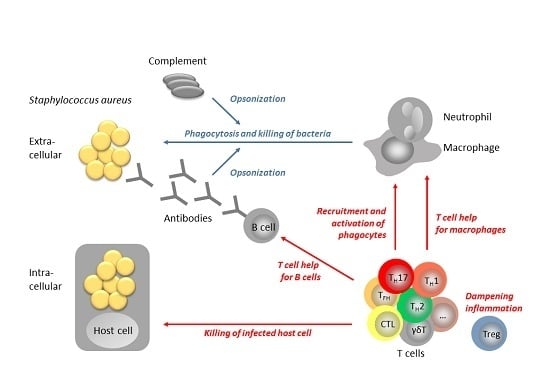
The T Cell Response to Staphylococcus aureus
Abstract
:1. Introduction
2. The role of T Cells in S. aureus Control
2.1. Evidence from Mouse Models
Vaccination Studies in Mice
2.2. Evidence from Livestock
Vaccination Studies in Cattle
2.3. Evidence from Humans
Vaccination Studies in Humans
2.4. A Special Case: S. aureus Persisting Inside Host Cells
2.5. The T Cell Response to S. aureus May Cause Harm
2.6. How S. aureus Manipulates T Cells
3. Conclusions and Future Directions of Research
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| ClfA | clumping factor A |
| CTL | cytotoxic T lymphocyte |
| DAMP | Damage-associated molecular pattern |
| DC | dendritic cell |
| ILC | innate lymphoid cell |
| i.v. | intravenous |
| MAIT | mucosa associated invariant T cell |
| MAMP | microbe-associated molecular pattern |
| MDSC | Myeloid derived suppressor cells |
| MHC | major histocompatibility complex |
| NK cell | natural killer cell |
| Sak | staphylokinase |
| SCID | severe combined immune deficiency |
| SCV | Small colony variant |
| SE | Staphylococcal enterotoxin |
| SEl | Staphylococcal enterotoxin-like |
| TCR | T cell receptor |
| TFH | T follicular helper cell |
| TH | T helper cell |
| Treg | regulatory T cell |
| TSST-1 | Toxic shock syndrome toxin-1 |
Appendix
A1. The World of T Cells and Innate Lymphocytes
A1.1. Signals 1, 2, 3—The Rules of the T Cell Game
| Inducing Cytokines | Lineage-Specific Transcription Factor | Control of … | Secreted Cytokines | Main Functions | |
|---|---|---|---|---|---|
| TH1 | IL-12 IFN-γ | Tbet | Intracellular vesicles | IFN-γ IL-2 | Activate macrophages Help CD8+ T cells |
| TH2 | IL-4 | Gata3 | Extracellular space | IL-4, IL-5, IL-9, IL-13 | Recruit eosinophils Orchestrate type 2 inflammation |
| TH17 | TGF-β IL1-β, IL-6, IL-21, IL-23 | RORγT | Extracellular space | IL-17 IL-6 | Enhance neutrophil response Help mucosal B cells (IgA) |
| TFH | IL-6, IL-21 TGF-β | Bcl6 | Extracellular space | IL-21 and others | Help B cells (antibody class switch to IgG, IgA and IgE; antibody affinity maturation) |
| Treg | TGF-β | Foxp3 | TGF-β, IL-10 | Suppress T cell responses Help mucosal B cells (IgA) | |
| CTL | Cytoplasm | IL-2, IFN-γ | Kill infected cells |
A1.2. Unconventional T Cells
| Innate Lymphoid Cells | T Cells | ||||||
|---|---|---|---|---|---|---|---|
| Inducing Cytokines | Lineage-Specific Transcription Factor | Secreted Cytokines | Inducing Cytokines | Lineage-Specific Transcription Factor | Secreted Cytokines | ||
| ILC1 | IL-12, IL-15, IL-18 | Tbet | IFN-γ TNF-α | TH1 | IL-12 IFN-γ | Tbet | IFN-γ IL-2 |
| ILC2 | IL-25, IL-33, TSLP | Gata3, RORα | IL-4, IL-5, IL-13, amphiregulin | TH2 | IL-4 | Gata3 | IL-4, IL-5, IL-9, IL-13 |
| ILC3 | IL-1β, IL-23 | RORγt, Ahr | IL-17, IL-22, LT, GM-CSF | TH17 | TGF-β IL1-β, IL-6, IL-21, IL-23 | RORγt | IL-17 IL-6 |
| NK | IL-12, IL-15, IL-18 | Eomes | IFN-γ | CTL | IL-2, IFN-γ | ||
A1.3. Innate Lymphoid Cells, Not T Cells But Similar?
References
- Fleischer, B.; Schrezenmeier, H. T-cell stimulation by staphylococcal enterotoxins—Clonally variable response and requirement for major histocompatibility complex class-II molecules on accessory or target-cells. J. Exp. Med. 1988, 167, 1697–1707. [Google Scholar] [CrossRef] [PubMed]
- Grumann, D.; Nübel, U.; Bröker, B.M. Staphylococcus aureus toxins-their functions and genetics. Infect. Genet. Evol. 2014, 21, 583–592. [Google Scholar] [CrossRef] [PubMed]
- Lowy, F. Staphylococcus aureus infections. N. Engl. J. Med. 1998, 339, 520–529. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.Y.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G., Jr. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef] [PubMed]
- Wertheim, H.F.L.; Melles, D.C.; Vos, M.C.; van Leeuwen, W.; van Belkum, A.; Verbrugh, H.A.; Nouwen, J.I. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect. Dis. 2005, 5, 751–762. [Google Scholar] [CrossRef]
- van Belkum, A.; Verkaik, N.J.; de Vogel, C.P.; Boelens, H.A.; Verveer, J.; Nouwen, J.L.; Verbrugh, H.A.; Wertheim, H.F. Reclassification of Staphylococcus aureus nasal carriage types. J. Infect. Dis. 2009, 199, 1820–1826. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.F.; Leech, J.M.; Rogers, T.R.; McLoughlin, R.M. Staphylococcus aureus Colonization: Modulation of Host Immune Response and Impact on Human Vaccine Design. Front. Immunol. 2014, 4, 507. [Google Scholar] [CrossRef] [PubMed]
- Laupland, K.B.; Lyytikainen, O.; Sogaard, M.; Kennedy, K.J.; Knudsen, J.D.; Ostergaard, C.; Galbraith, J.C.; Valiquette, L.; Jacobsson, G.; Collignon, P.; et al. The changing epidemiology of Staphylococcus aureus bloodstream infection: A multinational population-based surveillance study. Clin. Microbiol. Infect. 2013, 19, 465–471. [Google Scholar] [CrossRef] [PubMed]
- van Belkum, A. Staphylococcal colonization and infection: Homeostasis versus disbalance of human (innate) immunity and bacterial virulence. Curr. Opin. Infect. Dis. 2006, 19, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Foster, T.J. Immune evasion by staphylococci. Nature Rev. Microbiol. 2005, 3, 948–958. [Google Scholar] [CrossRef] [PubMed]
- Thammavongsa, V.; Kim, H.K.; Missiakas, D.; Schneewind, O. Staphylococcal manipulation of host immune responses. Nat. Rev. Microbiol. 2015, 13, 529–543. [Google Scholar] [CrossRef] [PubMed]
- Foster, T.J.; Geoghegan, J.A.; Ganesh, V.K.; Hook, M. Adhesion, invasion and evasion: The many functions of the surface proteins of Staphylococcus aureus. Nat. Rev. Microbiol. 2014, 12, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Spaan, A.N.; Surewaard, B.G.; Nijland, R.; van Strijp, J.A. Neutrophils versus Staphylococcus aureus: A biological tug of war. Annu. Rev. Microbiol. 2013, 67, 629–650. [Google Scholar] [CrossRef] [PubMed]
- Laarman, A.; Milder, F.; van Strijp, J.; Rooijakkers, S. Complement inhibition by gram-positive pathogens: Molecular mechanisms and therapeutic implications. J. Mol. Med. (Berl.) 2010, 88, 115–120. [Google Scholar] [CrossRef] [PubMed]
