadze, A.; Pink, J.; Court, R.; McCarthy, N.; Sutcliffe, P.; Clarke, A. Comparing interferon-gamma release assays with tuberculin skin test for identifying latent tuberculosis infection that progresses to active tuberculosis: Systematic review and meta-analysis. BMC Infect. Dis. 2017, 17, 200. [Google Scholar] [CrossRef] [PubMed]
- De la Rua-Domenech, R.; Goodchild, A.; Vordermeier, H.; Hewinson, R.; Christiansen, K.; Clifton-Hadley, R. Ante mortem diagnosis of tuberculosis in cattle: A review of the tuberculin tests, γ-interferon assay and other ancillary diagnostic techniques. Res. Vet. Sci. 2006, 81, 190–210. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Danelishvili, L.; Rose, S.J.; Bermudez, L.E. Mycobacterium bovis BCG Surface Antigens Expressed under the Granuloma-Like Conditions as Potential Inducers of the Protective Immunity. Int J. Microbiol. 2019, 2019. [Google Scholar] [CrossRef]
- European Community (Ed.) Commission Regulation (EC) 1226/2002 of 8 July 2002 amending Annex B to Council Directive 64/432/EEC. Off. J. Eur. Commun. 2002, 179, 13–18. [Google Scholar]
- EU Council. EU Council Directive 64/432/EEC. 1964. Available online: https //eur-lex.europa.eu/eli/dir/1964/432/oj/ (accessed on 15 October 2009).
- Clegg, T.A.; Good, M.; Doyle, M.; Duignan, A.; More, S.J.; Gormley, E. The performance of the interferon gamma assay when used as a diagnostic or quality assurance test in Mycobacterium bovis infected herds. Prev. Vet. Med. 2017, 140, 116–121. [Google Scholar] [CrossRef]
- World Organisation for Animal Health (OIE). Manual of Diagnostic Tests and Vaccines for Terrestrial Animals; Bovine Tuberculosis Chapter 3.4.6 (NB: Version adopted in May 2009); OIE: Paris, France, 2009; Available online: https://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/3.04.06_BOVINE_TB.pdf (accessed on 26 August 2019).
- Eirin, M.E.; Macias, A.; Magnano, G.; Morsella, C.; Mendez, L.; Blanco, F.C.; Bianco, M.V.; Severina, W.; Alito, A.; de los Angeles Pando, M.; et al. Identification and evaluation of new Mycobacterium bovis antigens in the in vitro interferon gamma release assay for bovine tuberculosis diagnosis. Tuberculosis Edinb. 2015, 95, 795–801. [Google Scholar] [CrossRef] [PubMed]
- Wood, P.R.; Jones, S.L. BOVIGAM: An in vitro cellular diagnostic test for bovine tuberculosis. Tuberculosis Edinb. 2001, 81, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Wood, P.R.; Rothel, J.S. In vitro immunodiagnostic assays for bovine tuberculosis. Vet. Microbiol. 1994, 40, 125–135. [Google Scholar] [CrossRef]
