Abstract
The continuously improving sophistication of molecular engineering techniques gives access to novel classes of bio-therapeutics and new challenges for their production in full respect of the strengthening regulations. Among these biologic agents are DNA based vaccines or gene therapy products and to a lesser extent genetically engineered live vaccines or delivery vehicles. The use of antibiotic-based selection, frequently associated with genetic manipulation of microorganism is currently undergoing a profound metamorphosis with the implementation and diversification of alternative selection means. This short review will present examples of alternatives to antibiotic selection and their context of application to highlight their ineluctable invasion of the bio-therapeutic world.
1. Introduction
The discovery of antibiotics, considered for several decades a major step forward in the global domain of therapy against some infectious agents, is still a very active domain of investigation. Unfortunately, the excessive and sometimes inappropriate use of antibiotics is responsible for a large-scale spreading of antibiotics in the environment. The first consequence is the emergence of resistant or even multi-resistant pathogenic bacterial strains that has become a general concern promise to even further increase [1]. The improvement of existing antibiotics and discovery of new molecules is, by definition, an endless quest [2,3]. Besides therapeutic applications, antibiotics are often used as a selection pressure to avoid bio-contamination in production processes such as fermentation. In this particular context, the problem can show two distinct facets: The antibiotic molecule itself, seen as a contaminant product in a given biological and the antibiotic resistance gene used as a selection marker.
The increasing regulatory requirements to which biological agents are subjected will hopefully have a great impact in the fields of industrial protein expression and production, live vectors for bio-therapeutic agent delivery and gene therapy. There is an expectation that in the near future, the rule becomes “zero tolerance” towards antibiotic-based selection in production and delivery systems. Besides the antibiotic itself, the antibiotic resistance gene is a major subject of consideration. The complete absence of antibiotic-resistance genes being the only way to ensure that propagation in the environment or transfer of resistance to pathogenic strains will not happen. In order to address these issues, different and complementary approaches can be applied. The first would be to design more stable host/vector couples allowing to set-up and conduct fermentation processes in the complete absence of antibiotics. A more achieved strategy would be to substitute the antibiotic-based selection by an alternative mean such as the complementation of an essential gene product, not expressed by the host, poison antidote mean of selection or sophisticated post-segregational killing mechanism.
For specific therapeutic agents or fields of application such as DNA vaccination, passive immunization using antibody gene transfer [4] or gene therapy, the presence of an antibiotic resistant gene in the vector backbone is rightly pointed out as undesirable by health authorities. The major issue associated with DNA delivery or immunization being a possibility of horizontal transfer of antibiotic resistance to circulating microbial population. Such a consideration is emphasized by the very long persistence of DNA constructs upon injection, some published data indicates several months. Armengol et al. have used in situ hybridization and PCR to document persistence of a plasmid harboring the cry11Bb gene of Bacillus up to two years after injection in mouse muscle [5]. Other studies based on bio-distribution and safety of antibiotic-free plasmid encoding somatostatin genes delivered by attenuated Salmonella enterica serovar Choleraesuis revealed detection in several examined tissues with the exception of ovary and brain, but a rapid clearance from the body (approximately 10 days) [6]. The situation can be seen as worthwhile for long term and multiple delivery protocols currently used in the field of gene therapy. Finally, another domain of application must be included in the discussion, this domain being the use of live bacteria as delivery vehicles containing extra chromosomal elements that need to be stably maintained. Obviously, in that category, we will find commensal bacterial strains expressing plasmid borne foreign antigens.
Regulatory Concern
To date, the use of the β-lactam antibiotic family is not acceptable, in order to avoid concerns for patients showing reactivity to β-lactam antibiotics. Therefore, Kanamycin and to a lesser extent, Tetracyclin, are commonly used and still acceptable to health authorities and the development of new drugs is always more challenging [7]. Susceptibility to the antibiotic compound is the visible part of the iceberg, the most critical issue being the horizontal genetic transfer of antibiotic resistant genes to prokaryotic organisms, present in the environment or in commensal flora. It becomes clear that antibiotic-resistant bacteria are causing a global health crisis and that mechanisms behind acquisition of resistance are more and more documented [8].
