The Evolutionary unZIPping of a Dimerization Motif—A Comparison of ZIP and PrP Architectures
Abstract
1. Introduction
2. Main
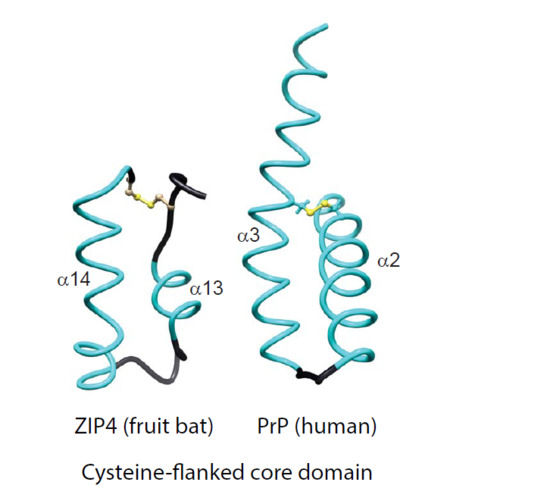
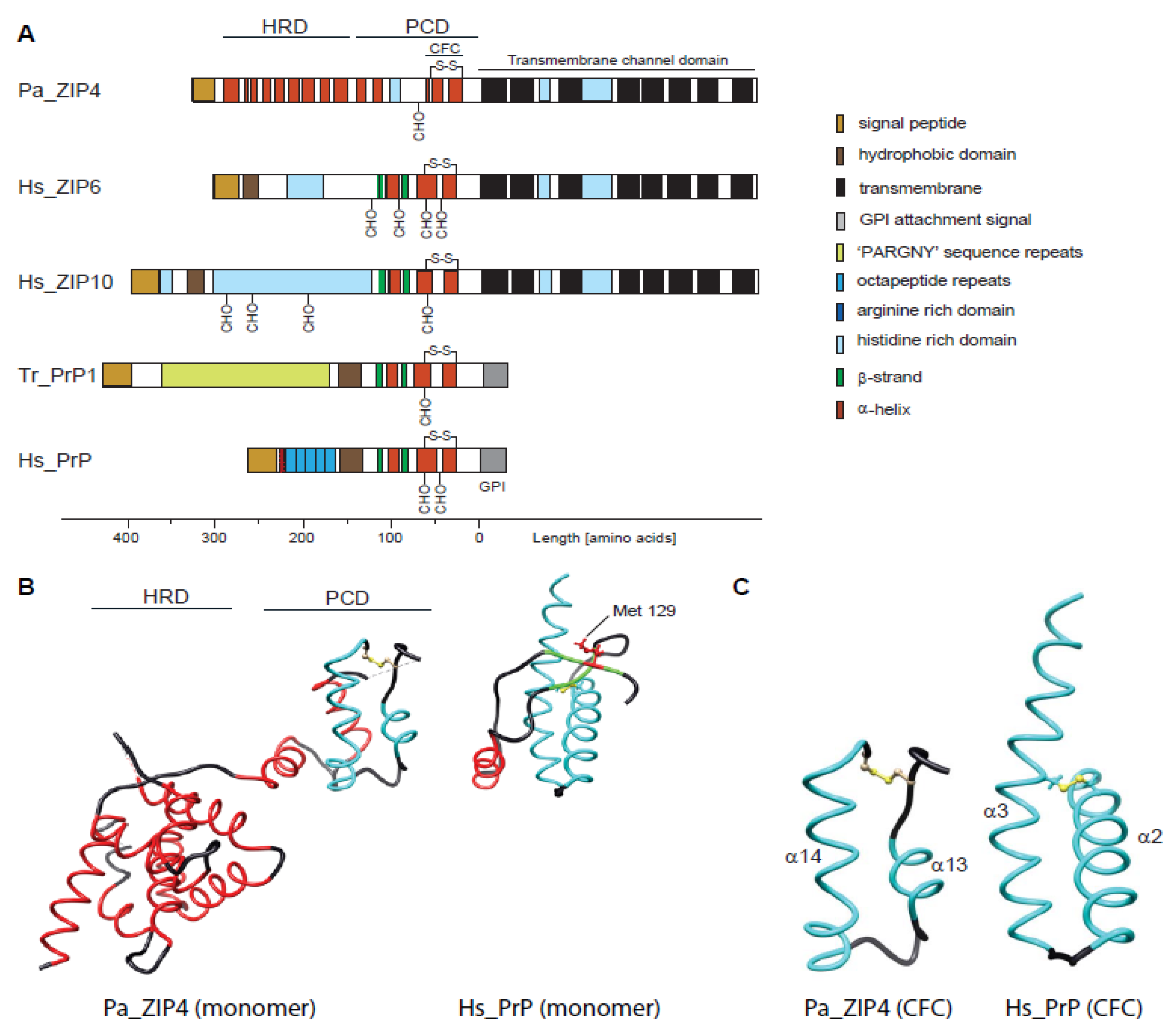
2.1. Comparison of Molecular Architectures of ZIP and Prion Proteins
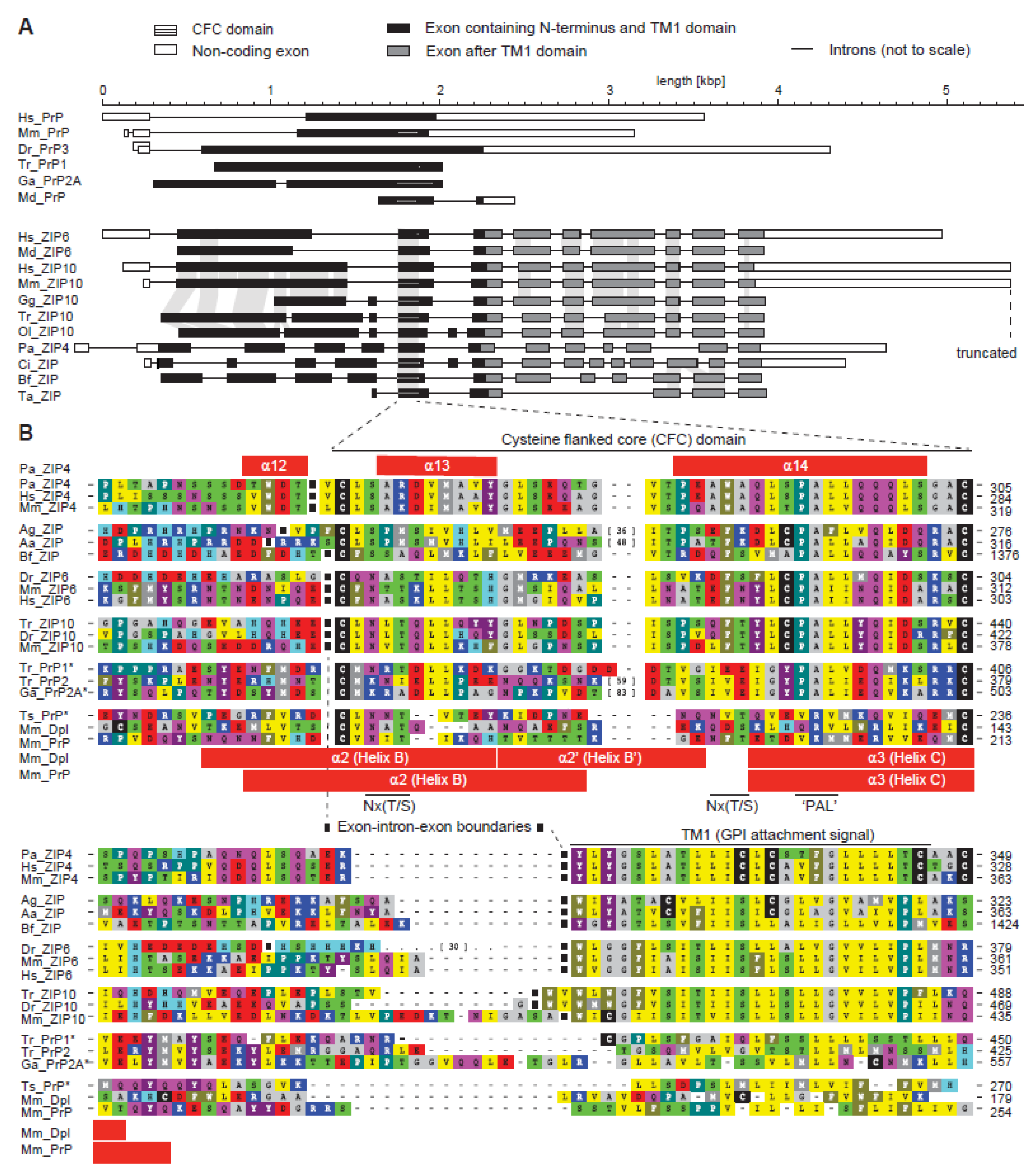
2.2. Disappearance of CPAL Motif in the Prion Protein Subbranch of ZIP-PrP Protein Family
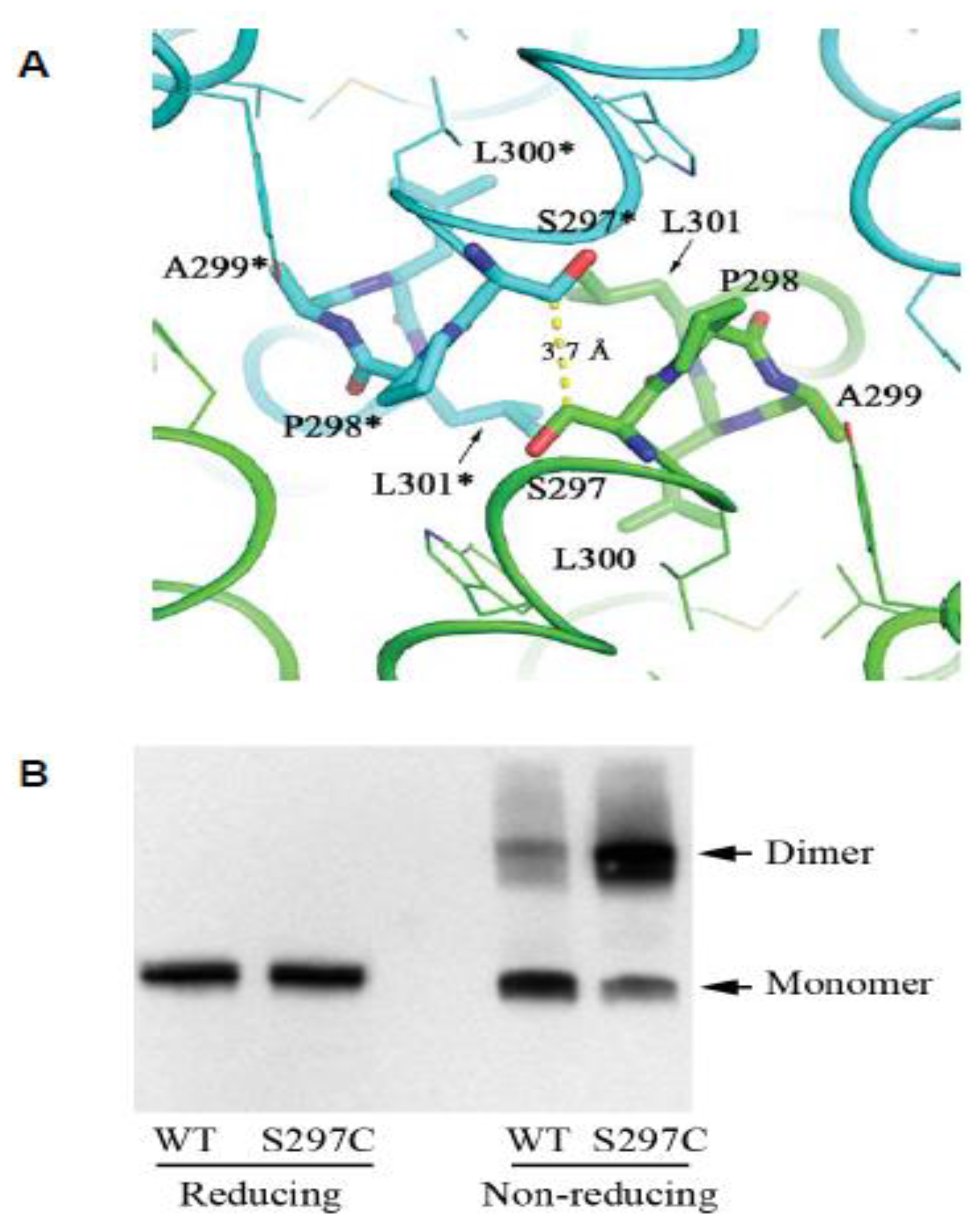
2.3. The CPAL Motif Represents a Dimerization Interface
3. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| Aa | Anopheles aegypti |
| Ag | Anopheles gambiae |
| AE | Acrodermatitis enteropathica |
| Bf | Branchiostoma florida |
| CDF | cation diffusion facilitator |
| CFC | cysteine-flanked core |
| Ci | Ciona intestinalis |
| CTD | carboxy-terminal domain |
| Dm | Drosophila melanogaster |
| Dr | Danio rerio |
| ECD | extracellular domain |
| Ga | Gasterosteus aculateus |
| Gg | Gallus gallus |
| GPI | glycosylphosphatidylinositol |
| HRD | helix-rich domain |
| Hs | Homo sapiens |
| LZT | LIV-1 subfamily of ZIP zinc transporters |
| Md | Monodelphis domestica |
| Mm | Mus musculus |
| Ol | Oryzias latipes |
| ORF | open reading frame |
| Pa | Pteropus alecto |
| PCD | PAL-motif containing domain |
| PL | prion-like |
| PrPC | cellular prion protein |
| SLC | solute carrier |
| Ta | Trichoplax adhaerens |
| TM | transmembrane |
| Tr | Takifugu rubripes |
| Ts | Trachemys scripta |
References
- Prusiner, S.B. Novel proteinaceous infectious particles cause scrapie. Science 1982, 216, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Prusiner, S.B. Prions. Proc. Natl. Acad. Sci. USA 1998, 95, 13363–13383. [Google Scholar] [CrossRef] [PubMed]
- Prusiner, S.B. Cell biology. A unifying role for prions in neurodegenerative diseases. Science 2012, 336, 1511–1513. [Google Scholar] [CrossRef] [PubMed]