- Wood, P.R.; Corner, L.A.; Rothel, J.S.; Baldock, C.; Jones, S.L.; Cousins, D.B.; McCormick, B.S.; Francis, B.R.; Creeper, J.; Tweddle, N.E. Field comparison of the interferon-gamma assay and the intradermal tuberculin test for the diagnosis of bovine tuberculosis. Aust. Vet. J. 1991, 68, 286–290. [Google Scholar] [CrossRef] [PubMed]
- Chiodini, R.J.; Chamberlin, W.M.; Sarosiek, J.; McCallum, R.W. Crohn’s disease and the mycobacterioses: A quarter century later. Causation or simple association? Crit. Rev. Microbiol. 2012, 38, 52–93. [Google Scholar] [CrossRef] [PubMed]
- Harris, N.B.; Barletta, R.G. Mycobacterium avium subsp. paratuberculosis in Veterinary Medicine. Clin. Microbiol. Rev. 2001, 14, 489–512. [Google Scholar] [CrossRef] [PubMed]
- Chiodini, R.J.; Van Kruiningen, H.J.; Merkal, R.S. Ruminant paratuberculosis (Johne’s disease): The current status and future prospects. Cornell Vet. 1984, 74, 218–262. [Google Scholar]
- Waddell, L.A.; Rajic, A.; Stark, K.D.; McEwen, S.A. The zoonotic potential of Mycobacterium avium ssp. paratuberculosis: A systematic review and meta-analyses of the evidence. Epidemiol. Infect. 2015, 143, 3135–3157. [Google Scholar] [CrossRef]
- Jungersen, G.; Mikkelsen, H.; Grell, S.N. Use of the johnin PPD interferon-gamma assay in control of bovine paratuberculosis. Vet. Immunol. Immunopathol. 2012, 148, 48–54. [Google Scholar] [CrossRef]
- Kalis, C.; Collins, M.; Hesselink, J.; Barkema, H. Specificity of two tests for the early diagnosis of bovine paratuberculosis based on cell-mediated immunity: The Johnin skin test and the gamma interferon assay. Vet. Microbiol. 2003, 97, 73–86. [Google Scholar] [CrossRef]
- Jungersen, G.; Huda, A.; Hansen, J.J.; Lind, P. Interpretation of the gamma interferon test for diagnosis of subclinical paratuberculosis in cattle. Clin. Diagn. Lab. Immunol. 2002, 9, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Kruh-Garcia, N.A.; Dobos, K.M. Purified protein derivatives of tuberculin--past, present, and future. FEMS Immunol. Med. Microbiol. 2012, 66, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Wynne, J.W.; Shiell, B.J.; Colgrave, M.L.; Vaughan, J.A.; Beddome, G.; Michalski, W.P. Production and proteomic characterisation of purified protein derivative from Mycobacterium avium subsp. paratuberculosis. Proteome Sci. 2012, 10, 22. [Google Scholar] [CrossRef] [PubMed]