Horizontal genetic transfer (HGT), an important driving force in bacterial evolution, is the passage of genetic elements between organisms [9,10]. HGT is influenced by the advantage conferred by the transferred gene, the toxicity of its product, the capacity of the transferred gene to be integrated into the host genome and to be stabilized, and a certain compatibility of codon usage between the transferred gene and the host. The probability of integration and stabilization of a HGT into a new host genome is correlated and increased by the similarity between the tRNA pools of the donor and recipient organism. Acquisition of antibiotic resistance is one example of this evolution by HGT. As early as in 1965, Datta and Kontomichalou [11] showed the widespread transfer of penicillin resistance across Enterobacteriaceae. A marked β-lactamase activity was also measured in various Pseudomonas aeruginosa resistant strains in the early seventies [12]. More recently, acquisition of the virulence factors that distinguish Salmonella from Escherichia coli has been clearly shown as the result of horizontal gene transfer [13]. Recent technological breakthroughs, such as the one that can be observed for next generation sequencing, offers unthinkable and extensive possibilities of detecting numerous HGT events, especially if the data acquisition is completed by new bioinformatics tools like nucleotide substitution rate matrixes [14]. Moreover, if we consider the situation at the microbiome level, it becomes obvious that a multitude of different bacterial species, exhibiting many types of complex interactions, are likely to use HGT as a preponderant mechanism for adaptation in difficult environment [15].
For the above mentioned reasons, health agencies and global organizations (FDA, EMEA, WHO) are all going in the same direction concerning recommendations towards the limitation in use of antibiotics and propagation of antibiotics resistance genes [16]. The principal rules to be applied can be summarized as follows:
- -
- It is strongly advised to avoid or minimize the use of any kind of antibiotics in cell or bacterial culture.
- -
- If antibiotics are nevertheless used, it is mandatory to minimize their amount and to monitor the presence of traces in the final product.
- -
- The rationale for the use of antibiotics must be clearly documented in the Common Technical Document (CTD).
- -
- Penicillin, and more generally β-lactams and streptomycin, must not be used in reason of potential concerns with hyper reactivity of some patients to antibiotics of the β-lactam family
- -
- Kanamycin and neomycin are the preferred choice and still tolerated.
- -
- The use of antibiotic resistance markers is generally discouraged, and if used, the in vivo outcome and effect needs to be evaluated.
There are specific mentions for the nature of the gene encoding resistance to kanamycin, as reviewed by Williams et al. [7] The gene neomycin phosphotransferase III [npt-III, aph(3')-III] should be avoided, since it also confers resistance to amikacin [17], a reserve antibiotic (EMEA, 2008).
As a final comment, it is easy to anticipate what the future requirements from health authorities might be: Constructs have to be completely devoid of antibiotic resistance genes in their final structure, even if in use at early stages of construction. Alternative solutions would be available and validated soon.
2. Vector Stabilization
Besides any consideration about pressure of selection obtained through antibiotic or antibiotic-free selection, natural stability of a given plasmid might be an additional source of investigation.
Several studies on natural plasmids, conducted in the 1970s or early 1980s, have highlighted that some plasmids naturally display regions necessary for their stability.
Plasmid F, which exists only as one to two copies per chromosome, is stably inherited to daughter cells during cell growth, contains stabilization sequences. Ogura; Hiraga [18], have shown that these sequences were independent from plasmid replication function.
Their work lead to the identification of three essential regions for plasmid maintenance:
SopA and SopB acting in trans and SopC acting in cis in order to stabilize the plasmid likely through interaction with, unidentified at that time, cellular components.
These authors also put in evidence that the combination SopA, SopB and SopC was not sufficient for a complete stabilization of mini-F plasmid, and thus identified the additional element ccd (control of cell death) region that seemed crucial to control cell division when copy number carrying ccd segment decreases [18]. The so-called ccd region can be split into two functional domains: ccdB, the product of which inhibits the host cell division and ccdA, able to act as an antagonist to the ccdB function. Two years after, Jaffé et al. [19], demonstrated that cell division is not immediately inhibited and that residual division could take place in the plasmid free-cells before finally being inhibited. Authors concluded that the ccd region guarantees that plasmid carrying cells could grow preferentially in a population by killing plasmid free daughter cells, introducing the concept of post-segregational killing. Plasmid R1 has also been shown to contain a stabilization system [20]. As for plasmid F, the stabilization system is based on post segregational killing due to the parAB + locus. This locus is composed of two genes Hok (Host killing) and Sok (suppression of killing). The translation of the Hok messenger, encoding a toxin lethal to the bacteria, is completely blocked by the anti-messenger Sok. In the absence of plasmid, Sok, which is less stable than Hok, is lost first, allowing the translation of the Hok mRNA and expression of the toxin lethal to the cell. Several hok mRNA paralogs have been identified in the genome of E. coli, and Hok protein orthologs are also found in the genomes of Enterobacteria [21,22,23].