- Sailer, A.; Bueler, H.; Fischer, M.; Aguzzi, A.; Weissmann, C. No propagation of prions in mice devoid of PrP. Cell 1994, 77, 967–968. [Google Scholar] [CrossRef]
- Moore, R.C.; Lee, I.Y.; Silverman, G.L.; Harrison, P.M.; Strome, R.; Heinrich, C.; Karunaratne, A.; Pasternak, S.H.; Chishti, M.A.; Liang, Y.; et al. Ataxia in prion protein (PrP)-deficient mice is associated with upregulation of the novel PrP-like protein doppel. J. Mol. Biol. 1999, 292, 797–817. [Google Scholar] [CrossRef] [PubMed]
- Silverman, G.L.; Qin, K.; Moore, R.C.; Yang, Y.; Mastrangelo, P.; Tremblay, P.; Prusiner, S.B.; Cohen, F.E.; Westaway, D. Doppel is an N-glycosylated, glycosylphosphatidylinositol-anchored protein. Expression in testis and ectopic production in the brains of Prnp(0/0) mice predisposed to Purkinje cell loss. J. Biol. Chem. 2000, 275, 26834–26841. [Google Scholar] [PubMed]
- Premzl, M.; Sangiorgio, L.; Strumbo, B.; Marshall Graves, J.A.; Simonic, T.; Gready, J.E. Shadoo, a new protein highly conserved from fish to mammals and with similarity to prion protein. Gene 2003, 314, 89–102. [Google Scholar] [CrossRef]
- Westaway, D.; Daude, N.; Wohlgemuth, S.; Harrison, P. The PrP-like proteins Shadoo and Doppel. Top. Curr. Chem. 2011, 305, 225–256. [Google Scholar] [PubMed]
- Watts, J.C.; Huo, H.; Bai, Y.; Ehsani, S.; Jeon, A.H.; Shi, T.; Daude, N.; Lau, A.; Young, R.; Xu, L.; et al. Interactome analyses identify ties of PrP and its mammalian paralogs to oligomannosidic N-glycans and endoplasmic reticulum-derived chaperones. PLoS Pathog. 2009, 5, e1000608. [Google Scholar] [CrossRef]
- Schmitt-Ulms, G.; Ehsani, S.; Watts, J.C.; Westaway, D.; Wille, H. Evolutionary descent of prion genes from the ZIP family of metal ion transporters. PLoS ONE 2009, 4, e7208. [Google Scholar] [CrossRef] [PubMed]
- Hatinen, T.; Holm, L.; Airaksinen, M.S. Loss of neurturin in frog—Comparative genomics study of GDNF family ligand-receptor pairs. Mol. Cell Neurosci. 2007, 34, 155–167. [Google Scholar] [CrossRef] [PubMed]