- Borsuk, S.; Newcombe, J.; Mendum, T.A.; Dellagostin, O.A.; McFadden, J. Identification of proteins from tuberculin purified protein derivative (PPD) by LC-MS/MS. Tuberculosis Edinb. 2009, 89, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Kuwabara, S.; Tsumita, T. Letter: Primary structure of tuberculin-active protein from tubercule bacilli. Jpn. J. Exp. Med. 1974, 44, 129–132. [Google Scholar] [PubMed]
- Seibert, F.; Glen, J. PPD-S was comprised of approximately 92.1% protein, 5.9% polysaccharides and 1.2% nucleic acid. Am. Rev. Tuberc. 1941, 44, 9–24. [Google Scholar]
- Cho, Y.S.; Dobos, K.M.; Prenni, J.; Yang, H.; Hess, A.; Rosenkrands, I.; Andersen, P.; Ryoo, S.W.; Bai, G.H.; Brennan, M.J.; et al. Deciphering the proteome of the in vivo diagnostic reagent “purified protein derivative” from Mycobacterium tuberculosis. Proteomics 2012, 12, 979–991. [Google Scholar] [CrossRef] [PubMed]
- Ho, M.M.; Kairo, S.K.; Corbel, M.J. Tuberculin purified protein derivative (PPD) immunoassay as an in vitro alternative assay for identity and confirmation of potency. Hum. Vaccin. 2006, 2, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Rowland, S.S.; Ruckert, J.L.; Cummings, P.J. Low molecular mass protein patterns in mycobacterial culture filtrates and purified protein derivatives. FEMS Immunol. Med. Microbiol. 1999, 23, 21–25. [Google Scholar] [CrossRef]
- Klausen, J.; Magnusson, M.; Andersen, A.B.; Koch, C. Characterization of purified protein derivative of tuberculin by use of monoclonal antibodies: Isolation of a delayed-type hypersensitivity reactive component from M. tuberculosis culture filtrate. Scand. J. Immunol. 1994, 40, 345–349. [Google Scholar] [CrossRef]
- Santema, W.; Overdijk, M.; Barends, J.; Krijgsveld, J.; Rutten, V.; Koets, A. Searching for proteins of Mycobacterium avium subspecies paratuberculosis with diagnostic potential by comparative qualitative proteomic analysis of mycobacterial tuberculins. Vet. Microbiol. 2009, 138, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Roscini, L.; Corte, L.; Antonielli, L.; Rellini, P.; Fatichenti, F.; Cardinali, G. Influence of cell geometry and number of replicas in the reproducibility of whole cell FTIR analysis. Analyst 2010, 135, 2099–2105. [Google Scholar] [CrossRef] [PubMed]
- Del Bove, M.; Lattanzi, M.; Rellini, P.; Pelliccia, C.; Fatichenti, F.; Cardinali, G. Comparison of molecular and metabolomic methods as characterization tools of Debaryomyces hansenii cheese isolates. Food Microbiol. 2009, 26, 453–459. [Google Scholar] [CrossRef] [PubMed]