Concerning plasmid maintenance, it has been shown that factors reducing multimerization of plasmid could increase plasmid stability [24]. ColE1 plasmid contains a region, cer that seems to be necessary for a recombination event converting multimers to monomers, allowing the plasmid to be more stable. Multimer resolution is achieved through action of the XerCD site-specific recombinase at the cer site. Precise characterization of the cer site by progressive deletions has shown the importance of its boundaries in terms of length and the apparent poorly conserved sequence among multimer resolution sites [25]. In recent work, Rcd was even more precisely described, a small regulatory RNA encoded within cer, which is also required. Rcd is transcribed from the Pcer promoter in plasmid dimers, but not in monomers, and is a key component of the checkpoint that delays the division of dimeror multimer-containing cells [26]. It was further demonstrated that a fifth chromosome-encoded protein, FIS, binds to cer in a sequence-specific manner. FIS appears to have little or no influence on cer-mediated recombination, but, along with XerC and XerD, is required for proper regulation of the Pcer promoter [27]. Cloning of the cer locus into various expression vectors has been extensively documented and the proof of principle largely established in high-cell density cultures [28].
Multimer Resolution
The multimer resolution rule applying for plasmids has some equivalent in chromosome duplication during cell division. In Prokaryotic microorganisms, cell division can be completed only after segregation of newly replicated monomeric chromosomes. The presence of dimeric chromosomes, often due to homologous recombination between replicated chromosomes must be precisely controlled and resolved before correct completion of the cell division.
In Escherichia coli, dimeric chromosome segregation has been shown to be mediated by a site-specific recombination between two small sequence elements termed dif sites (deletion-induced filamentation). Functional dif site was identified as contained into a DNA fragment of 32 bp which is sufficient to allow Xer-mediated plasmid multimerization and dimer resolution. A core recombination site containing two 11 bp half-sites, flanking a 6 bp central region is the current definition of the dif site, and dimer resolution is achieved through action of the XerCD site-specific recombinase at the dif site in a similar way as cer.
The structural organization of dif is similar to that of related lambda integrase family site-specific recombination loci, e.g., P1 loxP, and Saccharomyces cerevisiae FRT.
In the context of DNA vectorization for genetic immunization or gene therapy the productivity is a major consideration and it seems useful to consider the induced metabolic stress and plasmid stability during production. Silva et al. have suggested possible evolution of fermentation methods towards online bioprocess monitoring to circumvent metabolic burden and plasmid instability [29]. Process optimization is a way to improve productivity, nevertheless the nature and design of the construct and recent innovations in strain and vector engineering or alterations of the host genome or vector backbone are more likely to dramatically increase production of plasmid DNA [30].
An ultimate evolution or simplification can be provided by so-called minicircle vectors that are genetic constructs devoid of plasmid bacterial sequences. The intrinsic small size of minicircles, combines safety and improved resistance to the shearing forces associated delivery methods using pneumatic devices [31].
3. Antibiotic-Free Selection and Plasmid Maintenance
To fulfill new regulatory constraints, to develop marker-less expression hosts or to produce safer gene delivery vectors, antibiotic-free selection systems and plasmid addiction mechanisms have been developed.
All these systems can be classified according to their mechanism of action. The different types of systems, as described in Table 1, are based on (i) complementation of auxotrophic marker, (ii) post-segregational killing (PSK), (iii) RNA-interference, (iv) de-repression of an essential gene, (v) chromosomal integration, (vi) minicircles and (vii) non antibiotic resistance.

Table 1.
Antibiotic-free selection systems: Mechanisms of action; Applications. Abbreviations used in the table: R: Resistance; Tet: Tetracycline; Cm: Chloramphenicol; Amp: Ampicillin; Kan: Kanamycin resistance; bla: Beta-lactamase; Neo: Neomycin; Rif: Rifampicin.
