Molecular Epizootiology of Toxoplasma gondii and Cryptosporidium parvum in the Eastern Oyster (Crassostrea virginica) from Maine (USA)
Abstract
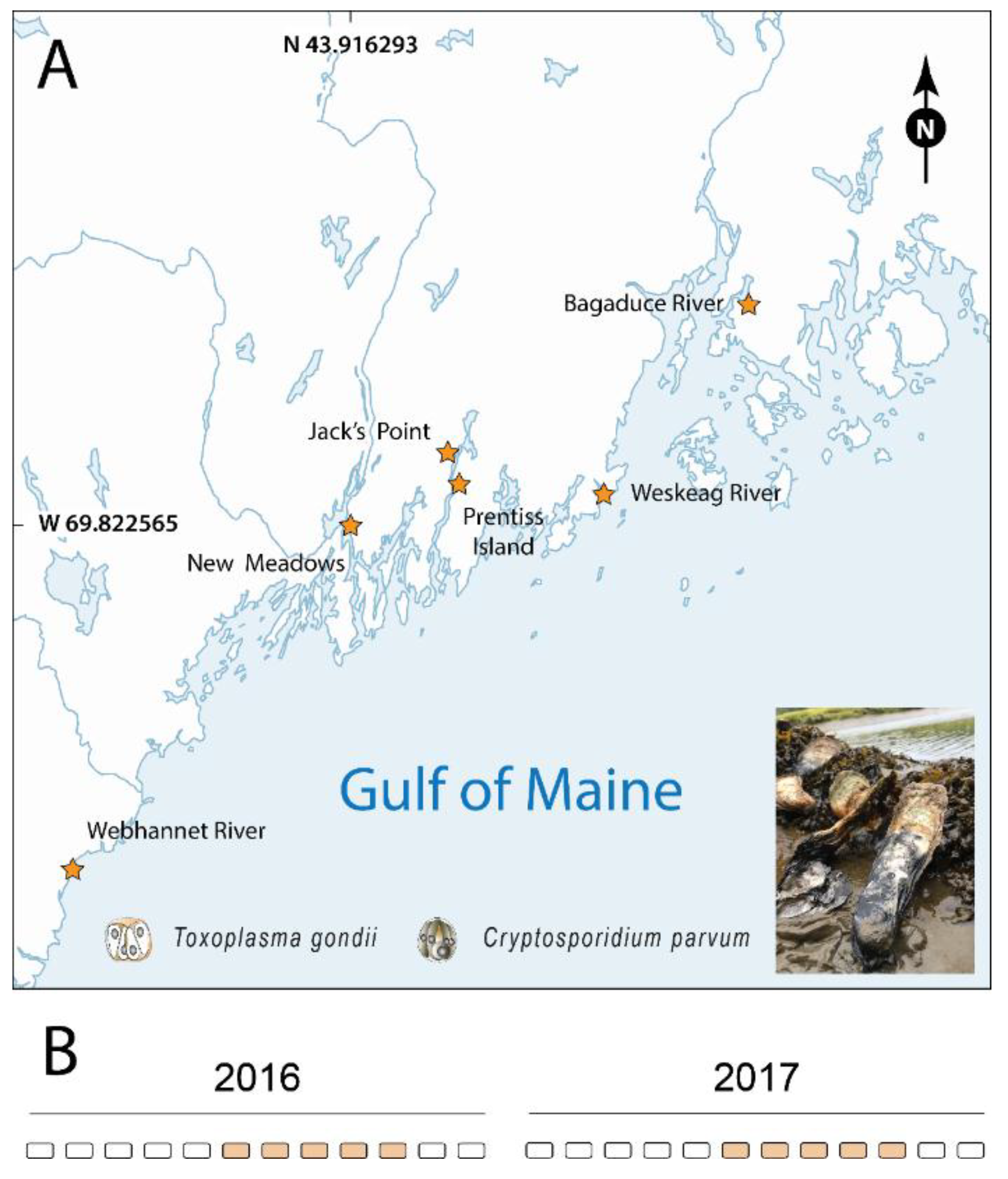
:1. Introduction
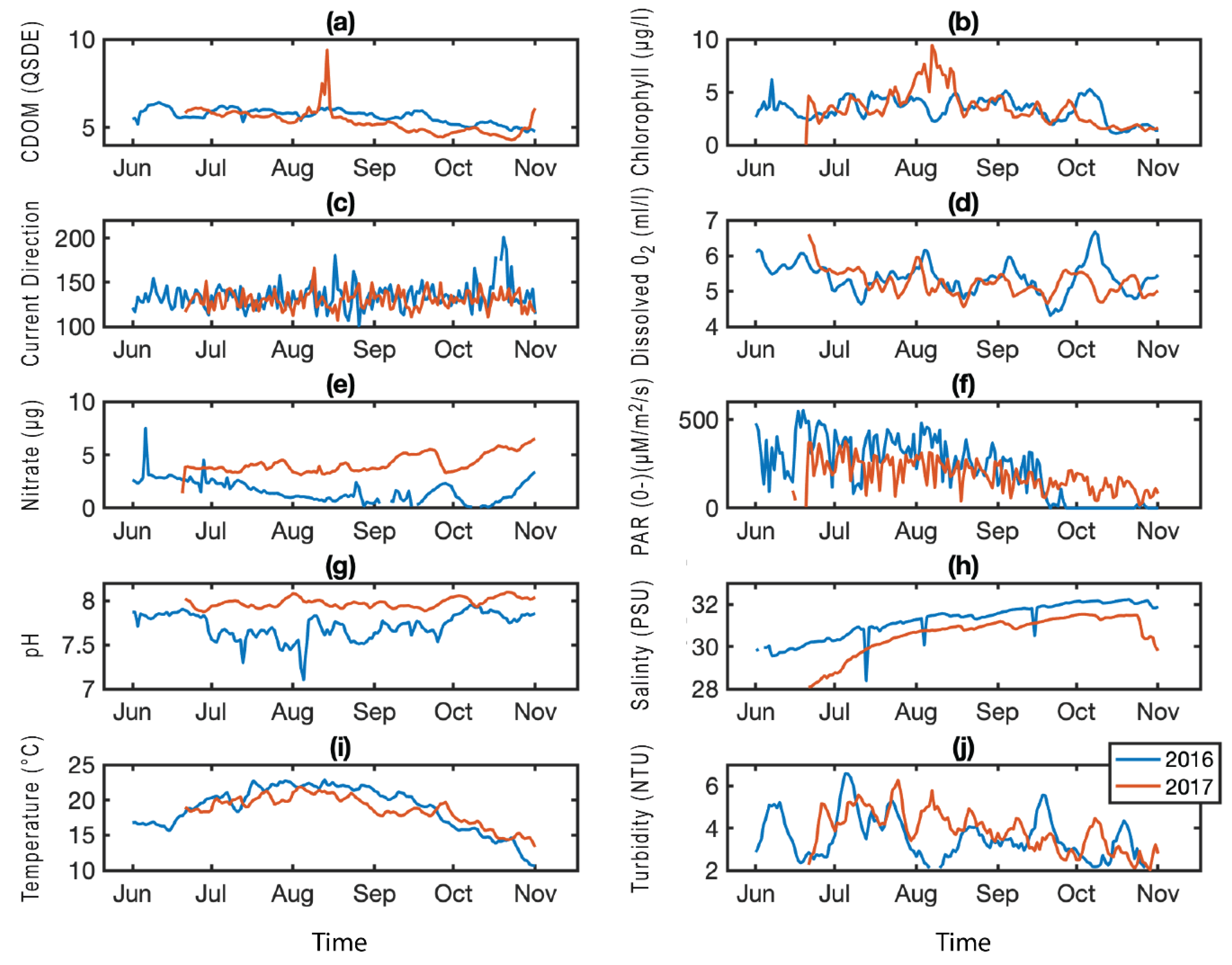
2. Results
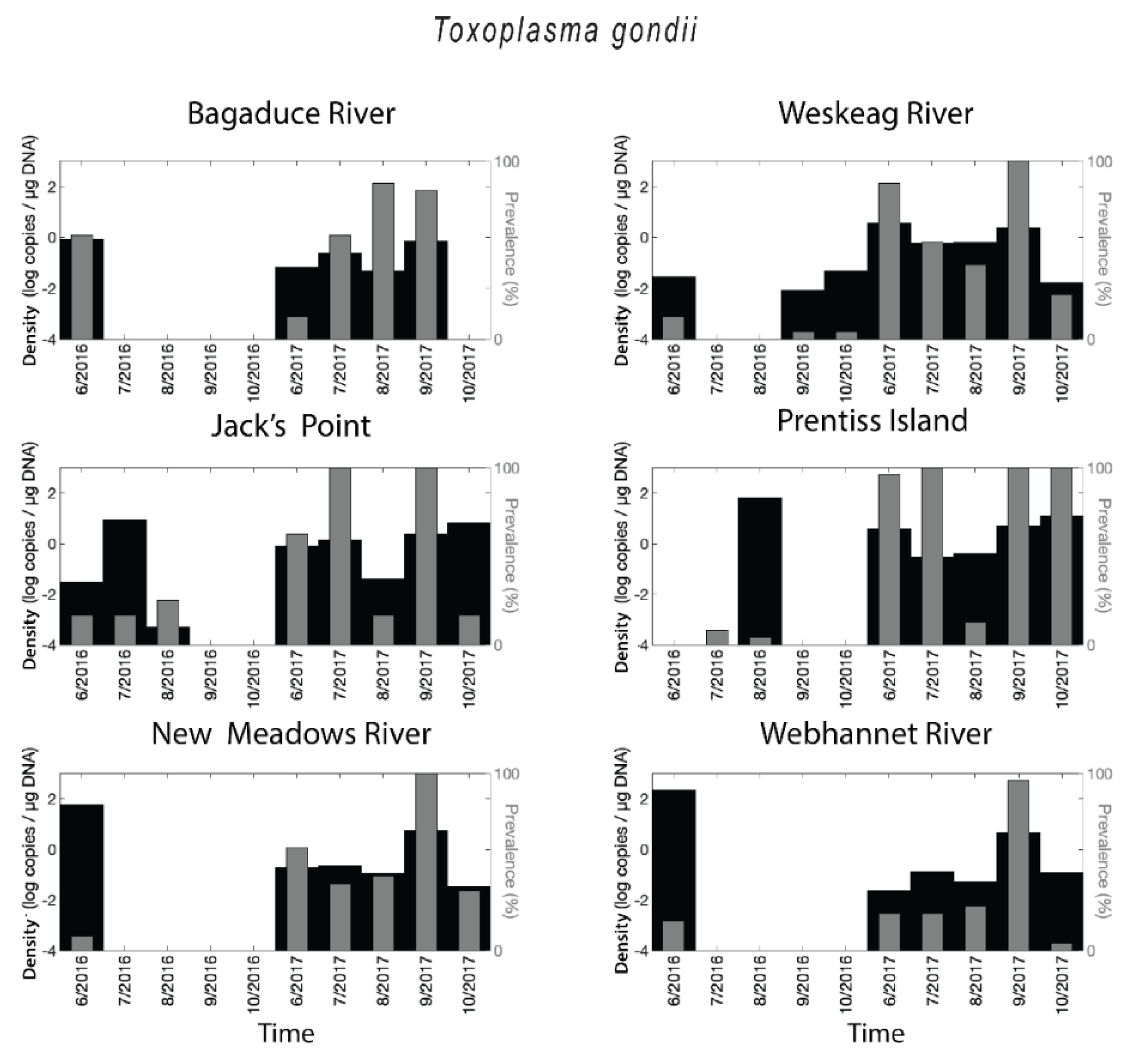
2.1. Toxoplasma gondii Prevalence and Densities
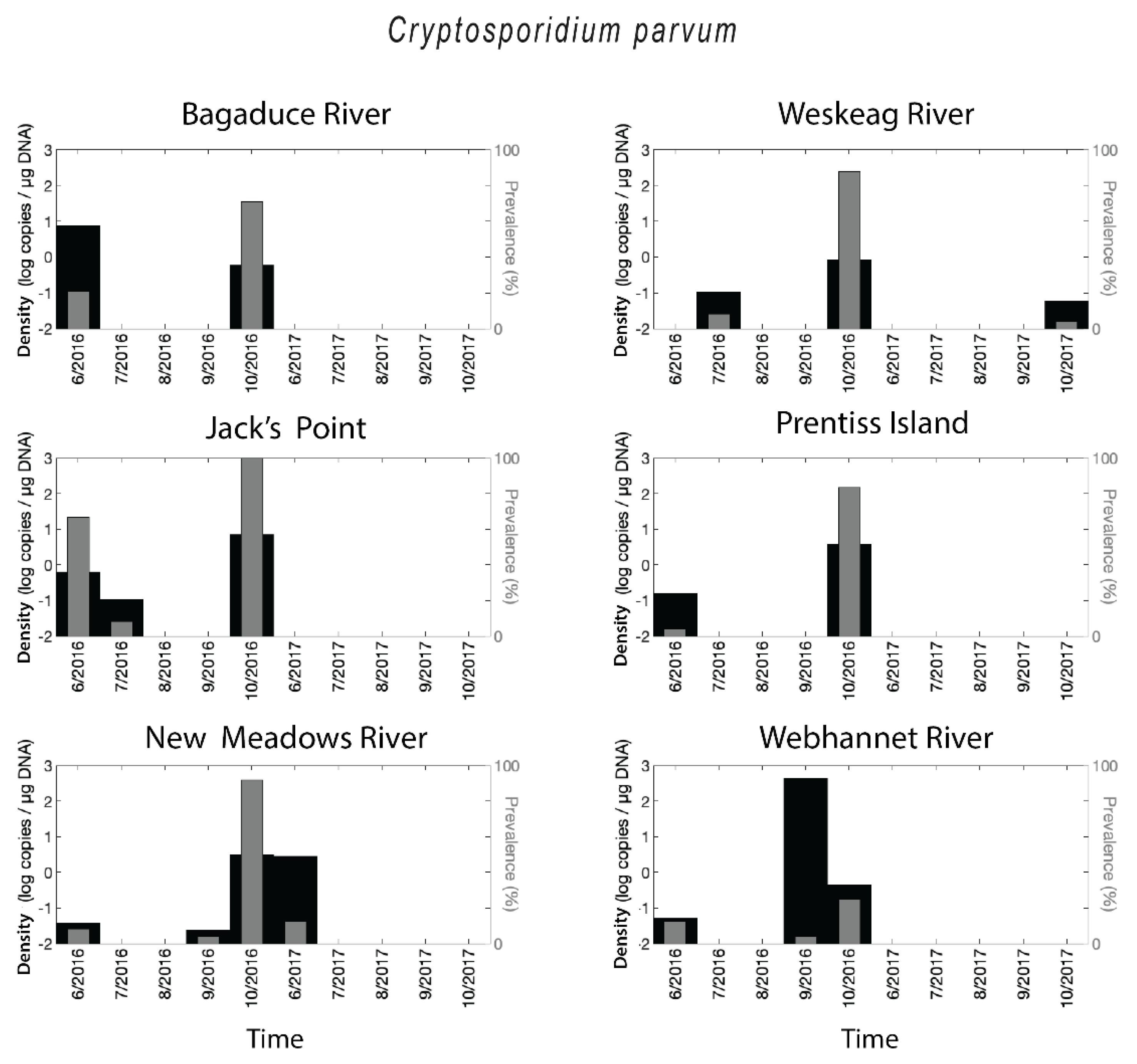
2.2. Cryptosporidium parvum Prevalence and Densities
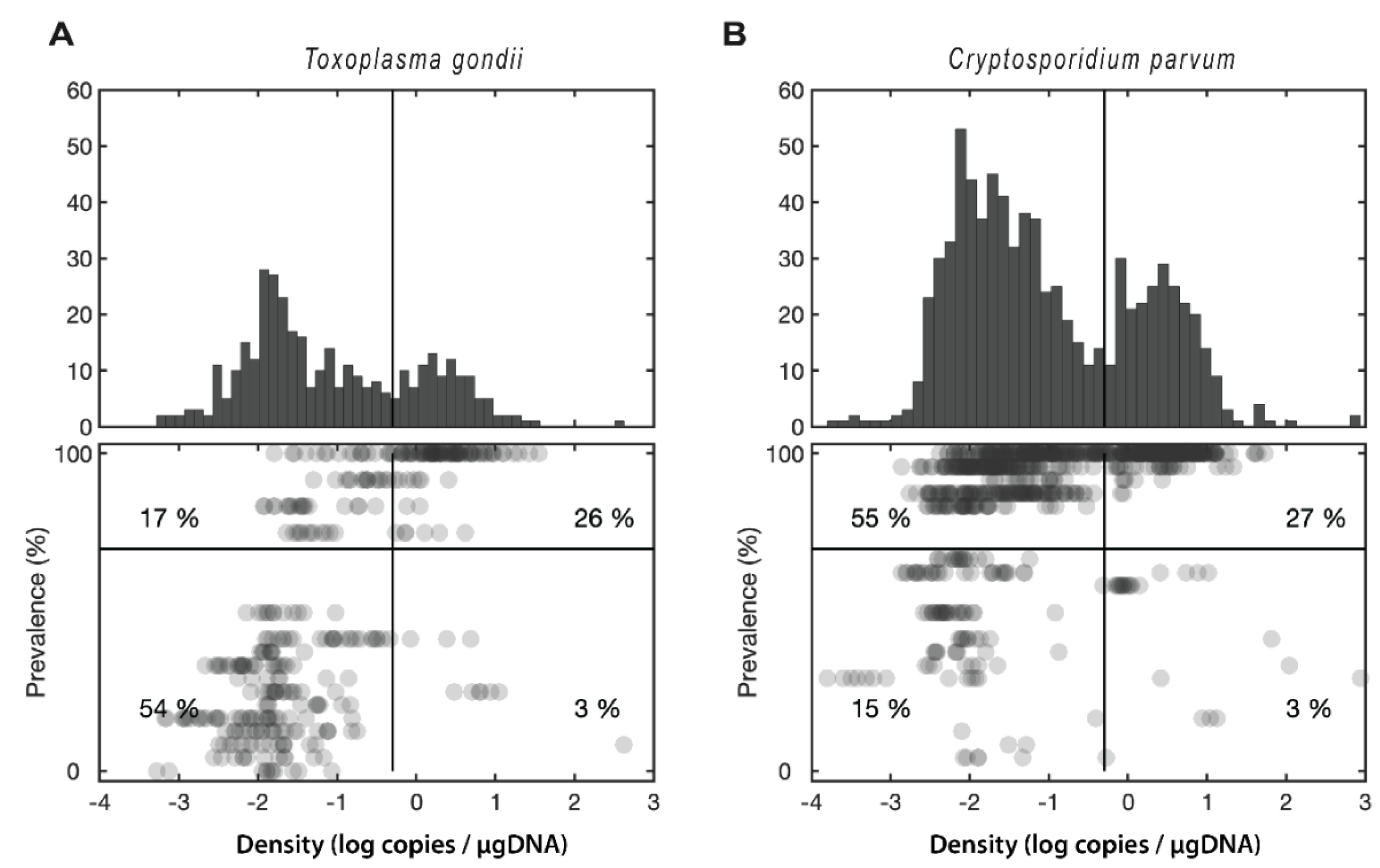
2.3. Statistical Analysis
3. Discussion
4. Materials and Methods
4.1. Collection of Tissue Specimens and DNA Extraction
4.2. qPCR Assays
4.3. Environmental Data
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Efstratiou, A.; Ongerth, J.E.; Karanis, P. Waterborne transmission of protozoan parasites: Review of worldwide outbreaks—An update 2011–2016. Water Res. 2017, 114, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Koutsoumanis, K.; Allende, A.; Alvarez-Ordóñez, A.; Bolton, D.; Bover-Cid, S.; Chemaly, M.; Davies, R.; De Cesare, A.; Herman, L.; Hilbert, F.; et al. Public health risks associated with food-borne parasites. EFSA J. 2018, 16, e05495. [Google Scholar]
- Rajapakse, S.; Weeratunga, P.; Rodrigo, C.; de Silva, N.L.; Fernando, S.D. Prophylaxis of human toxoplasmosis: A systematic review. Pathog. Glob. Health 2017, 111, 333–342. [Google Scholar] [CrossRef] [PubMed]
- Dickerson, F.; Wilcox, H.C.; Adamos, M.; Katsafanas, E.; Khushalani, S.; Origoni, A.; Savage, C.; Schweinfurth, L.; Stallings, C.; Sweeney, K.; et al. Suicide attempts and markers of immune response in individuals with serious mental illness. J. Psychiatr. Res. 2017, 87, 37–43. [Google Scholar] [CrossRef]
- Flegr, J.; Horacek, J. Toxoplasma-infected subjects report an Obsessive-Compulsive Disorder diagnosis more often and score higher in Obsessive-Compulsive Inventory. Eur. Psychiatry 2017, 40, 82–87. [Google Scholar] [CrossRef] [PubMed]
- Lindgren, M.; Torniainen-Holm, M.; Harkanen, T.; Dickerson, F.; Yolken, R.H.; Suvisaari, J. The association between Toxoplasma and the psychosis continuum in a general population setting. Schizophr. Res. 2018, 193, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Suvisaari, J.; Torniainen-Holm, M.; Lindgren, M.; Harkanen, T.; Yolken, R.H. Toxoplasma gondii infection and common mental disorders in the Finnish general population. J. Affect. Disord. 