- Szeghalmi, A.; Kaminskyj, S.; Gough, K.M. A synchrotron FTIR microspectroscopy investigation of fungal hyphae grown under optimal and stressed conditions. Anal. Bioanal. Chem. 2007, 387, 1779–1789. [Google Scholar] [CrossRef] [PubMed]
- Fischer, G.; Braun, S.; Thissen, R.; Dott, W. FT-IR spectroscopy as a tool for rapid identification and intra-species characterization of airborne filamentous fungi. J. Microbiol. Methods 2006, 64, 63–77. [Google Scholar] [CrossRef] [PubMed]
- Adt, I.; Toubas, D.; Pinon, J.M.; Manfait, M.; Sockalingum, G.D. FTIR spectroscopy as a potential tool to analyse structural modifications during morphogenesis of Candida albicans. Arch. Microbiol. 2006, 185, 277–285. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Irudayaraj, J. Spectroscopic characterization of microorganisms by Fourier transform infrared microspectroscopy. Biopolymers 2005, 77, 368–377. [Google Scholar] [CrossRef]
- Zhao, H.; Kassama, Y.; Young, M.; Kell, D.B.; Goodacre, R. Differentiation of Micromonospora isolates from a coastal sediment in Wales on the basis of Fourier transform infrared spectroscopy, 16S rRNA sequence analysis, and the amplified fragment length polymorphism technique. Appl. Environ. Microbiol. 2004, 70, 6619–6627. [Google Scholar] [CrossRef]
- Lai, S.; Goodacre, R.; Manchester, L.N. Whole-organism fingerprinting of the genus Carnobacterium using Fourier transform infrared spectroscopy (FT-IR). Syst. Appl. Microbiol. 2004, 27, 186–191. [Google Scholar] [CrossRef][Green Version]
- Wenning, M.; Seiler, H.; Scherer, S. Fourier-transform infrared microspectroscopy, a novel and rapid tool for identification of yeasts. Appl. Environ. Microbiol. 2002, 68, 4717–4721. [Google Scholar] [CrossRef]
- Oberreuter, H.; Seiler, H.; Scherer, S. Identification of coryneform bacteria and related taxa by Fourier-transform infrared (FT-IR) spectroscopy. Int. J. Syst. Evol. Microbiol. 2002, 52, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Kummerle, M.; Scherer, S.; Seiler, H. Rapid and reliable identification of food-borne yeasts by Fourier-transform infrared spectroscopy. Appl. Environ. Microbiol. 1998, 64, 2207–2214. [Google Scholar] [PubMed]