- Airaksinen, M.S.; Holm, L.; Hatinen, T. Evolution of the GDNF family ligands and receptors. Brain Behav. Evol. 2006, 68, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Bell, L.M.; Solomon, K.R.; Gold, J.P.; Tan, K.N. Cytoplasmic tail deletion of T cell receptor (TCR) beta-chain results in its surface expression as glycosylphosphatidylinositol-anchored polypeptide on mature T cells in the absence of TCR-alpha. J. Biol. Chem. 1994, 269, 22758–22763. [Google Scholar] [PubMed]
- Aguzzi, A.; Baumann, F.; Bremer, J. The Prion’s Elusive Reason for Being. Annu. Rev. Neurosci. 2008, 31, 439–477. [Google Scholar] [CrossRef] [PubMed]
- Castle, A.R.; Gill, A.C. Physiological Functions of the Cellular Prion Protein. Front. Mol. Biosci. 2017, 4, 19. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Kim, B.E.; Dufner-Beattie, J.; Petris, M.J.; Andrews, G.; Eide, D.J. Acrodermatitis enteropathica mutations affect transport activity, localization and zinc-responsive trafficking of the mouse ZIP4 zinc transporter. Hum. Mol. Genet. 2004, 13, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Giunta, C.; Elcioglu, N.H.; Albrecht, B.; Eich, G.; Chambaz, C.; Janecke, A.R.; Yeowell, H.; Weis, M.; Eyre, D.R.; Kraenzlin, M.; et al. Spondylocheiro dysplastic form of the Ehlers-Danlos syndrome—An autosomal-recessive entity caused by mutations in the zinc transporter gene SLC39A13. Am. J. Hum. Genet. 2008, 82, 1290–1305. [Google Scholar] [CrossRef] [PubMed]
- Kong, B.Y.; Duncan, F.E.; Que, E.L.; Kim, A.M.; O’Halloran, T.V.; Woodruff, T.K. Maternally-derived zinc transporters ZIP6 and ZIP10 drive the mammalian oocyte-to-egg transition. Mol. Hum. Reprod. 2014, 20, 1077–1089. [Google Scholar] [CrossRef] [PubMed]