2017, 223, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Gohardehi, S.; Sharif, M.; Sarvi, S.; Moosazadeh, M.; Alizadeh-Navaei, R.; Hosseini, S.A.; Amouei, A.; Pagheh, A.; Sadeghi, M.; Daryani, A. The potential risk of toxoplasmosis for traffic accidents: A systematic review and meta-analysis. Exp. Parasitol. 2018, 191, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.K.; Fitza, M.A.; Lerner, D.A.; Calhoun, D.M.; Beldon, M.A.; Chan, E.T.; Johnson, P.T.J. Risky business: Linking Toxoplasma gondii infection and entrepreneurship behaviours across individuals and countries. Proc. Biol. Sci. 2018, 285, 20180822. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.K.; Knowles, S.; Cerqueira-Cezar, C.K.; Kwok, O.C.; Jiang, T.; Su, C.; Dubey, J.P. An update on Toxoplasma gondii infections in northern sea otters (Enhydra lutris kenyoni) from Washington State, USA. Vet. Parasitol. 2018, 258, 133–137. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.A.; Miller, W.A.; Conrad, P.A.; James, E.R.; Melli, A.C.; Leutenegger, C.M.; Dabritz, H.A.; Packham, A.E.; Paradies, D.; Harris, M.; et al. Type X Toxoplasma gondii in a wild mussel and terrestrial carnivores from coastal California: New linkages between terrestrial mammals, runoff and toxoplasmosis of sea otters. Int. J. Parasitol. 2008, 38, 1319–1328. [Google Scholar] [CrossRef] [PubMed]
- Arkush, K.D.; Miller, M.A.; Leutenegger, C.M.; Gardner, I.A.; Packham, A.E.; Heckeroth, A.R.; Tenter, A.M.; Barr, B.C.; Conrad, P.A. Molecular and bioassay-based detection of Toxoplasma gondii oocyst uptake by mussels (Mytilus galloprovincialis). Int. J. Parasitol. 2003, 33, 1087–1097. [Google Scholar] [CrossRef]
- Chiang, T.Y.; Kuo, M.C.; Chen, C.H.; Yang, J.Y.; Kao, C.F.; Ji, D.D.; Fang, C.T. Risk factors for acute Toxoplasma gondii diseases in Taiwan: A population-based case-control study. PLoS ONE 2014, 9, e90880. [Google Scholar] [CrossRef] [PubMed]