- Goodacre, R.; Timmins, E.M.; Rooney, P.J.; Rowland, J.J.; Kell, D.B. Rapid identification of Streptococcus and Enterococcus species using diffuse reflectance-absorbance Fourier transform infrared spectroscopy and artificial neural networks. FEMS Microbiol. Lett. 1996, 140, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Helm, D.; Labischinski, H.; Schallehn, G.; Naumann, D. Classification and identification of bacteria by Fourier-transform infrared spectroscopy. J. Gen. Microbiol. 1991, 137, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Corte, L.; Antonielli, L.; Roscini, L.; Fatichenti, F.; Cardinali, G. Influence of cell parameters in Fourier transform infrared spectroscopy analysis of whole yeast cells. Analyst 2011, 136, 2339–2349. [Google Scholar] [CrossRef] [PubMed]
- Corte, L.; Rellini, P.; Roscini, L.; Fatichenti, F.; Cardinali, G. Development of a novel, FTIR (Fourier transform infrared spectroscopy) based, yeast bioassay for toxicity testing and stress response study. Anal. Chim. Acta 2010, 659, 258–265. [Google Scholar] [CrossRef] [PubMed]
- Winder, C.L.; Gordon, S.V.; Dale, J.; Hewinson, R.G.; Goodacre, R. Metabolic fingerprints of Mycobacterium bovis cluster with molecular type: Implications for genotype-phenotype links. Microbiology 2006, 152, 2757–2765. [Google Scholar] [CrossRef]
- Rebuffo-Scheer, C.A.; Kirschner, C.; Staemmler, M.; Naumann, D. Rapid species and strain differentiation of non-tubercoulous mycobacteria by Fourier-Transform Infrared microspectroscopy. J. Microbiol. Methods 2007, 68, 282–290. [Google Scholar] [CrossRef]
- Semret, M.; Bakker, D.; Smart, N.; Olsen, I.; Haslov, K.; Behr, M.A. Genetic analysis of Mycobacterium avium complex strains used for producing purified protein derivatives. Clin. Vacc. Immunol. CVI 2006, 13, 991–996. [Google Scholar] [CrossRef]
- Mazzone, P.; Corneli, S.; Vitale, N.; Di Paolo, A.; Biagetti, M.; Maresca, C.; Ricchi, M.; Mangili, P.; Papa, P.; Pezzotti, G.; et al. Use of new johnin in gamma-IFN test to detect Mycobacterium avium subsp. paratuberculosis (MAP) infected cattle: Preliminary data. In Proceedings of the XII International Colloquium on Paratuberculosis, Parma, Italy, 22–26 June 2014; p. 127. [Google Scholar]
- Rue-Albrecht, K.; Magee, D.A.; Killick, K.E.; Nalpas, N.C.; Gordon, S.V.; MacHugh, D.E. Comparative functional genomics and the bovine macrophage response to strains of the mycobacterium genus. Front. Immunol. 2014, 5, 536. [Google Scholar] [CrossRef]
- Dvorska, L.; Bartos, M.; Martin, G.; Erler, W.; Pavlik, I. Strategies for differentiation, identification and typing of medically important species of mycobacteria by molecular methods. Vet. Med. PRAHA 2001, 46, 309–328. [Google Scholar] [CrossRef]