- Taylor, K.M.; Muraina, I.; Brethour, D.; Schmitt-Ulms, G.; Nimmanon, T.; Ziliotto, S.; Kille, P.; Hogstrand, C. Zinc transporter ZIP10 forms a heteromer with ZIP6 which regulates embryonic development and cell migration. Biochem. J. 2016, 473, 2531–2544. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, S.; Miyagi, C.; Fukada, T.; Kagara, N.; Che, Y.S.; Hirano, T. Zinc transporter LIVI controls epithelial-mesenchymal transition in zebrafish gastrula organizer. Nature 2004, 429, 298–302. [Google Scholar] [CrossRef] [PubMed]
- Malaga-Trillo, E.; Solis, G.P.; Schrock, Y.; Geiss, C.; Luncz, L.; Thomanetz, V.; Stuermer, C.A. Regulation of embryonic cell adhesion by the prion protein. PLoS Biol. 2009, 7, e55. [Google Scholar] [CrossRef] [PubMed]
- Brethour, D.; Mehrabian, M.; Williams, D.; Wang, X.; Ghodrati, F.; Ehsani, S.; Rubie, E.A.; Woodgett, J.R.; Sevalle, J.; Xi, Z.; et al. A ZIP6-ZIP10 heteromer controls NCAM1 phosphorylation and integration into focal adhesion complexes during epithelial-to-mesenchymal transition. Sci. Rep. 2017, 7, 40313. [Google Scholar] [CrossRef] [PubMed]
- Schmitt-Ulms, G.; Legname, G.; Baldwin, M.A.; Ball, H.L.; Bradon, N.; Bosque, P.J.; Crossin, K.L.; Edelman, G.M.; DeArmond, S.J.; Cohen, F.E.; et al. Binding of neural cell adhesion molecules (N-CAMs) to the cellular prion protein. J. Mol. Biol. 2001, 314, 1209–1225. [Google Scholar] [CrossRef] [PubMed]
- Slapsak, U.; Salzano, G.; Amin, L.; Abskharon, R.N.; Ilc, G.; Zupancic, B.; Biljan, I.; Plavec, J.; Giachin, G.; Legname, G. The N Terminus of the Prion Protein Mediates Functional Interactions with the Neuronal Cell Adhesion Molecule (NCAM) Fibronectin Domain. J. Biol. Chem. 2016, 291, 21857–21868. [Google Scholar] [CrossRef] [PubMed]
- Santuccione, A.; Sytnyk, V.; Leshchyns’ka, I.; Schachner, M. Prion protein recruits its neuronal receptor NCAM to lipid rafts to activate p59fyn and to enhance neurite outgrowth. J. Cell Biol. 2005, 169, 341–354. [Google Scholar] [CrossRef] [PubMed]