- Ryan, U.; Hijjawi, N.; Xiao, L. Foodborne cryptosporidiosis. Int. J. Parasitol. 2018, 48, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Pumipuntu, N.; Piratae, S. Cryptosporidiosis: A zoonotic disease concern. Vet. World 2018, 11, 681–686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dumbauld, B.R.; Ruesink, J.L.; Rumrill, S.S. The ecological role of bivalve shellfish aquaculture in the estuarine environment: A review with application to oyster and clam culture in West Coast (USA) estuaries. Aquaculture 2009, 290, 196–223. [Google Scholar] [CrossRef]
- Rick, T.C.; Reeder-Myers, L.A.; Hofman, C.A.; Breitburg, D.; Lockwood, R.; Henkes, G.; Kellogg, L.; Lowery, D.; Luckenbach, M.W.; Mann, R.; et al. Millennial-scale sustainability of the Chesapeake Bay Native American oyster fishery. Proc. Natl. Acad. Sci. USA 2016, 113, 6568–6573. [Google Scholar] [CrossRef] [Green Version]
- Sussarellu, R.; Suquet, M.; Thomas, Y.; Lambert, C.; Fabioux, C.; Pernet, M.E.; Le Goic, N.; Quillien, V.; Mingant, C.; Epelboin, Y.; et al. Oyster reproduction is affected by exposure to polystyrene microplastics. Proc. Natl. Acad. Sci. USA 2016, 113, 2430–2435. [Google Scholar] [CrossRef] [Green Version]
- Chèvre, N.; Gagné, F.; Gagnon, P.; Blaise, C. Application of rough sets analysis to identify polluted aquatic sites based on a battery of biomarkers: A comparison with classical methods. Chemosphere 2003, 51, 13–23. [Google Scholar] [CrossRef]
- Cajaraville, M.P.; Bebianno, M.J.; Blasco, J.; Porte, C.; Sarasquete, C.; Viarengo, A. The use of biomarkers to assess the impact of pollution in coastal environments of the Iberian Peninsula: A practical approach. Sci. Total Environ. 2000, 247, 295–311. [Google Scholar] [CrossRef]
- Larsen, P.F.; Wilson, K.A.; Morse, D. Observations on the expansion of a relict population of eastern oysters (Crassostrea virginica) in a Maine estuary: Implications for climate change and restoration. Northeastern Nat. 2013, 20, N28–N32. [Google Scholar] [CrossRef]
- Fernández Robledo, J.A.; Marquis, N.D.; Countway, P.D.; Record, N.R.; Irish, E.L.; Schuldt, M.M.; Kingston, S.E.; Bishop, T.J.; Messerman, N.A.; Bowden, T.J. Pathogens of marine bivalves in Maine (USA): A historical perspective. Aquaculture 2018, 493, 9–17. [Google Scholar] [CrossRef]
- Marquis, N.D.; Record, N.R.; Fernández Robledo, J.A. Survey for protozoan parasites in Eastern oysters (Crassostrea virginica) from the Gulf of Maine using PCR-based assays. Parasitol. Int. 2015, 64, 299–302. [Google Scholar] [CrossRef]