| Strain | Strain Modification | Plasmid Maintenance Gene | Antibiotic Resistance Gene Used for Selection | Application | Reference |
|---|---|---|---|---|---|
| Complementation of an essential gene | |||||
| Escherichia coli | dapD mutation → DAP auxotrohpy | dapD | No | DNA and protein expression | Degryse, [32] |
| Salmonella typhimurium | Δasd → DAP auxotrophy | asd | No | Live vaccine | Galan et al. [33] |
| Salmonella typhi | Δasd → DAP auxotrophy | asd | No | Vaccin delivery | Tacket et al. [34] |
| Salmonella typhimurium and S.typhi | thyA mutation → Thymidine auxotrophy | ThyA | No | Live vaccine | Morona et al. [35] |
| Vibrio cholerae | glnA with an internal deletion → Glutamine auxotrophy | glnA | No but presence of TetR and CmR genes on plasmid | Vaccine delivery | Ryan et al. [36] |
| Salmonella enterica serovar Choleraesuis | Δasd → DAP auxotrophy | asd | No | Live vaccine expressing somatostatin | Liang, et al. [6] |
| Escherichia coli | ΔProBA → Proline auxotrophy | proBA | Yes, CmR | protein expression | Fiedler et al. [37] |
| Escherichia coli | ΔTpiA →no growth on glycerol | TpiA | No but presence of AmpR and KanaR genes on plasmid | protein expression | Velur Selvamani, et al. [38] |
| Escherichia coli | ArgE amber mutation + pir116 → arginine auxotrophy | Phe sup tRNA + ori γ | No | DNA (pCOR) | Soubrier et al. [39] |
| Escherichia coli | ThyA amber mutation → thymidine auxotrophy | His sup tRNA | No | DNA (pFAR) | Marie, et al. [40] |
| Salmonella enteritidis | Δasd → DAP auxotrophy | asd | No | Ghost vaccine, live vaccine | Jawale lee, [41] |
| Escherichia coli | ΔglyA → Glycine auxotrohpy | glyA | protein expression | Vidal et al. [42] | |
| Escherichia coli | ΔQAPRTase → NAD auxotrophy | QPARTase | protein expression | Dong, et al. [43] | |
| Bacillus amyloliquefaciens | ΔgspA → G3P auxotrophy | glycerine-3-phosphate dehydrogenase | No | protein expression | US2010/0248306A1 |
| Escherichia coli | ΔaraD | araD | No | protein expression | US2007/0036822 |
| Post Segregational Killing | |||||
| Escherichia coli | - | hok/sok (parB locus) | Yes, bla | DNA and protein expression | Gerdes et al. [44] |
| Escherichia coli | - | hok/sok, parDE | Yes, KanR, AmpR | DNA and protein expression | Pecota et al. [45] |
| Escherichia coli | - | hok/sok (parB locus) | DNA and protein expression | Schweder et al. [46] | |
| Salmonella typhi | - | hok/sok and parA | Yes, bla | Live vaccine | Galen et al. [47] |
| \Escherichia coli | - | ccdA/ccdB | Yes, AmpR | protein expression | Wegerer, et al. [48] |
| Escherichia coli | ccdB gene insertion | ccdA | No but AmpR or spectinomycinR gene present on plasmid | DNA and protein expression | Szpirer et al. [49]. |
| Streptomyces lividans | YoeBsl gene | YefMsl gene | No, but NeoR gene present on plasmid | protein production | Sevilliano et al. [50] |
| Chinese Ovary Hamster cell line (CHO) | Kid | Kis | No but G418R gene present on plasmid | protein production | Nehlsen, et al. [51] |
| Repression of toxic gene | |||||
| Escherichia coli | cI-sacB | cI repressor | No but CmR gene present on plasmid | DNA production | WO2010/135742 |
| Non-antibiotic resistance | |||||
| Pseudomonas putida | Rifr Cmr hsdR1, prototrophic | tellurite resistance gene | RifR and CmR strain but no resistance on plasmid | Bioconversion of toluene | Sanchez-romero [52] |
| Escherichia coli | - | Fabl | no | DNA and protein production | Goh and Good [53] |
| Pseudomonas putida/Klebsiella pneumoniae | - | Biophalos (herbicide), mercury or arsenic resistance gene | No but KanR gene present on plasmid | Strain engineering | Herrero et al. [54] |
| RNA-based drugless systems | |||||