- van Kessel, K.P.; Bestebroer, J.; van Strijp, J.A. Neutrophil-mediated phagocytosis of Staphylococcus aureus. Front. Immunol. 2014, 5, 467. [Google Scholar] [PubMed]
- Paul, W.E. The immune system. In Fundamental Immunology, 7th ed.; Paul, W.E., Ed.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; pp. 1–21. [Google Scholar]
- McHeyzer-Williams, M.; Okitsu, S.; Wang, N.; McHeyzer-Williams, L. Molecular programming of B cell memory. Nat. Rev. Immunol. 2012, 12, 24–34. [Google Scholar] [CrossRef] [PubMed]
- O´Shea, J. Helper T-cell differentiation and plasticity. In Fundamental Immunology, 7th ed.; Paul, W.E., Ed.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; pp. 708–717. [Google Scholar]
- Lowy, F.D. Is Staphylococcus aureus an intracellular pathogen? Trends Microbiol. 2000, 8, 341–343. [Google Scholar] [CrossRef]
- Lieberman, J. Cell-mediated cytotoxicity. In Fundamental Immunology, 7th ed.; Paul, W.E., Ed.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; pp. 891–909. [Google Scholar]
- Spellberg, B.; Daum, R. Development of a vaccine against Staphylococcus aureus. Semin. Immunopathol. 2012, 34, 335–348. [Google Scholar] [CrossRef] [PubMed]
- Proctor, R.A. Is there a future for a Staphylococcus aureus vaccine? Vaccine 2012, 30, 2921–2927. [Google Scholar] [CrossRef] [PubMed]
- von Köckritz-Blickwede, M.; Rohde, M.; Oehmcke, S.; Miller, L.S.; Cheung, A.L.; Herwald, H.; Foster, S.; Medina, E. Immunological mechanisms underlying the genetic predisposition to severe Staphylococcus aureus infection in the mouse model. Am. J. Pathol. 2008, 173, 1657–1668. [Google Scholar] [CrossRef] [PubMed]
- Nippe, N.; Varga, G.; Holzinger, D.; Löffler, B.; Medina, E.; Becker, K.; Roth, J.; Ehrchen, J.M.; Sunderkötter, C. Subcutaneous infection with S. aureus in mice reveals association of resistance with influx of neutrophils and Th2 response. J. Invest. Dermatol. 2011, 131, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, C.P.; Daniels, M.; Zhao, F.; Alegre, M.L.; Chong, A.S.; Daum, R.S. Protective immunity against recurrent Staphylococcus aureus skin infection requires antibody and interleukin-17A. Infect. Immun. 2014, 82, 2125–2134. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, C.; Goldmann, O.; Hobeika, E.; Geffers, R.; Peters, G.; Medina, E. The dynamics of T cells during persistent Staphylococcus aureus infection: From antigen-reactivity to in vivo anergy. EMBO Mol. Med. 2011, 3, 652–666. [Google Scholar] [CrossRef] [PubMed]
- Tebartz, C.; Horst, S.A.; Sparwasser, T.; Huehn, J.; Beineke, A.; Peters, G.; Medina, E. A major role for myeloid-derived suppressor cells and a minor role for regulatory T cells in immunosuppression during Staphylococcus aureus infection. J. Immunol. 2015, 194, 1100–1111. [Google Scholar] [CrossRef] [PubMed]
- Archer, N.K.; Harro, J.M.; Shirtliff, M.E. Clearance of Staphylococcus aureus nasal carriage is T cell dependent and mediated through interleukin-17A expression and neutrophil influx. Infect. Immun. 2013, 81, 2070–2075. [Google Scholar] [CrossRef] [PubMed]
- Dong, C. TH17 cells in development: An updated view of their molecular identity and genetic programming. Nat. Rev. Immunol. 2008, 8, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Shibata, K.; Yamada, H.; Hara, H.; Kishihara, K.; Yoshikai, Y. Resident Vdelta1+ gammadelta T cells control early infiltration of neutrophils after Escherichia coli infection via IL-17 production. J. Immunol. 2007, 178, 4466–4472. [Google Scholar] [CrossRef] [PubMed]
- Happel, K.I.; Zheng, M.; Young, E.; Quinton, L.J.; Lockhart, E.; Ramsay, A.J.; Shellito, J.E.; Schurr, J.R.; Bagby, G.J.; Nelson, S.; et al. Cutting edge: Roles of Toll-like receptor 4 and IL-23 in IL-17 expression in response to Klebsiella pneumoniae infection. J. Immunol. 2003, 170, 4432–4436. [Google Scholar] [CrossRef] [PubMed]
- Chien, Y.H.; Zeng, X.; Prinz, I. The natural and the inducible: Interleukin (IL)-17-producing gammadelta T cells. Trends Immunol. 2013, 34, 151–154. [Google Scholar] [CrossRef] [PubMed]
- Ishigame, H.; Kakuta, S.; Nagai, T.; Kadoki, M.; Nambu, A.; Komiyama, Y.; Fujikado, N.; Tanahashi, Y.; Akitsu, A.; Kotaki, H.; et al. Differential roles of interleukin-17A and -17F in host defense against mucoepithelial bacterial infection and allergic responses. Immunity 2009, 30, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.S.; Pietras, E.M.; Garcia, N.C.; Ramos, R.I.; Farzam, D.M.; Monroe, H.R.; Magorien, J.E.; Blauvelt, A.; Kolls, J.K.; Cheung, A.L.; et al. IL-17 is essential for host defense against cutaneous Staphylococcus aureus infection in mice. J. Clin. Invest. 2010. [Google Scholar] [CrossRef] [PubMed]
- Maher, B.M.; Mulcahy, M.E.; Murphy, A.G.; Wilk, M.; O’Keeffe, K.M.; Geoghegan, J.A.; Lavelle, E.C.; McLoughlin, R.M. Nlrp-3-driven interleukin 17 production by γδT cells controls infection outcomes during Staphylococcus aureus surgical site infection. Infect. Immun. 2013, 81, 4478–4489. [Google Scholar] [CrossRef] [PubMed]
- Molne, L.; Corthay, A.; Holmdahl, R.; Tarkowski, A. Role of gamma/delta T cell receptor-expressing lymphocytes in cutaneous infection caused by Staphylococcus aureus. Clin. Exp. Immunol. 2003, 132, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Murphy, A.G.; O’Keeffe, K.M.; Lalor, S.J.; Maher, B.M.; Mills, K.H.; McLoughlin, R.M. Correction: Staphylococcus aureus Infection of mice expands a population of memory γδ T cells that are protective against subsequent infection. J. Immunol. 2015, 194, 4588. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Ibrahim, A.S.; Xu, X.; Farber, J.M.; Avanesian, V.; Baquir, B.; Fu, Y.; French, S.W.; Edwards, J.E., Jr.; Spellberg, B. Th1-Th17 cells mediate protective adaptive immunity against Staphylococcus aureus and Candida albicans infection in mice. PLoS Pathog. 2009, 5, e1000703. [Google Scholar] [CrossRef] [PubMed]
- Narita, K.; Hu, D.L.; Mori, F.; Wakabayashi, K.; Iwakura, Y.; Nakane, A. Role of interleukin-17A in cell-mediated protection against Staphylococcus aureus infection in mice immunized with the fibrinogen-binding domain of clumping factor A. Infect. Immun. 2010, 78, 4234–4242. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Kamath, A.; Das, H.; Li, L.; Bukowski, J.F. Antibacterial effect of human V gamma 2V delta 2 T cells in vivo. J. Clin. Invest. 2001, 108, 1349–1357. [Google Scholar] [CrossRef] [PubMed]