- Graczyk, T.K.; Lewis, E.J.; Glass, G.; Dasilva, A.J.; Tamang, L.; Girouard, A.S.; Curriero, F.C. Quantitative assessment of viable Cryptosporidium parvum load in commercial oysters (Crassostrea virginica) in the Chesapeake Bay. Parasitol. Res. 2007, 100, 247–253. [Google Scholar] [CrossRef]
- Tei, F.F.; Kowalyk, S.; Reid, J.A.; Presta, M.A.; Yesudas, R.; Mayer, D.C. Assessment and molecular characterization of human intestinal parasites in bivalves from Orchard Beach, NY, USA. Int. J. Environ. Res. Public Health 2016, 13, 381. [Google Scholar] [CrossRef]
- Lévesque, B.; Gagnon, F.; Valentin, A.; Cartier, J.F.; Chevalier, P.; Cardinal, P.; Cantin, P.; Gingras, S. A study to assess the microbial contamination of Mya arenaria clams from the north shore of the St Lawrence River estuary, (Quebec, Canada). Can. J. Microbiol. 2006, 52, 984–991. [Google Scholar] [CrossRef]
- Iqbal, A.; Measures, L.; Lair, S.; Dixon, B. Toxoplasma gondii infection in stranded St. Lawrence Estuary beluga Delphinapterus leucas in Quebec, Canada. Dis. Aquat. Organ. 2018, 130, 165–175. [Google Scholar] [CrossRef]
- Souza, D.S.; Ramos, A.P.; Nunes, F.F.; Moresco, V.; Taniguchi, S.; Leal, D.A.; Sasaki, S.T.; Bicego, M.C.; Montone, R.C.; Durigan, M.; et al. Evaluation of tropical water sources and mollusks in southern Brazil using microbiological, biochemical, and chemical parameters. Ecotoxicol. Environ. Saf. 2012, 76, 153–161. [Google Scholar] [CrossRef]
- Hussain, M.A.; Stitt, V.; Szabo, E.A.; Nelan, B. Toxoplasma gondii in the Food Supply. Pathogens 2017, 6, 21. [Google Scholar] [CrossRef]
- Xiao, L.; Alderisio, K.; Limor, J.; Royer, M.; Lal, A.A. Identification of species and sources of Cryptosporidium oocysts in storm waters with a small-subunit rRNA-based diagnostic and genotyping tool. Appl. Environ. Microbiol. 2000, 66, 5492–5498. [Google Scholar] [CrossRef]
- Miller, W.A.; Lewis, D.J.; Pereira, M.D.; Lennox, M.; Conrad, P.A.; Tate, K.W.; Atwill, E.R. Farm factors associated with reducing Cryptosporidium loading in storm runoff from dairies. J. Environ. Qual. 2008, 37, 1875–1882. [Google Scholar] [CrossRef]