| Escherichia coli | murA under control of TetR (including a RNAI complementary seq.) | RNAi (ColE1 ori) | no | DNA or protein production | Pfaffenzeller et al. [55] |
| Escherichia coli | murA under control of TetR (including a RNAI complementary seq.) | RNAi (ColE1 ori) | no | DNA production | Mairhofer et al. [56] |
| Escherichia coli | RNA-IN sacB | RNA-OUT | no | DNA production | Luke, et al. [57] |
| Operator Repressor Titration systems | |||||
| Escherichia coli | lac-DapD | lacO | no | DNA or protein production | Nuttall, et al. [58], Cranenburgh, et al. [59] |
| Salmonella enterica serovar Typhimurium | lac-DapD | lacO | no | Live vaccine expressing Yersinia pestis F1 antigen | Garmory, et al. [60] |
| Chromosomal integration (plasmid free) | |||||
| Bacillus subtilis | Coat gene in fusion with heterologous peptide | No plasmid | - | Protein display on spores | Iwanicki et al. [61] |
| Escherichia coli | heterologous gene to be expressed | No plasmid | - | protein expression | Striedner, et al. [62] |
| Escherichia coli | GDP-l-fucose de novo pathway genes and H. pylori futC | No plasmid | - | protein production by metabolism modification | Baumgartner et al. [64] |
| Minicircles | |||||
| Escherichia coli | - | Amp resistance gene in producer plasmid | Yes, AmpR | DNA production for gene therapy | Chen et al. [65] |
| Escherichia coli | - | Amp resistance gene in producer plasmid | Yes, AmpR | DNA production for gene therapy | Chang, et al. [66] |
| Escherichia coli | - | Amp resistance gene in producer plasmid | Yes, AmpR | DNA production for gene therapy | Huang, et al. [67] |
| Escherichia coli | ΔendA + PhiC31 integrase gene multiple insertion | Kan resistance gene in producer plasmid | Yes, KanR | DNA production for gene therapy | Kay, et al. [68] |
| Escherichia coli | ΔendA + PhiC31 integrase gene multiple insertion | Kan resistance gene in producer plasmid | Yes, KanR | DNA production for gene therapy | Yi, et al. [69] |
| Escherichia coli | - | Amp resistance gene in producer plasmid | Yes, AmpR | DNA production for gene therapy | Zhang, et al. [70] |
The first systems investigated were based on the complementation of auxotrophic markers. Among these the one established by Degryse et al. [32] uses an E.coli strain mutated in the DAP (Diaminopimelic acid) pathway due to a mutation or deletion in DapD gene. The plasmid carries DapD gene to complement the stain DAP auxotrophy. As a consequence, only bacteria containing the plasmid will grow when cultivated without DAP supplementation in the culture medium (Scheme 1A). As DAP is implicated in cell wall synthesis and mutation in the DAP pathway are lethal, several systems based on DAP complementation were investigated in other gram negative bacteria, especially Salmonella strains. Avirulent strains of Salmonella with a deletion of asd gene were developed to be used as live vaccines expressing a recombinant protein using a plasmid maintained with the asd complementation. Tacket et al. [34] constructed a S.typhi vaccine strain expressing the core protein of Hepatitis B virus. More recently, Liang, et al. [6], adopted the same strategy to express somatostatin in a S.enterica serovar Choleraesius. in a particular application of the same complementation, an antibiotic-free system to generate “ghost” Salmonella enteritidis bacteria as a vaccine. To do so, they express PhiX174 lysis gene E in Salmonella enteritidis. By using another auxotrophic marker in combination with DAP auxotrophy, Wang, et al. [71] propose a recombinant attenuated salmonella vaccine with dual plasmid delivery system.
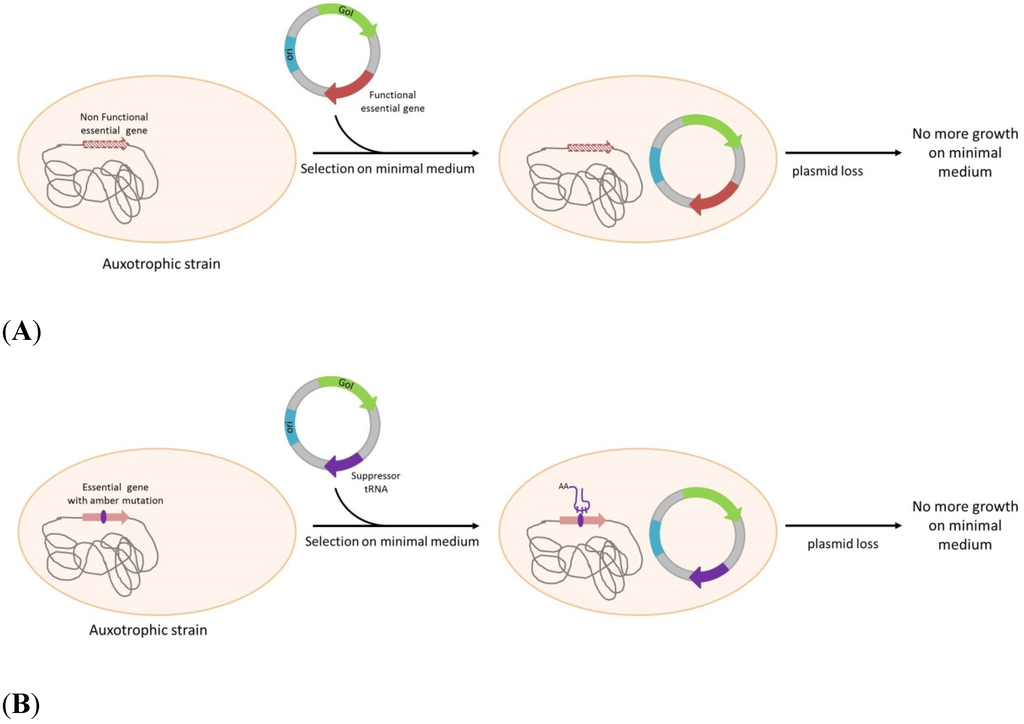
Scheme 1.