- Rogall, T.; Wolters, J.; Flohr, T.; Bottger, E.C. Towards a phylogeny and definition of species at the molecular level within the genus Mycobacterium. Int. J. Syst. Bacteriol. 1990, 40, 323–330. [Google Scholar] [CrossRef] [PubMed]
- Brosch, R.; Gordon, S.V.; Marmiesse, M.; Brodin, P.; Buchrieser, C.; Eiglmeier, K.; Garnier, T.; Gutierrez, C.; Hewinson, G.; Kremer, K.; et al. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. USA 2002, 99, 3684–3689. [Google Scholar] [CrossRef] [PubMed]
- Schiller, I.; Vordermeier, H.M.; Waters, W.R.; Kyburz, A.; Cagiola, M.; Whelan, A.; Palmer, M.V.; Thacker, T.C.; Meijlis, J.; Carter, C.; et al. Comparison of tuberculin activity using the interferon-gamma assay for the diagnosis of bovine tuberculosis. Vet. Rec. 2010, 167, 322–326. [Google Scholar] [CrossRef] [PubMed]
- Capsel, R.T.; Thoen, C.O.; Reinhardt, T.A.; Lippolis, J.D.; Olsen, R.; Stabel, J.R.; Bannantine, J.P. Composition and Potency Characterization of Mycobacterium avium subsp. paratuberculosis Purified Protein Derivatives. PLoS ONE 2016, 11, e0154685. [Google Scholar] [CrossRef]
- Prasad, T.S.; Verma, R.; Kumar, S.; Nirujogi, R.S.; Sathe, G.J.; Madugundu, A.K.; Sharma, J.; Puttamallesh, V.N.; Ganjiwale, A.; Myneedu, V.P.; et al. Proteomic analysis of purified protein derivative of Mycobacterium tuberculosis. Clin. Proteomics 2013, 10, 8. [Google Scholar] [CrossRef] [PubMed]
- Wiker, H.G. Unmatched sequences in public databases—Exemplified by tuberculin-active protein. Scand. J. Immunol. 2004, 59, 607–608. [Google Scholar] [CrossRef]
- Harboe, M.; Nagai, S.; Wiker, H.G.; Sletten, K.; Haga, S. Homology between the MPB70 and MPB83 proteins of Mycobacterium bovis BCG. Scand. J. Immunol. 1995, 42, 46–51. [Google Scholar] [CrossRef]
- Harboe, M.; Wiker, H.G.; Lachmann, P.J. Carrier effect of concanavalin A-reactive and -non-reactive material in tuberculin PPD. Scand. J. Immunol. 1990, 32, 263–271. [Google Scholar] [CrossRef]
- Roperto, S.; Varano, M.; Russo, V.; Luca, R.; Cagiola, M.; Gaspari, M.; Ceccarelli, D.M.; Cuda, G.; Roperto, F. Proteomic analysis of protein purified derivative of Mycobacterium bovis. J. Transl. Med. 2017, 15, 68. [Google Scholar] [CrossRef]
- Colabella, C.; Corte, L.; Roscini, L.; Shapaval, V.; Kohler, A.; Tafintseva, V.; Tascini, C.; Cardinali, G. Merging FT-IR and NGS for simultaneous phenotypic and genotypic identification of pathogenic Candida species. PLoS ONE 2017, 12, e0188104. [Google Scholar] [CrossRef] [PubMed]