- Graczyk, T.K.; Conn, D.B.; Lucy, F.; Minchin, D.; Tamang, L.; Moura, L.N.; DaSilva, A.J. Human waterborne parasites in zebra mussels (Dreissena polymorpha) from the Shannon River drainage area, Ireland. Parasitol. Res. 2004, 93, 385–391. [Google Scholar] [CrossRef]
- Fayer, R.; Graczyk, T.K.; Lewis, E.J.; Trout, J.M.; Farley, C.A. Survival of infectious Cryptosporidium parvum oocysts in seawater and eastern oysters (Crassostrea virginica) in the Chesapeake Bay. Appl. Environ. Microbiol. 1998, 64, 1070–1074. [Google Scholar]
- Izumi, T.; Yagita, K.; Izumiyama, S.; Endo, T.; Itoh, Y. Depletion of Cryptosporidium parvum oocysts from contaminated sewage by using freshwater benthic pearl clams (Hyriopsis schlegeli). Appl. Environ. Microbiol. 2012, 78, 7420–7428. [Google Scholar] [CrossRef]
- Nappier, S.P.; Graczyk, T.K.; Tamang, L.; Schwab, K.J. Co-localized Crassostrea virginica and Crassostrea ariakensis oysters differ in bioaccumulation, retention and depuration of microbial indicators and human enteropathogens. J. Appl. Microbiol. 2010, 108, 736–744. [Google Scholar] [CrossRef]
- Gómez-Couso, H.; Mendez-Hermida, F.; Castro-Hermida, J.A.; Ares-Mazas, E. Cryptosporidium contamination in harvesting areas of bivalve molluscs. J. Food Prot. 2006, 69, 185–190. [Google Scholar] [CrossRef]
- Tryland, I.; Myrmel, M.; Ostensvik, O.; Wennberg, A.C.; Robertson, L.J. Impact of rainfall on the hygienic quality of blue mussels and water in urban areas in the Inner Oslofjord, Norway. Mar. Pollut. Bull. 2014, 85, 42–49. [Google Scholar] [CrossRef]
- Graczyk, T.K.; Fayer, R.; Lewis, E.J.; Farley, C.A.; Trout, J.M. In vitro interactions between hemocytes of the eastern oyster, Crassostrea virginica Gmelin, 1791 and Cryptosporidium parvum oocysts. J. Parasitol. 1997, 83, 949–952. [Google Scholar] [CrossRef]
- Lindsay, D.S.; Phelps, K.K.; Smith, S.A.; Flick, G.; Sumner, S.S.; Dubey, J.P. Removal of Toxoplasma gondii oocysts from sea water by eastern oysters (Crassostrea virginica). J. Eukaryot. Microbiol. 2001, 48, 197S–198S. [Google Scholar] [CrossRef]
- Lindsay, D.S.; Collins, M.V.; Mitchell, S.M.; Wetch, C.N.; Rosypal, A.C.; Flick, G.J.; Zajac, A.M.; Lindquist, A.; Dubey, J.P. Survival of Toxoplasma gondii oocysts in Eastern oysters (Crassostrea virginica). J. Parasitol. 2004, 90, 1054–1057. [Google Scholar] [CrossRef]