- Parker, D.; Ryan, C.L.; Alonzo, F., 3rd; Torres, V.J.; Planet, P.J.; Prince, A.S. CD4+ T cells promote the pathogenesis of Staphylococcus aureus pneumonia. J. Infect. Dis. 2015, 211, 835–845. [Google Scholar] [CrossRef] [PubMed]
- Cheng, P.; Liu, T.; Zhou, W.Y.; Zhuang, Y.; Peng, L.S.; Zhang, J.Y.; Yin, Z.N.; Mao, X.H.; Guo, G.; Shi, Y.; et al. Role of gamma-delta T cells in host response against Staphylococcus aureus-induced pneumonia. BMC Immunol. 2012, 13, 38. [Google Scholar] [CrossRef] [PubMed]
- Kudva, A.; Scheller, E.V.; Robinson, K.M.; Crowe, C.R.; Choi, S.M.; Slight, S.R.; Khader, S.A.; Dubin, P.J.; Enelow, R.I.; Kolls, J.K.; et al. Influenza A inhibits Th17-mediated host defense against bacterial pneumonia in mice. J. Immunol. 2011, 186, 1666–1674. [Google Scholar] [CrossRef] [PubMed]
- Ohlsen, K.; Lorenz, U. Immunotherapeutic strategies to combat staphylococcal infections. Int. J. Med. Microbiol. 2010, 300, 402–410. [Google Scholar] [CrossRef] [PubMed]
- Joshi, A.; Pancari, G.; Cope, L.; Bowman, E.P.; Cua, D.; Proctor, R.A.; McNeely, T. Immunization with Staphylococcus aureus iron regulated surface determinant B (IsdB) confers protection via Th17/IL17 pathway in a murine sepsis model. Hum. Vaccines Immunother. 2012, 8, 336–346. [Google Scholar] [CrossRef] [PubMed]
- Misstear, K.; McNeela, E.A.; Murphy, A.G.; Geoghegan, J.A.; O’Keeffe, K.M.; Fox, J.; Chan, K.; Heuking, S.; Collin, N.; Foster, T.J.; et al. Targeted nasal vaccination provides antibody-independent protection against Staphylococcus aureus. J. Infect. Dis. 2014, 209, 1479–1484. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.F.; Murphy, A.G.; Lalor, S.J.; Leech, J.M.; O’Keeffe, K.M.; Mac Aogain, M.; O’Halloran, D.P.; Lacey, K.A.; Tavakol, M.; Hearnden, C.H.; et al. Memory Th1 cells are protective in invasive Staphylococcus aureus infection. PLoS Pathog. 2015, 11, e1005226. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Y.; Huang, Z.X.; Chen, Y.G.; Lu, X.; Zhu, P.; Wen, K.; Fu, N.; Liu, B.Y. A multiple antigenic peptide mimicking peptidoglycan induced T cell responses to protect mice from systemic infection with Staphylococcus aureus. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Murphy, A.G.; O’Keeffe, K.M.; Lalor, S.J.; Maher, B.M.; Mills, K.H.; McLoughlin, R.M. Staphylococcus aureus infection of mice expands a population of memory gd T cells that are protective against subsequent infection. J. Immunol. 2014, 192, 3697–3708. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, J.W. Mastitis control programs: Bovine mastitis and milking management. In Document AS-1129; Service, North Dacota State University Extension Service, Publ.: Fargo, ND, USA, 2012. [Google Scholar]
- Barkema, H.W.; Green, M.J.; Bradley, A.J.; Zadoks, R.N. Invited review: The role of contagious disease in udder health. J. Dairy Sci. 2009, 92, 4717–4729. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.Y.; Bartlett, P.C.; Lance, S.E.; Anderson, J.; Heider, L.E. Costs of clinical mastitis and mastitis prevention in dairy herds. J. Am. Vet. Med. Assoc. 1993, 202, 1230–1236. [Google Scholar] [PubMed]
- Fabres-Klein, M.H.; Aguilar, A.P.; Silva, M.P.; Silva, D.M.; Ribon, A.O. Moving towards the immunodiagnosis of staphylococcal intramammary infections. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 2095–2104. [Google Scholar] [CrossRef] [PubMed]
- Seegers, H.; Fourichon, C.; Beaudeau, F. Production effects related to mastitis and mastitis economics in dairy cattle herds. Vet. Res. 2003, 34, 475–491. [Google Scholar] [CrossRef] [PubMed]
- Soltys, J.; Quinn, M.T. Selective recruitment of T-cell subsets to the udder during staphylococcal and streptococcal mastitis: Analysis of lymphocyte subsets and adhesion molecule expression. Infect. Immun. 1999, 67, 6293–6302. [Google Scholar] [PubMed]
- Gronlund, U.; Johannisson, A.; Persson Waller, K. Changes in blood and milk lymphocyte sub-populations during acute and chronic phases of Staphylococcus aureus induced bovine mastitis. Res. Vet. Sci. 2006, 80, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Rivas, A.L.; Quimby, F.W.; Coksaygan, O.; Olmstead, L.; Lein, D.H. Longitudinal evaluation of CD4+ and CD8+ peripheral blood and mammary gland lymphocytes in cows experimentally inoculated with Staphylococcus aureus. Can. J. Vet. Res. 2000, 64, 232–237. [Google Scholar] [PubMed]
- Park, Y.H.; Joo, Y.S.; Park, J.Y.; Moon, J.S.; Kim, S.H.; Kwon, N.H.; Ahn, J.S.; Davis, W.C.; Davies, C.J. Characterization of lymphocyte subpopulations and major histocompatibility complex haplotypes of mastitis-resistant and susceptible cows. J. Vet. Sci. 2004, 5, 29–39. [Google Scholar] [PubMed]
- Chang, B.S.; Bohach, G.A.; Lee, S.U.; Davis, W.C.; Fox, L.K.; Ferens, W.A.; Seo, K.S.; Koo, H.C.; Kwon, N.H.; Park, Y.H. Immunosuppression by T regulatory cells in cows infected with Staphylococcal superantigen. J. Vet. Sci. 2005, 6, 247–250. [Google Scholar] [PubMed]
- Riollet, C.; Rainard, P.; Poutrel, B. Cell subpopulations and cytokine expression in cow milk in response to chronic Staphylococcus aureus infection. J. Dairy Sci. 2001, 84, 1077–1084. [Google Scholar] [CrossRef]
- Bharathan, M.; Mullarky, I.K. Targeting mucosal immunity in the battle to develop a mastitis vaccine. J. Mammary Gland Biol. Neoplasia 2011, 16, 409–419. [Google Scholar] [CrossRef] [PubMed]
- Pereira, U.P.; Oliveira, D.G.S.; Mesquita, L.R.; Costa, G.M.; Pereira, L.J. Efficacy of Staphylococcus aureus vaccines for bovine mastitis: A systematic review. Vet. Microbiol. 2011, 148, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Péton, V.; Le Loir, Y. Staphylococcus aureus in veterinary medicine. Infect. Genet. Evol. 2014, 21, 602–615. [Google Scholar] [CrossRef] [PubMed]