- Corte, L.; Dell’abate, M.T.; Magini, A.; Migliore, M.; Felici, B.; Roscini, L.; Sardella, R.; Tancini, B.; Emiliani, C.; Cardinali, G.; et al. Assessment of safety and efficiency of nitrogen organic fertilizers from animal-based protein hydrolysates--a laboratory multidisciplinary approach. J. Sci. Food Agric. 2014, 94, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Corte, L.; di Cagno, R.; Groenewald, M.; Roscini, L.; Colabella, C.; Gobbetti, M.; Cardinali, G. Phenotypic and molecular diversity of Meyerozyma guilliermondii strains isolated from food and other environmental niches, hints for an incipient speciation. Food Microbiol. 2015, 48, 206–215. [Google Scholar] [CrossRef] [PubMed]
- Ricchi, M.; Barbieri, G.; Taddei, R.; Belletti, G.L.; Carra, E.; Cammi, G.; Garbarino, C.A.; Arrigoni, N. Effectiveness of combination of Mini-and Microsatellite loci to sub-type Mycobacterium avium subsp. paratuberculosis Italian type C isolates. BMC Vet. Res. 2011, 7, 54. [Google Scholar] [CrossRef]
- Helrich, K. Official Methods of Analysis of the AOAC; AOAC 2275 Research Blvd, Ste 300 (7146,04 km) 20850–3250; Association of Official Analytical Chemists Inc.: Rockville, MD, USA, 1990; Volume 2. [Google Scholar]
- Essendoubi, M.; Toubas, D.; Bouzaggou, M.; Pinon, J.M.; Manfait, M.; Sockalingum, G.D. Rapid identification of Candida species by FT-IR microspectroscopy. Biochim. Biophys. Acta 2005, 1724, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Naumann, D.; Helm, D.; Labischinski, H. Microbiological characterizations by FT-IR spectroscopy. Nature 1991, 351, 81–82. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; McEwen, G.D.; Wu, Y.; Miller, C.D.; Zhou, A. Characterization and analysis of mycobacteria and Gram-negative bacteria and co-culture mixtures by Raman microspectroscopy, FTIR, and atomic force microscopy. Anal. Bioanal. Chem. 2013, 405, 1577–1591. [Google Scholar] [CrossRef] [PubMed]
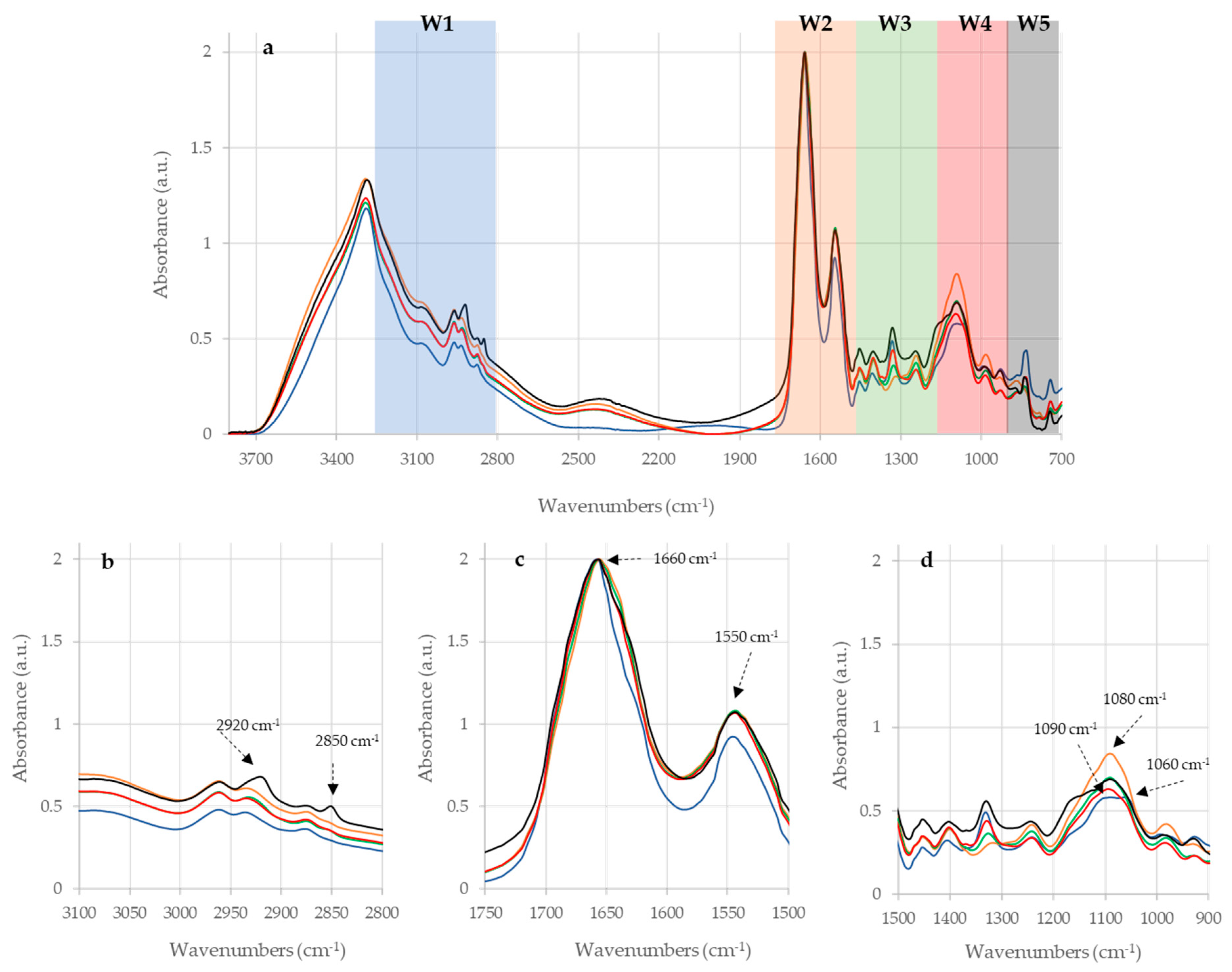
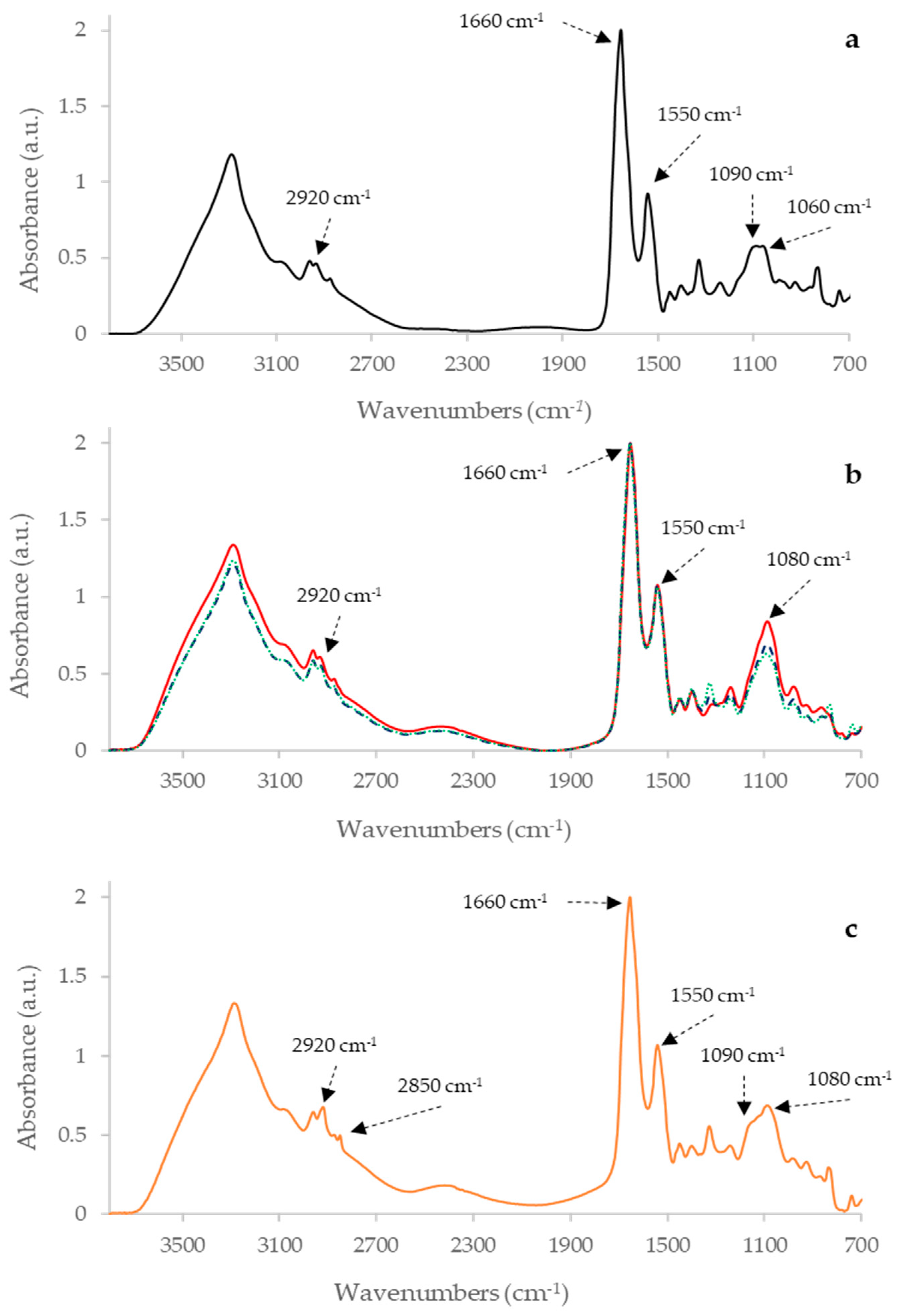
| Sample | Wavenumbers (cm−1) | Definition of Band Assignments |
|---|---|---|
| PPDA | 2920 | Asymmetric stretching of CH2 in fatty acids 1 |
| 2850 | Symmetric stretching of CH2 in fatty acids 1 | |
| 1660 | Amide I of α-helical structure 1 | |
| 1550 | Amide II 1 | |
| 1160 | stretching of CC and bending of COP and COH in DNA and RNA backbone 1 | |
| 1080 | DNA and RNA backbones C-C symmetric stretching 1 | |
| PPDB | 2920 | Asymmetric stretching of CH2 in fatty acids 1 |
| 1660 | α-helix of Amide I 1 | |
| 1550 | Amide II 1 | |
| 1090 | Symmetric stretching of P = O of nucleic acids and phospholipids 1 | |