Plasmid selection and maintenance by auxotrophy complementation. (A): Strain auxotrophy is due to an essential gene mutation or deletion. The auxotrophy is complemented by the plasmid-born expression of the functional gene. (B): Strain auxotrophy is due to an amber mutation in an essential gene. Expression of the suppressor tRNA correcting amber mutation can complement the auxotrophy. GoI: Gene of Interest; ori: Origin of replication.
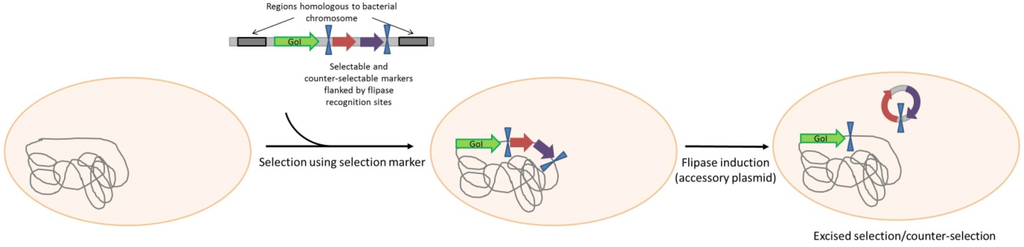
The ORT® technology (Scheme 2) uses also DAP as a central element, but in this case the regulation is based on a plasmid-mediated repressor titration driving the expression of the chromosomal dapD gene. The engineered host strain contains dapD under control of the lac operator/promoter (lacO/P). In the absence of IPTG as inducer, cell growth is blocked by dapD repression. The presence, in the host, of a high copy number plasmid containing the lac operator (lacO) releases the repression of dapD expression through titration of LacI from the operator [59]. In addition to plasmid DNA production, the ORT® system has been adapted for vaccine delivery using Salmonella enterica live bacterial vectors [72]. The system has been completed by a very elegant mean of excision of the selectable marker used for the vector cloning steps and strain engineering. The antibiotic resistance gene is flanked by dif sites that are the recognition sequence for the native Xer recombinases responsible for chromosome dimer resolution. Upon transformation and chromosomal insertion, the recombinases resolve the two directly repeated dif sites to a single site, excising the antibiotic resistance gene [73].
DAP auxotrophy and more generally the use of auxotrophic markers should have some influence on the overall productivity of a given host strain. It is nevertheless difficult to evaluate the real cost of compensating for auxotrophy [74]. The analysis would have to include the impact on specific amino acid biosynthesis pathways, the composition of the media and the additional metabolic burden associated with the expression of the plasmid-borne essential gene.
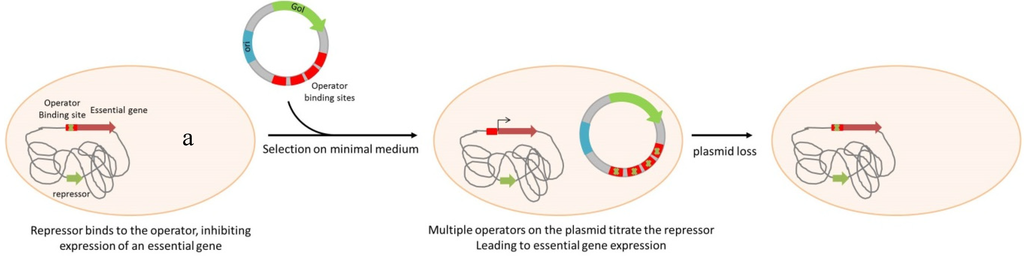
Scheme 2.
Plasmid selection and maintenance by Operator Repressor Titration. Note of a: An essential gene is repressed by a genome encoded repressor. Plasmid containing several sequences of the operator can titrate this repressor by competition, allowing essential gene expression. GoI: Gene of Interest; ori: Origin of replication.
The most recently developed complementation system is based on tpiA auxotrophy in Escherichia coli [38]). Authors use a strain ΔTpiA, which presents a low growth rate with glucose as carbon source and no growth with glycerol. Only the strain carrying the plasmid expressing tpiA gene can restore normal growth behavior to a medium with glycerol as sole carbon source. Even if the plasmid used in the study still carries ampicillin and kanamycin resistance genes, authors show that the TpiA auxotrophy complementation is efficient enough to allow bacterial selection, plasmid stability and recombinant protein production in a 2L-batch fermentation process.
More elegant systems express a tRNA suppressor to restore an amber mutation in an essential gene. The pFAR plasmids (Marie, et al. [40]) allows the expression of histidine suppressor tRNA to suppress an amber mutation in the thyA gene. Only bacteria carrying a plasmid can grow in absence of thymidine (Scheme 1B). Plasmids called pCOR [39] use the same mechanism with a phenylalanine suppressor tRNA that suppress an amber mutation in ArgE. The plasmid maintenance of pCOR is improved by a conditional replication origin on the plasmid that needs pir116, added on the E.coli chromosome. Both plasmid and chromosome are in a mutually dependent, leading to a safer DNA vector that is unable to replicate in environmental bacterial strains [39].