- Camussone, C.M.; Pujato, N.; Renna, M.S.; Veaute, C.M.; Morein, B.; Marcipar, I.S.; Calvinho, L.F. Immune response and functional role of antibodies raised in heifers against a Staphylococcus aureus CP5 lysate and recombinant antigens vaccine formulated with Iscom Matrix adjuvant. Vet. Immunol. Immunopathol. 2014, 162, 96–107. [Google Scholar] [CrossRef] [PubMed]
- Schukken, Y.H.; Bronzo, V.; Locatelli, C.; Pollera, C.; Rota, N.; Casula, A.; Testa, F.; Scaccabarozzi, L.; March, R.; Zalduendo, D.; et al. Efficacy of vaccination on Staphylococcus aureus and coagulase-negative staphylococci intramammary infection dynamics in 2 dairy herds. J. Dairy Sc. 2014, 97, 5250–5264. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-W.; O’Brien, C.N.; Guidry, A.J.; Paape, M.J.; Shafer-Weaver, K.A.; Zhao, X. Effect of a trivalent vaccine against Staphylococcus aureus mastitis lymphocyte subpopulations, antibody production, and neutrophil phagocytosis. Can. J. Vet. Res. 2005, 69, 11–18. [Google Scholar] [PubMed]
- Prenafeta, A.; March, R.; Foix, A.; Casals, I.; Costa, L. Study of the humoral immunological response after vaccination with a Staphylococcus aureus biofilm-embedded bacterin in dairy cows: Possible role of the exopolysaccharide specific antibody production in the protection from Staphylococcus aureus induced mastitis. Vet. Immunol. Immunopathol. 2010, 134, 208–217. [Google Scholar] [PubMed]
- Watson, D.; McCOLL, M.; Davies, H. Field trial of a staphylococcal mastitis vaccine in dairy herds: Clinical, subclinical and microbiological assessments. Aust. Vet. J. 1996, 74, 447–450. [Google Scholar] [CrossRef] [PubMed]
- Middleton, J.R.; Ma, J.; Rinehart, C.L.; Taylor, V.N.; Luby, C.D.; Steevens, B.J. Efficacy of different Lysigin formulations in the prevention of Staphylococcus aureus intramammary infection in dairy heifers. J. Dairy Res. 2006, 73, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Middleton, J.R. Staphylococcus aureus antigens and challenges in vaccine development. Expert Rev. Vaccines 2008, 7, 805–815. [Google Scholar] [CrossRef] [PubMed]
- Middleton, J.R.; Luby, C.D.; Adams, D.S. Efficacy of vaccination against staphylococcal mastitis: A review and new data. Vet. Microbiol. 2009, 134, 192–198. [Google Scholar] [CrossRef] [PubMed]
- Pankey, J.W.; Boddie, N.T.; Watts, J.L.; Nickerson, S.C. Evaluation of Protein A and a commercial bacterin as vaccines against Staphylococcus aureus mastitis by experimental challenge. J. Dairy Sci. 1985, 68, 726–731. [Google Scholar] [CrossRef]
- Buzzola, F.R.; Alvarez, L.P.; Tuchscherr, L.P.N.; Barbagelata, M.S.; Lattar, S.M.; Calvinho, L.; Sordelli, D.O. Differential abilities of capsulated and noncapsulated Staphylococcus aureus isolates from diverse agr groups to invade mammary epithelial cells. Infect. Immun. 2007, 75, 886–891. [Google Scholar] [CrossRef] [PubMed]
- Pellegrino, M.; Giraudo, J.; Raspanti, C.; Nagel, R.; Odierno, L.; Primo, V.; Bogni, C. Experimental trial in heifers vaccinated with Staphylococcus aureus avirulent mutant against bovine mastitis. Vet. Microbiol. 2008, 127, 186–190. [Google Scholar] [CrossRef] [PubMed]
- Pellegrino, M.; Giraudo, J.; Raspanti, C.; Odierno, L.; Bogni, C. Efficacy of immunization against bovine mastitis using a Staphylococcus aureus avirulent mutant vaccine. Vaccine 2010, 28, 4523–4528. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, P.K.; Rokbi, B.; Arnaud-Barbe, N.; Sutten, E.L.; Norimine, J.; Lahmers, K.K.; Brown, W.C. CD4 T cell antigens from Staphylococcus aureus Newman strain identified following immunization with heat-killed bacteria. Clin. Vaccine Immunol. 2012, 19, 477–489. [Google Scholar] [CrossRef] [PubMed]
- Pujol, J.; Bouillenne, F.; Farnir, F.; Dufrasne, I.; Mainil, J.; Galleni, M.; Lekeux, P.; Bureau, F.; Fievez, L. Generation of a soluble recombinant trimeric form of bovine CD40L and its potential use as a vaccine adjuvant in cows. Vet. Immunol. Immunopathol. 2015, 168, 1–13. [Google Scholar] [CrossRef] [PubMed]
- van Belkum, A.; Melles, D.C.; Nouwen, J.; van Leeuwen, W.B.; van Wamel, W.; Vos, M.C.; Wertheim, H.F.; Verbrugh, H.A. Co-evolutionary aspects of human colonisation and infection by Staphylococcus aureus. Infect. Genet. Evol. 2009, 9, 32–47. [Google Scholar] [CrossRef] [PubMed]
- Chandesris, M.O.; Melki, I.; Natividad, A.; Puel, A.; Fieschi, C.; Yun, L.; Thumerelle, C.; Oksenhendler, E.; Boutboul, D.; Thomas, C.; et al. Autosomal dominant STAT3 deficiency and hyper-IgE syndrome: Molecular, cellular, and clinical features from a French national survey. Medicine (Baltimore) 2012, 91, e1–e19. [Google Scholar] [CrossRef] [PubMed]
- Cook, M.C.; Tangye, S.G. Primary immune deficiencies affecting lymphocyte differentiation: Lessons from the spectrum of resulting infections. Int. Immunol. 2009, 21, 1003–1011. [Google Scholar] [CrossRef] [PubMed]
- Milner, J.D.; Brenchley, J.M.; Laurence, A.; Freeman, A.F.; Hill, B.J.; Elias, K.M.; Kanno, Y.; Spalding, C.; Elloumi, H.Z.; Paulson, M.L.; et al. Impaired T(H)17 cell differentiation in subjects with autosomal dominant hyper-IgE syndrome. Nature 2008, 452, 773–776. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.S.; Chew, G.Y.; Simpson, N.; Priyadarshi, A.; Wong, M.; Grimbacher, B.; Fulcher, D.A.; Tangye, S.G.; Cook, M.C. Deficiency of Th17 cells in hyper IgE syndrome due to mutations in STAT3. J. Exp. Med. 2008, 205, 1551–1557. [Google Scholar] [CrossRef] [PubMed]
- Renner, E.D.; Rylaarsdam, S.; Anover-Sombke, S.; Rack, A.L.; Reichenbach, J.; Carey, J.C.; Zhu, Q.; Jansson, A.F.; Barboza, J.; Schimke, L.F.; et al. Novel signal transducer and activator of transcription 3 (STAT3) mutations, reduced T(H)17 cell numbers, and variably defective STAT3 phosphorylation in hyper-IgE syndrome. J. Allergy Clin. Immunol. 2008, 122, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Holland, S.M.; DeLeo, F.R.; Elloumi, H.Z.; Hsu, A.P.; Uzel, G.; Brodsky, N.; Freeman, A.F.; Demidowich, A.; Davis, J.; Turner, M.L.; et al. STAT3 mutations in the hyper-IgE syndrome. N. Engl. J. Med. 2007, 357, 1608–1619. [Google Scholar] [CrossRef] [PubMed]
- Craven, D.E. Staphylococcus aureus colonisation and bacteraemia in persons infected with human immunodeficiency virus: A dynamic interaction with the host. J. Chemother. 1995, 7, 19–28. [Google Scholar] [PubMed]