- Mehrabian, M.; Brethour, D.; Wang, H.; Xi, Z.; Rogaeva, E.; Schmitt-Ulms, G. The Prion Protein Controls Polysialylation of Neural Cell Adhesion Molecule 1 during Cellular Morphogenesis. PLoS ONE 2015, 10, e0133741. [Google Scholar] [CrossRef] [PubMed]
- Riek, R.; Hornemann, S.; Wider, G.; Billeter, M.; Glockshuber, R.; Wuthrich, K. NMR structure of the mouse prion protein domain PrP(121-321). Nature 1996, 382, 180–182. [Google Scholar] [CrossRef] [PubMed]
- Wuthrich, K.; Riek, R. Three-dimensional structures of prion proteins. Adv. Protein Chem. 2001, 57, 55–82. [Google Scholar] [PubMed]
- Calzolai, L.; Lysek, D.A.; Perez, D.R.; Guntert, P.; Wuthrich, K. Prion protein NMR structures of chickens, turtles, and frogs. Proc. Natl. Acad. Sci. USA 2005, 102, 651–655. [Google Scholar] [CrossRef] [PubMed]
- Christen, B.; Wuthrich, K.; Hornemann, S. Putative prion protein from Fugu (Takifugu rubripes). FEBS J. 2008, 275, 263–270. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Sui, D.; Hu, J. Structural insights of ZIP4 extracellular domain critical for optimal zinc transport. Nat. Commun. 2016, 7, 11979. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Liu, J.; Fellner, M.; Zhang, C.; Sui, D.; Hu, J. Crystal structures of a ZIP zinc transporter reveal a binuclear metal center in the transport pathway. Sci. Adv. 2017, 3, e1700344. [Google Scholar] [CrossRef] [PubMed]
- Ehsani, S.; Tao, R.; Pocanschi, C.L.; Ren, H.; Harrison, P.M.; Schmitt-Ulms, G. Evidence for retrogene origins of the prion gene family. PLoS ONE 2011, 6, e26800. [Google Scholar] [CrossRef] [PubMed]
- Rudd, P.M.; Endo, T.; Colominas, C.; Groth, D.; Wheeler, S.F.; Harvey, D.J.; Wormald, M.R.; Serban, H.; Prusiner, S.B.; Kobata, A.; et al. Glycosylation differences between the normal and pathogenic prion protein isoforms. Proc. Natl. Acad. Sci. USA 1999, 96, 13044–13049. [Google Scholar] [CrossRef] [PubMed]