| 1060 | P=O absorption band of nucleic acids and phospholipids 2 | |
| PPDJ (A, B and C) | 2920 | Asymmetric stretching of CH2 in fatty acids 1 |
| 1660 | α-helix of Amide I 1 | |
| 1550 | Amide II 1 | |
| 1080 | DNA and RNA backbones C-C symmetric stretching 1 |
| PPD-A | PPD-B | PPD-JA | PPD-JB | PPD-JC | |
|---|---|---|---|---|---|
| Average W1 Peak | 14.859 | 11.844 | 12.096 | 13.187 | 12.421 |
| Average W2 Peak | 175.482 | 183.900 | 189.907 | 191.089 | 189.649 |
| Average W3 Peak | 6.329 | 3.339 | 13.418 | 11.288 | 8.394 |
| Average W4 Peak | 36.162 | 35.787 | 59.996 | 50.583 | 45.902 |
| Average W5 Peak | 0.621 | 0.556 | 0.359 | 0.303 | 0.435 |
| Average WS Sum | 233.453 | 235.426 | 275.775 | 266.449 | 256.801 |
| Average W1/WS | 0.064 | 0.050 | 0.044 | 0.049 | 0.048 |
| Average W2/WS | 0.752 | 0.781 | 0.689 | 0.717 | 0.739 |
| Average W3/WS | 0.027 | 0.014 | 0.049 | 0.042 | 0.033 |
| Average W4/WS | 0.155 | 0.152 | 0.218 | 0.190 | 0.179 |
| Average W5/WS | 0.003 | 0.002 | 0.001 | 0.001 | 0.002 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Corneli, S.; Corte, L.; Roscini, L.; Di Paolo, A.; Colabella, C.; Petrucci, L.; Severi, G.; Cagiola, M.; Mazzone, P. Spectroscopic Characterization of Bovine, Avian and Johnin Purified Protein Derivative (PPD) with High-Throughput Fourier Transform InfraRed-Based Method. Pathogens 2019, 8, 136. https://doi.org/10.3390/pathogens8030136
Corneli S, Corte L, Roscini L, Di Paolo A, Colabella C, Petrucci L, Severi G, Cagiola M, Mazzone P. Spectroscopic Characterization of Bovine, Avian and Johnin Purified Protein Derivative (PPD) with High-Throughput Fourier Transform InfraRed-Based Method. Pathogens. 2019; 8(3):136. https://doi.org/10.3390/pathogens8030136
Chicago/Turabian StyleCorneli, Sara, Laura Corte, Luca Roscini, Antonella Di Paolo, Claudia Colabella, Linda Petrucci, Giulio Severi, Monica Cagiola, and Piera Mazzone. 2019. "Spectroscopic Characterization of Bovine, Avian and Johnin Purified Protein Derivative (PPD) with High-Throughput Fourier Transform InfraRed-Based Method" Pathogens 8, no. 3: 136. https://doi.org/10.3390/pathogens8030136
APA StyleCorneli, S., Corte, L., Roscini, L., Di Paolo, A., Colabella, C., Petrucci, L., Severi, G., Cagiola, M., & Mazzone, P. (2019). Spectroscopic Characterization of Bovine, Avian and Johnin Purified Protein Derivative (PPD) with High-Throughput Fourier Transform InfraRed-Based Method. Pathogens, 8(3), 136. https://doi.org/10.3390/pathogens8030136