- Reddy, E.A.; Shaw, A.V.; Crump, J.A. Community-acquired bloodstream infections in Africa: A systematic review and meta-analysis. Lancet Infect. Dis. 2010, 10, 417–432. [Google Scholar] [CrossRef]
- Laupland, K.B.; Ross, T.; Gregson, D.B. Staphylococcus aureus bloodstream infections: Risk factors, outcomes, and the influence of methicillin resistance in Calgary, Canada, 2000–2006. J. Infect. Dis. 2008, 198, 336–343. [Google Scholar] [CrossRef] [PubMed]
- Wiese, L.; Mejer, N.; Schonheyder, H.C.; Westh, H.; Jensen, A.G.; Larsen, A.R.; Skov, R.; Benfield, T.; Danish Staphylococcal Bacteraemia Study Group. A nationwide study of comorbidity and risk of reinfection after Staphylococcus aureus bacteraemia. J. Infect. 2013, 67, 199–205. [Google Scholar] [CrossRef] [PubMed]
- Bröker, B.M.; Holtfreter, S.; Bekeredjian-Ding, I. Immune control of Staphylocuccus aureus—Regulation and counter-regulation of the adaptive immune response. Int. J. Med. Microbiol. 2014, 304, 204–214. [Google Scholar] [CrossRef] [PubMed]
- Bröker, B.M.; van Belkum, A. Immune proteomics of Staphylococcus aureus. Proteomics 2011, 11, 3221–3231. [Google Scholar] [CrossRef] [PubMed]
- Vytvytska, O.; Nagy, E.; Bluggel, M.; Meyer, H.; Kurzbauer, R.; Huber, L.; Klade, C. Identification of vaccine candidate antigens of Staphylococcus aureus by serological proteome analysis. Proteomics 2002, 2, 580–590. [Google Scholar] [CrossRef]
- Kolata, J.; Bode, L.G.; Holtfreter, S.; Steil, L.; Kusch, H.; Holtfreter, B.; Albrecht, D.; Hecker, M.; Engelmann, S.; van Belkum, A.; et al. Distinctive patterns in the human antibody response to Staphylococcus aureus bacteremia in carriers and non-carriers. Proteomics 2011, 11, 3914–3927. [Google Scholar] [CrossRef] [PubMed]
- Holtfreter, S.; Roschack, K.; Eichler, P.; Eske, K.; Holtfreter, B.; Kohler, C.; Engelmann, S.; Hecker, M.; Greinacher, A.; Bröker, B.M. Staphylococcus aureus carriers neutralize superantigens by antibodies specific for their colonizing strain: A potential explanation for their improved prognosis in severe sepsis. J. Infect. Dis. 2006, 193, 1275–1278. [Google Scholar] [CrossRef] [PubMed]
- Crotty, S. A brief history of T cell help to B cells. Nat. Rev. Immunol. 2015, 15, 185–189. [Google Scholar] [CrossRef] [PubMed]
- Crotty, S. Follicular helper CD4 T cells (TFH). Annu. Rev. Immunol. 2011, 29, 621–663. [Google Scholar] [CrossRef] [PubMed]
- Zielinski, C.E.; Corti, D.; Mele, F.; Pinto, D.; Lanzavecchia, A.; Sallusto, F. Dissecting the human immunologic memory for pathogens. Immunol. Rev. 2011, 240, 40–51. [Google Scholar] [CrossRef] [PubMed]
- Kolata, J.; Kühbandner, I.; Link, C.; Normann, N.; Weidenmaier, C.; Bröker, B. The fall of a dogma? Unexpectedly high T cell memory response to Staphylococcus aureus in humans. J. Infect. Dis. 2015. [Google Scholar] [CrossRef] [PubMed]
- Zielinski, C.E.; Mele, F.; Aschenbrenner, D.; Jarrossay, D.; Ronchi, F.; Gattorno, M.; Monticelli, S.; Lanzavecchia, A.; Sallusto, F. Pathogen-induced human TH17 cells produce IFN-gamma or IL-10 and are regulated by IL-1beta. Nature 2012, 484, 514–518. [Google Scholar] [CrossRef] [PubMed]
- Stentzel, S.; Teufelberger, A.; Nordengrün, M.; Kolata, J.; Schmidt, F.; van Crombruggen, K.; Michalik, S.; Kumpfmüller, J.; Tischer, S.; Schweder, T.; et al. Spls are pacemakers of allergic airway reactions to Staphylococcus aureus. Unpublished work. 2016. [Google Scholar]
- Frodermann, V.; Chau, T.A.; Sayedyahossein, S.; Toth, J.M.; Heinrichs, D.E.; Madrenas, J. A modulatory interleukin-10 response to staphylococcal peptidoglycan prevents Th1/Th17 adaptive immunity to Staphylococcus aureus. J. Infect. Dis. 2011, 204, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Belkaid, Y.; Tarbell, K. Regulatory T cells in the control of host-microorganism interactions (*). Annu. Rev. Immunol. 2009, 27, 551–589. [Google Scholar] [CrossRef] [PubMed]
- Belkaid, Y.; Rouse, B.T. Natural regulatory T cells in infectious disease. Nat. Immunol. 2005, 6, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Fowler, V.G.; Allen, K.B.; Moreira, E.D.; Moustafa, M.; Isgro, F.; Boucher, H.W.; Corey, G.R.; Carmeli, Y.; Betts, R.; Hartzel, J.S.; et al. Effect of an investigational vaccine for preventing Staphylococcus aureus infections after cardiothoracic surgery: A randomized trial. JAMA 2013, 309, 1368–1378. [Google Scholar] [CrossRef] [PubMed]
- Niebuhr, M.; Gathmann, M.; Scharonow, H.; Mamerow, D.; Mommert, S.; Balaji, H.; Werfel, T. Staphylococcal alpha-toxin is a strong inducer of interleukin-17 in humans. Infect. Immun. 2011, 79, 1615–1622. [Google Scholar] [CrossRef] [PubMed]
- Breuer, K.; Wittmann, M.; Kempe, K.; Kapp, A.; Mai, U.; Dittrich-Breiholz, O.; Kracht, M.; Mrabet-Dahbi, S.; Werfel, T. Alpha-toxin is produced by skin colonizing Staphylococcus aureus and induces a T helper type 1 response in atopic dermatitis. Clin. Exp. Allergy 2005, 35, 1088–1095. [Google Scholar] [CrossRef] [PubMed]
- Warmerdam, P.A.; Vanderlick, K.; Vandervoort, P.; De Smedt, H.; Plaisance, S.; De Maeyer, M.; Collen, D. Staphylokinase-specific cell-mediated immunity in humans. J. Immunol. 2002, 168, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Weidenmaier, C.; McLoughlin, R.M.; Lee, J.C. The zwitterionic cell wall teichoic acid of Staphylococcus aureus provokes skin abscesses in mice by a novel CD4+ T-cell-dependent mechanism. PLoS ONE 2010, 5, e13227. [Google Scholar] [CrossRef] [PubMed]
- Levy, J.; Licini, L.; Haelterman, E.; Moris, P.; Lestrate, P.; Damaso, S.; Van Belle, P.; Boutriau, D. Safety and immunogenicity of an investigational 4-component Staphylococcus aureus vaccine with or without AS03B adjuvant: Results of a randomized phase I trial. Hum. Vaccines Immunother. 2015, 11, 620–631. [Google Scholar] [CrossRef] [PubMed]
- Nissen, M.; Marshall, H.; Richmond, P.; Shakib, S.; Jiang, Q.; Cooper, D.; Rill, D.; Baber, J.; Eiden, J.; Gruber, W.; et al. A randomized phase I study of the safety and immunogenicity of three ascending dose levels of a 3-antigen Staphylococcus aureus vaccine (SA3Ag) in healthy adults. Vaccine 2015, 33, 1846–1854. [Google Scholar] [CrossRef] [PubMed]