- Fayer, R.; Farley, C.A.; Lewis, E.J.; Trout, J.M.; Graczyk, T.K. Potential role of the eastern oyster, Crassostrea virginica, in the epidemiology of Cryptosporidium parvum. Appl. Environ. Microbiol. 1997, 63, 2086–2088. [Google Scholar]
- Sutthikornchai, C.; Popruk, S.; Chumpolbanchorn, K.; Sukhumavasi, W.; Sukthana, Y. Oyster is an effective transmission vehicle for Cryptosporidium infection in human. Asian Pac. J. Trop. Med. 2016, 9, 562–566. [Google Scholar] [CrossRef]
- Freire-Santos, F.; Gomez-Couso, H.; Ortega-Inarrea, M.R.; Castro-Hermida, J.A.; Oteiza-Lopez, A.M.; Garcia-Martin, O.; Ares-Mazas, M.E. Survival of Cryptosporidium parvum oocysts recovered from experimentally contaminated oysters (Ostrea edulis) and clams (Tapes decussatus). Parasitol. Res. 2002, 88, 130–133. [Google Scholar] [CrossRef]
- Tamburrini, A.; Pozio, E. Long-term survival of Cryptosporidium parvum oocysts in seawater and in experimentally infected mussels (Mytilus galloprovincialis). Int. J. Parasitol. 1999, 29, 711–715. [Google Scholar] [CrossRef]
- Freire-Santos, F.; Oteiza-Lopez, A.M.; Castro-Hermida, J.A.; Garcia-Martin, O.; Ares-Mazas, M.E. Viability and infectivity of oocysts recovered from clams, Ruditapes philippinarum, experimentally contaminated with Cryptosporidium parvum. Parasitol. Res. 2001, 87, 428–430. [Google Scholar]
- Gómez-Bautista, M.; Ortega-Mora, L.M.; Tabares, E.; Lopez-Rodas, V.; Costas, E. Detection of infectious Cryptosporidium parvum oocysts in mussels (Mytilus galloprovincialis) and cockles (Cerastoderma edule). Appl. Environ. Microbiol. 2000, 66, 1866–1870. [Google Scholar] [CrossRef]
- Aksoy, U.; Marangi, M.; Papini, R.; Ozkoc, S.; Bayram Delibas, S.; Giangaspero, A. Detection of Toxoplasma gondii and Cyclospora cayetanensis in Mytilus galloprovincialis from Izmir Province coast (Turkey) by Real Time PCR/High-Resolution Melting analysis (HRM). Food Microbiol. 2014, 44, 128–135. [Google Scholar] [CrossRef]
- Coupe, A.; Howe, L.; Burrows, E.; Sine, A.; Pita, A.; Velathanthiri, N.; Vallee, E.; Hayman, D.; Shapiro, K.; Roe, W.D. First report of Toxoplasma gondii sporulated oocysts and Giardia duodenalis in commercial green-lipped mussels (Perna canaliculus) in New Zealand. Parasitol. Res. 2018, 117, 1453–1463. [Google Scholar] [CrossRef]