An improved version of the system, with increased copy number, was obtained through genetic engineering in Escherichia coli. The bacterial gene encoding the pi initiator-protein, which plays a pivotal role in pCOR replication, was mutagenized to identify novel copy-up mutations. A particular combination of copy-up mutations was shown to produce a 3-5-fold increase in plasmid DNA per biomass unit [75].
These two systems have been mainly used for DNA vector preparation, and their applications in pre-clinical and clinical studies have been reviewed by Vandermeulen, et al. [76].
The second kind of system is based on a mechanism initially discovered by studying natural plasmids, called PSK. PSK consist of a plasmid locus that encodes a toxin and its own antidote, or antitoxin. In most cases, the toxin expressed is generally more stable than the antitoxin, the bacteria losing the plasmid after segregation will therefore be killed. Among the numerous toxin/antitoxin identified and recently referenced into the TADB database [77], two have been investigated for applications in biotechnology. the parB locus, also identified as hok/sok, is used for plasmid maintenance since 1986 [44] and have also been combined with parDE [45]. Hok is a messenger encoding a toxic protein, inhibited in its expression by the Sok anti-messenger. In case of plasmid loss, Sok due to shorter half-life disappears first and allow translation of the Hok protein inducing cell death (Scheme 3A). Combination of parB and parA, also showed a real improvement in plasmid maintenance [78], for a live vaccine application.
The ccdB/ccdA toxin/antitoxin pair is also used by several authors [48,79], by cloning the entire locus on the plasmid to allow its stabilization in E.coli. If this system is efficient for maintenance, it cannot allow a real antibiotic-free selection. For this reason, Szpirer et al. developed a “separate component” system where toxin is carried by the chromosome and the antidote is brought by the plasmid [49] (Scheme3B). This system, using the same genes, allows for an efficient selection of transformed bacteria. To further improve this, Peubez, et al. [28] proposed a way to completely eliminate the antibiotic resistant gene necessary for the construction of the vector, with a positive selection. In their strategy, the antibiotic resistant gene is flanked by both 5' and 3' ends of the ccdA gene containing a small homologous region. Upon transformation the two ccdA moieties associate through homologous recombination and give a fully functional gene, ensuring complete elimination of the antibiotic resistance gene (patent WO2010007246).
Toxin/antitoxin pairs that play an important role in plasmid maintenance and evolution in the bacterial world are numerous, and still need to be explored for future biotechnological applications [80], adjustable killing activity can even be transferred to higher eukaryotic systems [81].
Interestingly, separated components toxin/antitoxin stabilization systems have been developed for protein production in Streptomces lividans, using YoeBsl gene on the chromosome and YefMsl gene on the plasmid [50].
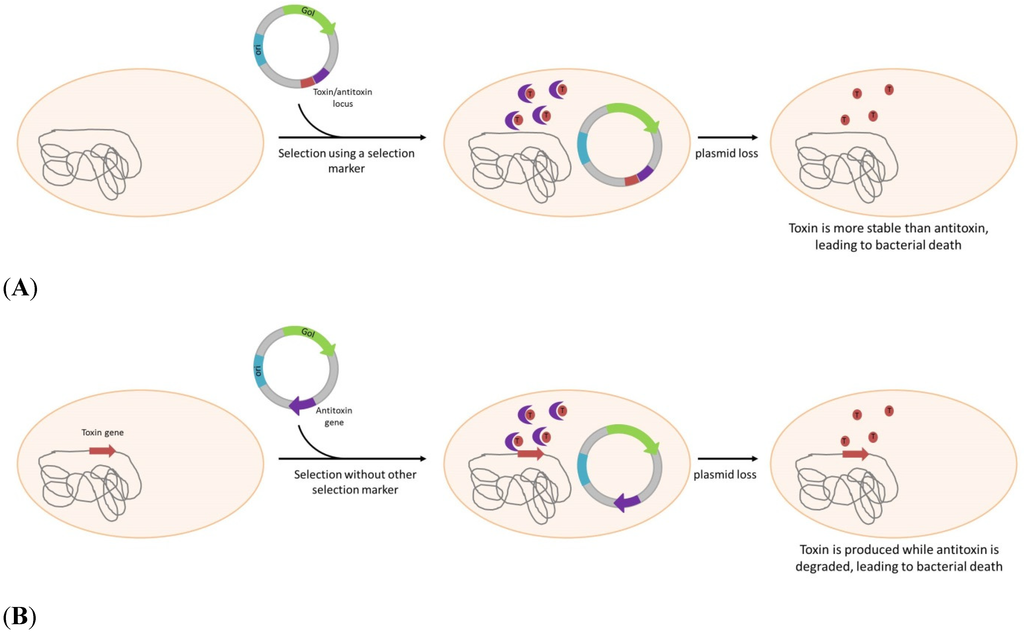
Scheme 3.