- Projan, S.J.; Nesin, M.; Dunman, P.M. Staphylococcal vaccines and immunotherapy: To dream the impossible dream? Curr. Opin. Pharmacol. 2006, 6, 473–479. [Google Scholar] [CrossRef] [PubMed]
- Verkaik, N.J.; van Wamel, W.J.; van Belkum, A. Immunotherapeutic approaches against Staphylococcus aureus. Immunotherapy 2011, 3, 1063–1073. [Google Scholar] [CrossRef] [PubMed]
- Fowler, V.G., Jr.; Proctor, R.A. Where does a Staphylococcus aureus vaccine stand? Clin. Microbiol. Infect. 2014, 20 Suppl 5, 66–75. [Google Scholar] [CrossRef] [PubMed]
- Bagnoli, F.; Bertholet, S.; Grandi, G. Inferring reasons for the failure of Staphylococcus aureus vaccines in clinical trials. Front. Cell. Infect. Microbiol. 2012, 2, 16. [Google Scholar] [CrossRef] [PubMed]
- Fattom, A.; Matalon, A.; Buerkert, J.; Taylor, K.; Damaso, S.; Boutriau, D. Efficacy profile of a bivalent Staphylococcus aureus glycoconjugated vaccine in adults on hemodialysis: Phase III randomized study. Hum. Vaccines Immunother. 2015, 11, 632–641. [Google Scholar] [CrossRef] [PubMed]
- Jansen, K.U.; Girgenti, D.Q.; Scully, I.L.; Anderson, A.S. Vaccine review: “Staphyloccocus aureus vaccines: Problems and prospects”. Vaccine 2013, 31, 2723–2730. [Google Scholar] [CrossRef] [PubMed]
- McNeely, T.B.; Shah, N.A.; Fridman, A.; Joshi, A.; Hartzel, J.S.; Keshari, R.S.; Lupu, F.; DiNubile, M.J. Mortality among recipients of the Merck V710 Staphylococcus aureus vaccine after postoperative S. aureus infections: An analysis of possible contributing host factors. Hum. Vaccines Immunother. 2014, 10, 3513–3516. [Google Scholar] [CrossRef] [PubMed]
- Löffler, B.; Tuchscherr, L.; Niemann, S.; Peters, G. Staphylococcus aureus persistence in non-professional phagocytes. Int. J. Med. Microbiol. 2014, 304, 170–176. [Google Scholar] [CrossRef] [PubMed]
- Grosz, M.; Kolter, J.; Paprotka, K.; Winkler, A.C.; Schafer, D.; Chatterjee, S.S.; Geiger, T.; Wolz, C.; Ohlsen, K.; Otto, M.; et al. Cytoplasmic replication of Staphylococcus aureus upon phagosomal escape triggered by phenol-soluble modulin alpha. Cell. Microbiol. 2014, 16, 451–465. [Google Scholar] [CrossRef] [PubMed]
- Thwaites, G.E.; Gant, V. Are bloodstream leukocytes Trojan Horses for the metastasis of Staphylococcus aureus? Nat. Rev. Microbiol. 2011, 9, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Tuchscherr, L.; Medina, E.; Hussain, M.; Volker, W.; Heitmann, V.; Niemann, S.; Holzinger, D.; Roth, J.; Proctor, R.A.; Becker, K.; et al. Staphylococcus aureus phenotype switching: An effective bacterial strategy to escape host immune response and establish a chronic infection. EMBO Mol. Med. 2011, 3, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Tuchscherr, L.; Heitmann, V.; Hussain, M.; Viemann, D.; Roth, J.; von Eiff, C.; Peters, G.; Becker, K.; Loffler, B. Staphylococcus aureus small-colony variants are adapted phenotypes for intracellular persistence. J. Infect. Dis. 2010, 202, 1031–1040. [Google Scholar] [CrossRef] [PubMed]
- Surmann, K.; Simon, M.; Hildebrandt, P.; Pförtner, H.; Michalik, S.; Stentzel, S.; Steil, L.; Dhople, V.M.; Bernhardt, J.; Schlüter, R.; et al. A proteomic perspective of the interplay of Staphylococcus aureus and human alveolar epithelial cells during infection. J. Proteomics 2015. [Google Scholar] [CrossRef] [PubMed]
- Surmann, K.; Michalik, S.; Hildebrandt, P.; Gierok, P.; Depke, M.; Brinkmann, L.; Bernhardt, J.; Salazar, M.G.; Sun, Z.; Shteynberg, D.; et al. Comparative proteome analysis reveals conserved and specific adaptation patterns of Staphylococcus aureus after internalization by different types of human non-professional phagocytic host cells. Front. Microbiol. 2014, 5, 392. [Google Scholar] [CrossRef] [PubMed]
- Holzinger, D.; Gieldon, L.; Mysore, V.; Nippe, N.; Taxman, D.J.; Duncan, J.A.; Broglie, P.M.; Marketon, K.; Austermann, J.; Vogl, T.; et al. Staphylococcus aureus Panton-Valentine leukocidin induces an inflammatory response in human phagocytes via the NLRP3 inflammasome. J. Leukoc. Biol. 2012, 92, 1069–1081. [Google Scholar] [CrossRef] [PubMed]
- Kubica, M.; Guzik, K.; Koziel, J.; Zarebski, M.; Richter, W.; Gajkowska, B.; Golda, A.; Maciag-Gudowska, A.; Brix, K.; Shaw, L.; et al. A potential new pathway for Staphylococcus aureus dissemination: The silent survival of S. aureus phagocytosed by human monocyte-derived macrophages. PLoS ONE 2008, 3, e1409. [Google Scholar] [CrossRef] [PubMed]
- Smith, R.P.; Baltch, A.L.; Ritz, W.J.; Michelsen, P.B.; Bopp, L.H. IFN-gamma enhances killing of methicillin-resistant Staphylococcus aureus by human monocytes more effectively than GM-CSF in the presence of daptomycin and other antibiotics. Cytokine 2010, 51, 274–277. [Google Scholar] [CrossRef] [PubMed]
- Penaloza-MacMaster, P.; Barber, D.L.; Wherry, E.J.; Provine, N.M.; Teigler, J.E.; Parenteau, L.; Blackmore, S.; Borducchi, E.N.; Larocca, R.A.; Yates, K.B.; et al. Vaccine-elicited CD4 T cells induce immunopathology after chronic LCMV infection. Science 2015, 347, 278–282. [Google Scholar] [CrossRef] [PubMed]
- McLoughlin, R.M.; Lee, J.C.; Kasper, D.L.; Tzianabos, A.O. IFN-gamma regulated chemokine production determines the outcome of Staphylococcus aureus infection. J. Immunol. 2008, 181, 1323–1332. [Google Scholar] [CrossRef] [PubMed]
- Barnes, P.J. Intrinsic asthma: Not so different from allergic asthma but driven by superantigens? Clin. Exp. Allergy 2009, 39, 1145–1151. [Google Scholar] [CrossRef] [PubMed]
- Bachert, C.; Zhang, N. Chronic rhinosinusitis and asthma: Novel understanding of the role of IgE ‘above atopy’. J. Intern. Med. 2012, 272, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.F.; Peng, R.D.; McCormack, M.C.; Matsui, E.C. Staphylococcus aureus colonization is associated with wheeze and asthma among US children and young adults. J. Allergy Clin. Immunol. 2015, 135, 811–813.e5. [Google Scholar] [CrossRef] [PubMed]
- Wills-Karp, M.; Lewkowich, I.P. Immunologic mechanism of allergic disorders. In Fundamental Immunology, 7th ed.; Paul, W.E., Ed.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; pp. 1113–1153. [Google Scholar]