- Clode, P.L.; Koh, W.H.; Thompson, R.C.A. Life without a Host Cell: What is Cryptosporidium? Trends Parasitol. 2015, 31, 614–624. [Google Scholar] [CrossRef]
- Feng, Y.; Ryan, U.M.; Xiao, L. Genetic diversity and population structure of Cryptosporidium. Trends Parasitol. 2018, 34, 997–1011. [Google Scholar] [CrossRef]
- Nader, J.L.; Mathers, T.C.; Ward, B.J.; Pachebat, J.A.; Swain, M.T.; Robinson, G.; Chalmers, R.M.; Hunter, P.R.; van Oosterhout, C.; Tyler, K.M. Evolutionary genomics of anthroponosis in Cryptosporidium. Nat. Microbiol. 2019, 4, 826–836. [Google Scholar] [CrossRef]
- Dardé, M.L. Genetic analysis of the diversity in Toxoplasma gondii. Ann. Ist. Super Sanita 2004, 40, 57–63. [Google Scholar]
- Sharif, M.; Amouei, A.; Sarvi, S.; Mizani, A.; Aarabi, M.; Hosseini, S.A.; Daryani, A. Genetic diversity of Toxoplasma gondii isolates from ruminants: A systematic review. Int. J. Food Microbiol. 2017, 258, 38–49. [Google Scholar] [CrossRef]
- Zhang, X.; Jian, Y.; Li, X.; Ma, L.; Karanis, G.; Qigang, C.; Karanis, P. Molecular detection and prevalence of Cryptosporidium spp. infections in two types of domestic farm animals in the Qinghai-Tibetan Plateau Area (QTPA) in China. Parasitol. Res. 2018, 117, 233–239. [Google Scholar] [CrossRef]
- Wang, G.; Wang, G.; Li, X.; Zhang, X.; Karanis, G.; Jian, Y.; Ma, L.; Karanis, P. Prevalence and molecular characterization of Cryptosporidium spp. and Giardia duodenalis in 1–2-month-old highland yaks in Qinghai Province, China. Parasitol. Res. 2018, 117, 1793–1800. [Google Scholar] [CrossRef]
- Balch, W.M.; Drapeau, D.T.; Bowler, B.C.; Huntington, T.G. Step-changes in the physical, chemical and biological characteristics of the Gulf of Maine, as documented by the GNATS time series. Mar. Ecol. Prog. Ser. 2012, 450, 11–35. [Google Scholar] [CrossRef] [Green Version]
- Pershing, A.J.; Alexander, M.A.; Hernandez, C.M.; Kerr, L.A.; Le Bris, A.; Mills, K.E.; Nye, J.A.; Record, N.R.; Scannell, H.A.; Scott, J.D.; et al. Slow adaptation in the face of rapid warming leads to collapse of the Gulf of Maine cod fishery. Science 2015, 350, 809–812. [Google Scholar] [CrossRef] [Green Version]