- Ehsani, S.; Salehzadeh, A.; Huo, H.; Reginold, W.; Pocanschi, C.L.; Ren, H.; Wang, H.; So, K.; Sato, C.; Mehrabian, M.; et al. LIV-1 ZIP ectodomain shedding in prion-infected mice resembles cellular response to transition metal starvation. J. Mol. Biol. 2012, 422, 556–574. [Google Scholar] [CrossRef] [PubMed]
- Pocanschi, C.L.; Ehsani, S.; Mehrabian, M.; Wille, H.; Reginold, W.; Trimble, W.S.; Wang, H.; Yee, A.; Arrowsmith, C.H.; Bozoky, Z.; et al. The ZIP5 Ectodomain Co-Localizes with PrP and May Acquire a PrP-Like Fold That Assembles into a Dimer. PLoS ONE 2013, 8, e72446. [Google Scholar] [CrossRef] [PubMed]
- Bin, B.H.; Fukada, T.; Hosaka, T.; Yamasaki, S.; Ohashi, W.; Hojyo, S.; Miyai, T.; Nishida, K.; Yokoyama, S.; Hirano, T. Biochemical characterization of human ZIP13 protein: A homo-dimerized zinc transporter involved in the Spondylocheiro dysplastic Ehlers-Danlos syndrome. J. Biol. Chem. 2011, 286, 40255–40265. [Google Scholar] [CrossRef] [PubMed]
- Priola, S.A.; Caughey, B.; Wehrly, K.; Chesebro, B. A 60-kDa prion protein (PrP) with properties of both the normal and scrapie-associated forms of PrP. J. Biol. Chem. 1995, 270, 3299–3305. [Google Scholar] [CrossRef] [PubMed]
- Knaus, K.J.; Morillas, M.; Swietnicki, W.; Malone, M.; Surewicz, W.K.; Yee, V.C. Crystal structure of the human prion protein reveals a mechanism for oligomerization. Nat. Struct. Biol. 2001, 8, 770–774. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Antony, L.; Hartmann, R.; Knaus, K.J.; Surewicz, K.; Surewicz, W.K.; Yee, V.C. Conformational diversity in prion protein variants influences intermolecular beta-sheet formation. EMBO J. 2010, 29, 251–262. [Google Scholar] [CrossRef] [PubMed]
- Meyer, R.K.; Lustig, A.; Oesch, B.; Fatzer, R.; Zurbriggen, A.; Vandevelde, M. A monomer-dimer equilibrium of a cellular prion protein (PrPC) not observed with recombinant PrP. J. Biol. Chem. 2000, 275, 38081–38087. [Google Scholar] [CrossRef] [PubMed]