- Berube, B.J.; Bubeck Wardenburg, J. Staphylococcus aureus alpha-toxin: Nearly a century of intrigue. Toxins 2013, 5, 1140–1166. [Google Scholar] [CrossRef] [PubMed]
- Inoshima, I.; Inoshima, N.; Wilke, G.A.; Powers, M.E.; Frank, K.M.; Wang, Y.; Wardenburg, J.B. A Staphylococcus aureus pore-forming toxin subverts the activity of ADAM10 to cause lethal infection in mice. Nat. Med. 2011, 17, 1310–1314. [Google Scholar] [CrossRef] [PubMed]
- Brauweiler, A.M.; Bin, L.; Kim, B.E.; Oyoshi, M.K.; Geha, R.S.; Goleva, E.; Leung, D.Y. Filaggrin-dependent secretion of sphingomyelinase protects against staphylococcal alpha-toxin-induced keratinocyte death. J. Allergy Clin. Immunol. 2012, 131, 421–427.e2. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Oscherwitz, J.; Cease, K.B.; Chan, S.M.; Munoz-Planillo, R.; Hasegawa, M.; Villaruz, A.E.; Cheung, G.Y.C.; McGavin, M.J.; Travers, J.B.; et al. Staphylococcus δ-toxin induces allergic skin disease by activating mast cells. Nature 2013, 503, 397–401. [Google Scholar] [CrossRef] [PubMed]
- Huvenne, W.; Hellings, P.W.; Bachert, C. Role of staphylococcal superantigens in airway disease. Int. Arch. Allergy Immunol. 2013, 161, 304–314. [Google Scholar] [CrossRef] [PubMed]
- Bachert, C.; Gevaert, P.; van Cauwenberge, P. Staphylococcus aureus enterotoxins: A key in airway disease? Allergy 2002, 57, 480–487. [Google Scholar] [CrossRef] [PubMed]
- Wilke, G.A.; Bubeck Wardenburg, J. Role of a disintegrin and metalloprotease 10 in Staphylococcus aureus alpha-hemolysin-mediated cellular injury. Proc. Natl. Acad. Sci. USA 2010, 107, 13473–13478. [Google Scholar] [CrossRef] [PubMed]
- Alonzo, F., 3rd; Kozhaya, L.; Rawlings, S.A.; Reyes-Robles, T.; DuMont, A.L.; Myszka, D.G.; Landau, N.R.; Unutmaz, D.; Torres, V.J. CCR5 is a receptor for Staphylococcus aureus leukotoxin ED. Nature 2013, 493, 51–55. [Google Scholar] [PubMed]
- Marrack, P.; Kappler, J. The staphylococcal enterotoxins and their relatives. Science 1990, 248, 1066. [Google Scholar] [CrossRef] [PubMed]
- Spaulding, A.R.; Salgado-Pabon, W.; Kohler, P.L.; Horswill, A.R.; Leung, D.Y.; Schlievert, P.M. Staphylococcal and streptococcal superantigen exotoxins. Clin. Microbiol. Rev. 2013, 26, 422–447. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.X.; McCormick, J.K. Staphylococcal superantigens in colonization and disease. Front. Cell. Infect. Microbiol. 2012, 2, 52. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.L.; Zhu, G.; Mori, F.; Omoe, K.; Okada, M.; Wakabayashi, K.; Kaneko, S.; Shinagawa, K.; Nakane, A. Staphylococcal enterotoxin induces emesis through increasing serotonin release in intestine and it is downregulated by cannabinoid receptor 1. Cell. Microbiol. 2007, 9, 2267–2277. [Google Scholar] [CrossRef] [PubMed]
- Holtfreter, S.; Grumann, D.; Schmudde, M.; Nguyen, H.T.; Eichler, P.; Strommenger, B.; Kopron, K.; Kolata, J.; Giedrys-Kalemba, S.; Steinmetz, I.; et al. Clonal distribution of superantigen genes in clinical Staphylococcus aureus isolates. J. Clin. Microbiol. 2007, 45, 2669–2680. [Google Scholar] [CrossRef] [PubMed]
- Baba, T.; Takeuchi, F.; Kuroda, M.; Yuzawa, H.; Aoki, K.; Oguchi, A.; Nagai, Y.; Iwama, N.; Asano, K.; Naimi, T.; et al. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 2002, 359, 1819–1827. [Google Scholar] [CrossRef]
- Becker, K.; Friedrich, A.W.; Lubritz, G.; Weilert, M.; Peters, G.; von Eiff, C. Prevalence of genes encoding pyrogenic toxin superantigens and exfoliative toxins among strains of Staphylococcus aureus isolated from blood and nasal specimens. J. Clin. Microbiol. 2003, 41. [Google Scholar] [CrossRef]
- Holtfreter, S.; Bauer, K.; Thomas, D.; Feig, C.; Lorenz, V.; Roschack, K.; Friebe, E.; Selleng, K.; Lövenich, S.; Greve, T.; et al. egc-Encoded superantigens from Staphylococcus aureus are neutralized by human sera much less efficiently than are classical staphylococcal enterotoxins or toxic shock syndrome toxin. Infect. Immun. 2004, 72, 4061–4071. [Google Scholar] [CrossRef] [PubMed]
- Jarraud, S.; Peyrat, M.A.; Lim, A.; Tristan, A.; Bes, M.; Mougel, C.; Etienne, J.; Vandenesch, F.; Bonneville, M.; Lina, G. egc, a highly prevalent operon of enterotoxin gene, forms a putative nursery of superantigens in Staphylococcus aureus. J. Immunol. 2001, 166, 669–677. [Google Scholar] [CrossRef] [PubMed]
- Grumann, D.; Scharf, S.S.; Holtfreter, S.; Kohler, C.; Steil, L.; Engelmann, S.; Hecker, M.; Völker, U.; Bröker, B.M. Immune cell activation by enterotoxin gene cluster (egc)-encoded and non-egc superantigens from Staphylococcus aureus. J. Immunol. 2008, 181, 5054–5061. [Google Scholar] [CrossRef] [PubMed]
- Fraser, J.D.; Proft, T. The bacterial superantigen and superantigen-like proteins. Immunol Rev 2008, 225, 226–243. [Google Scholar] [CrossRef] [PubMed]
- Proft, T.; Fraser, J. Bacterial Superantigens. Clin. Exp. Immunol. 2003, 133, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Fraser, J.; Arcus, V.; Kong, P.; Baker, E.; Proft, T. Superantigens—powerful modifiers of the immune system. Mol Med Today 2000, 6, 125–132. [Google Scholar] [CrossRef]
- Fast, D.J.; Schlievert, P.M.; Nelson, R.D. Nonpurulent response to toxic shock syndrome toxin 1-producing Staphylococcus aureus. Relationship to toxin-stimulated production of tumor necrosis factor. J. Immunol. 1988, 140, 949–953. [Google Scholar] [PubMed]
- DeVries, A.S.; Lesher, L.; Schlievert, P.M.; Rogers, T.; Villaume, L.G.; Danila, R.; Lynfield, R. Staphylococcal toxic shock syndrome 2000–2006: Epidemiology, clinical features, and molecular characteristics. PLoS ONE 2011, 6, e22997. [Google Scholar] [CrossRef] [PubMed]