- Record, N.R.; Runge, J.A.; Pendleton, D.E.; Balch, W.M.; Davies, K.T.A.; Pershing, A.J.; Johnson, C.L.; Stamieszkin, K.; Ji, R.; Feng, X.; et al. Rapid climate-driven circulation changes threaten conservation of endangered North Atlantic right whales. Oceanography 2019, 32, 163–169. [Google Scholar] [CrossRef]
- Jauregui, L.H.; Higgins, J.; Zarlenga, D.; Dubey, J.P.; Lunney, J.K. Development of a real-time PCR assay for detection of Toxoplasma gondii in pig and mouse tissues. J. Clin. Microbiol. 2001, 39, 2065–2071. [Google Scholar] [CrossRef]
- Fontaine, M.; Guillot, E. Development of a TaqMan quantitative PCR assay specific for Cryptosporidium parvum. FEMS Microbiol. Lett. 2002, 214, 13–17. [Google Scholar] [CrossRef]
- Hohweyer, J.; Dumetre, A.; Aubert, D.; Azas, N.; Villena, I. Tools and methods for detecting and characterizing Gardia, Cryptosporidium, and Toxoplasma parasites in marine mollusks. J. Food Prot. 2013, 76, 1649–1657. [Google Scholar] [CrossRef]
- Efstratiou, A.; Ongerth, J.; Karanis, P. Evolution of monitoring for Giardia and Cryptosporidium in water. Water Res. 2017, 123, 96–112. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marquis, N.D.; Bishop, T.J.; Record, N.R.; Countway, P.D.; Fernández Robledo, J.A. Molecular Epizootiology of Toxoplasma gondii and Cryptosporidium parvum in the Eastern Oyster (Crassostrea virginica) from Maine (USA). Pathogens 2019, 8, 125. https://doi.org/10.3390/pathogens8030125
Marquis ND, Bishop TJ, Record NR, Countway PD, Fernández Robledo JA. Molecular Epizootiology of Toxoplasma gondii and Cryptosporidium parvum in the Eastern Oyster (Crassostrea virginica) from Maine (USA). Pathogens. 2019; 8(3):125. https://doi.org/10.3390/pathogens8030125
Chicago/Turabian StyleMarquis, Nicholas D., Theodore J. Bishop, Nicholas R. Record, Peter D. Countway, and José A. Fernández Robledo. 2019. "Molecular Epizootiology of Toxoplasma gondii and Cryptosporidium parvum in the Eastern Oyster (Crassostrea virginica) from Maine (USA)" Pathogens 8, no. 3: 125. https://doi.org/10.3390/pathogens8030125
APA StyleMarquis, N. D., Bishop, T. J., Record, N. R., Countway, P. D., & Fernández Robledo, J. A. (2019). Molecular Epizootiology of Toxoplasma gondii and Cryptosporidium parvum in the Eastern Oyster (Crassostrea virginica) from Maine (USA). Pathogens, 8(3), 125. https://doi.org/10.3390/pathogens8030125