- Hundt, C.; Gauczynski, S.; Leucht, C.; Riley, M.L.; Weiss, S. Intra- and interspecies interactions between prion proteins and effects of mutations and polymorphisms. Biol. Chem. 2003, 384, 791–803. [Google Scholar] [CrossRef] [PubMed]
- Beland, M.; Roucou, X. Homodimerization as a molecular switch between low and high efficiency PrP C cell surface delivery and neuroprotective activity. Prion 2013, 7, 170–174. [Google Scholar] [CrossRef] [PubMed]
- Beland, M.; Motard, J.; Barbarin, A.; Roucou, X. PrP(C) homodimerization stimulates the production of PrPC cleaved fragments PrPN1 and PrPC1. J. Neurosci. 2012, 32, 13255–13263. [Google Scholar] [CrossRef] [PubMed]
- Supattapone, S.; Bosque, P.; Muramoto, T.; Wille, H.; Aagaard, C.; Peretz, D.; Nguyen, H.-O.B.; Heinrich, C.; Torchia, M.; Safar, J.; et al. Prion protein of 106 residues creates an artificial transmission barrier for prion replication in transgenic mice. Cell 1999, 96, 869–878. [Google Scholar] [CrossRef]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, J.; Wille, H.; Schmitt-Ulms, G. The Evolutionary unZIPping of a Dimerization Motif—A Comparison of ZIP and PrP Architectures. Pathogens 2018, 7, 4. https://doi.org/10.3390/pathogens7010004
Hu J, Wille H, Schmitt-Ulms G. The Evolutionary unZIPping of a Dimerization Motif—A Comparison of ZIP and PrP Architectures. Pathogens. 2018; 7(1):4. https://doi.org/10.3390/pathogens7010004
Chicago/Turabian StyleHu, Jian, Holger Wille, and Gerold Schmitt-Ulms. 2018. "The Evolutionary unZIPping of a Dimerization Motif—A Comparison of ZIP and PrP Architectures" Pathogens 7, no. 1: 4. https://doi.org/10.3390/pathogens7010004
APA StyleHu, J., Wille, H., & Schmitt-Ulms, G. (2018). The Evolutionary unZIPping of a Dimerization Motif—A Comparison of ZIP and PrP Architectures. Pathogens, 7(1), 4. https://doi.org/10.3390/pathogens7010004