- Grumann, D.; Ruotsalainen, E.; Kolata, J.; Kuusela, P.; Jarvinen, A.; Kontinen, V.P.; Bröker, B.M.; Holtfreter, S. Characterization of infecting strains and superantigen-neutralizing antibodies in Staphylococcus aureus bacteremia. Clin. Vaccine Immunol. 2011, 18, 487–493. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.X.; Kasper, K.J.; Zeppa, J.J.; McCormick, J.K. Superantigens modulate bacterial density during Staphylococcus aureus nasal colonization. Toxins 2015, 7, 1821–1836. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.X.; Gilmore, K.J.; Szabo, P.A.; Zeppa, J.J.; Baroja, M.L.; Haeryfar, S.M.; McCormick, J.K. Superantigens subvert the neutrophil response to promote abscess formation and enhance Staphylococcus aureus survival in vivo. Infect. Immun. 2014, 82, 3588–3598. [Google Scholar] [CrossRef] [PubMed]
- Chau, T.A.; McCully, M.L.; Brintnell, W.; An, G.; Kasper, K.J.; Vines, E.D.; Kubes, P.; Haeryfar, S.M.; McCormick, J.K.; Cairns, E.; et al. Toll-like receptor 2 ligands on the staphylococcal cell wall downregulate superantigen-induced T cell activation and prevent toxic shock syndrome. Nat. Med. 2009, 15, 641–648. [Google Scholar] [CrossRef] [PubMed]
- Schreiner, J.; Kretschmer, D.; Klenk, J.; Otto, M.; Buhring, H.J.; Stevanovic, S.; Wang, J.M.; Beer-Hammer, S.; Peschel, A.; Autenrieth, S.E. Staphylococcus aureus phenol-soluble modulin peptides modulate dendritic cell functions and increase in vitro priming of regulatory T cells. J. Immunol. 2013, 190, 3417–3426. [Google Scholar] [CrossRef] [PubMed]
- Davies, M.M.; Chien, Y.-H. T-lymphocytes. In Fundamental Immunology, 7th ed.; Paul, W.E., Ed.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; pp. 279–305. [Google Scholar]
- Blattman, J.N.; Antia, R.; Sourdive, D.J.; Wang, X.; Kaech, S.M.; Murali-Krishna, K.; Altman, J.D.; Ahmed, R. Estimating the precursor frequency of naive antigen-specific CD8 T cells. J. Exp. Med. 2002, 195, 657–664. [Google Scholar] [CrossRef] [PubMed]
- Geiger, R.; Duhen, T.; Lanzavecchia, A.; Sallusto, F. Human naive and memory CD4+ T cell repertoires specific for naturally processed antigens analyzed using libraries of amplified T cells. J. Exp. Med. 2009, 206, 1525–1534. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.D.; van der Most, R.G.; Akondy, R.S.; Glidewell, J.T.; Albott, S.; Masopust, D.; Murali-Krishna, K.; Mahar, P.L.; Edupuganti, S.; Lalor, S.; et al. Human effector and memory CD8+ T cell responses to smallpox and yellow fever vaccines. Immunity 2008, 28, 710–722. [Google Scholar] [CrossRef] [PubMed]
- Trombetta, E.S.; Mellman, I. Cell biology of antigen processing in vitro and in vivo. Annu. Rev. Immunol. 2005, 23, 975–1028. [Google Scholar] [CrossRef] [PubMed]
- Crotty, S.; Kaech, S.M.; Schoenberger, S.P. Immunologic memory. In Fundamental Immunology, 7th ed.; Paul, W.E., Ed.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; pp. 741–764. [Google Scholar]
- Schmitt, N.; Ueno, H. Regulation of human helper T cell subset differentiation by cytokines. Curr. Opin. Immunol. 2015, 34, 130–136. [Google Scholar] [CrossRef] [PubMed]
- Stavnezer, J.; Guikema, J.E.; Schrader, C.E. Mechanism and regulation of class switch recombination. Annu. Rev. Immunol. 2008, 26, 261–292. [Google Scholar] [CrossRef] [PubMed]
- Kaech, S.M.; Cui, W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat. Rev. Immunol. 2012, 12, 749–761. [Google Scholar] [CrossRef] [PubMed]
- Hale, J.S.; Ahmed, R. Memory T follicular helper CD4 T cells. Front. Immunol. 2015, 6, 16. [Google Scholar] [CrossRef] [PubMed]
- Liuzzi, A.R.; McLaren, J.E.; Price, D.A.; Eberl, M. Early innate responses to pathogens: Pattern recognition by unconventional human T-cells. Curr. Opin. Immunol. 2015, 36, 31–37. [Google Scholar] [CrossRef] [PubMed]
- van Schaik, B.; Klarenbeek, P.; Doorenspleet, M.; van Kampen, A.; Moody, D.B.; de Vries, N.; Van Rhijn, I. Discovery of invariant T cells by next-generation sequencing of the human TCR alpha-chain repertoire. J. Immunol. 2014, 193, 5338–5344. [Google Scholar] [CrossRef] [PubMed]
- Eberl, G.; Colonna, M.; Di Santo, J.P.; McKenzie, A.N. Innate lymphoid cells. Innate lymphoid cells: A new paradigm in immunology. Science 2015, 348, aaa6566. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Williams, A.P. Role of Innate T Cells in Anti-Bacterial Immunity. Front. Immunol. 2015, 6, 302. [Google Scholar] [CrossRef] [PubMed]
- Bendelac, A.; Savage, P.B.; Teyton, L. The biology of NKT cells. Annu. Rev. Immunol. 2007, 25, 297–336. [Google Scholar] [CrossRef] [PubMed]
- Doisne, J.M.; Soulard, V.; Becourt, C.; Amniai, L.; Henrot, P.; Havenar-Daughton, C.; Blanchet, C.; Zitvogel, L.; Ryffel, B.; Cavaillon, J.M.; et al. Cutting edge: Crucial role of IL-1 and IL-23 in the innate IL-17 response of peripheral lymph node NK1.1- invariant NKT cells to bacteria. J. Immunol. 2011, 186, 662–666. [Google Scholar] [CrossRef] [PubMed]
- Napier, R.J.; Adams, E.J.; Gold, M.C.; Lewinsohn, D.M. The Role of Mucosal Associated Invariant T Cells in Antimicrobial Immunity. Front. Immunol. 2015, 6, 344. [Google Scholar] [CrossRef] [PubMed]
- Gold, M.C.; Cerri, S.; Smyk-Pearson, S.; Cansler, M.E.; Vogt, T.M.; Delepine, J.; Winata, E.; Swarbrick, G.M.; Chua, W.J.; Yu, Y.Y.; et al. Human mucosal associated invariant T cells detect bacterially infected cells. PLoS Biol 2010, 8, e1000407. [Google Scholar] [CrossRef] [PubMed]
- Hepworth, M.R.; Monticelli, L.A.; Fung, T.C.; Ziegler, C.G.; Grunberg, S.; Sinha, R.; Mantegazza, A.R.; Ma, H.L.; Crawford, A.; Angelosanto, J.M.; et al. Innate lymphoid cells regulate CD4+ T-cell responses to intestinal commensal bacteria. Nature 2013, 498, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Hazenberg, M.D.; Spits, H. Human innate lymphoid cells. Blood 2014, 124, 700–709. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bröker, B.M.; Mrochen, D.; Péton, V. The T Cell Response to Staphylococcus aureus. Pathogens 2016, 5, 31. https://doi.org/10.3390/pathogens5010031
Bröker BM, Mrochen D, Péton V. The T Cell Response to Staphylococcus aureus. Pathogens. 2016; 5(1):31. https://doi.org/10.3390/pathogens5010031
Chicago/Turabian StyleBröker, Barbara M., Daniel Mrochen, and Vincent Péton. 2016. "The T Cell Response to Staphylococcus aureus" Pathogens 5, no. 1: 31. https://doi.org/10.3390/pathogens5010031
APA StyleBröker, B. M., Mrochen, D., & Péton, V. (2016). The T Cell Response to Staphylococcus aureus. Pathogens, 5(1), 31. https://doi.org/10.3390/pathogens5010031