Plasmid selection or/and maintenance by post-segregational killing. (A): Both toxin and antitoxin genes are on the plasmid allowing plasmid maintenance. Another marker (i.e., antibiotic resistant gene) is still needed to allow strain selection. (B): As the toxin gene is inserted into bacterial genome and antitoxin is plasmid-born, this system allows both selection and plasmid maintenance in absence of any antibiotic. GoI: Gene of Interest; ori: Origin of replication.
An alternative and novel plasmid addiction model, based on ColE1 derived RNAI properties was suggested by Pfaffenzeller et al. [55]. The replication inhibitor RNAI of ColE1 based plasmid can serve as an antisense to bind mRNA of an engineered toxic gene, encoded by the bacterial chromosome. This system was proposed in a fine-tuned version by using the same “selection marker” serving as an antisense to alleviate the repression of an essential gene. The essential gene murA, implicated in cell wall synthesis, is under control of the Tet operator, repressed by the Tet repressor that integrate a complementary sequence of RNAI. In absence of plasmid, murA expression is repressed by TetR, leading to cell death (Scheme 4A). In bacteria carrying a replicating ColE1 based plasmid, RNAI interfere with its complementary sequence, repressing TetR, resulting in de-repression, expression of murA and cell growth [56]. By integrating other RNA/AS-RNA sequences, a system not restricted to ColE1-derived plasmids, was developed. In this system, Vectors express a RNA-OUT antisense RNA. RNA-OUT represses the expression of a chromosomally integrated, constitutively expressed counter-selectable marker (sacB). The expression of sacB, derived from Bacillus subtilis, induce a susceptibility to sucrose. In a culture medium containing sucrose, bacteria that lack the RNA-out plasmid will not keep growing (Scheme 4B) (Luke, et al. [57,82]).
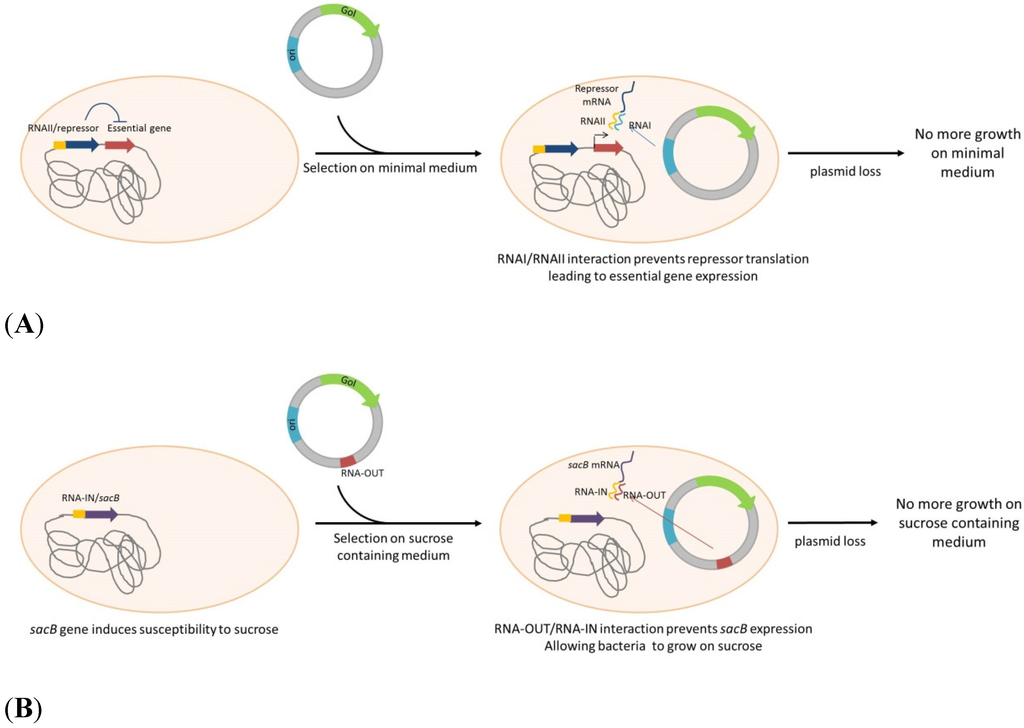
Scheme 4.
Plasmid selection and maintenance by RNA/RNA interaction. (A): An essential gene is under control of a repressor fused to RNAI homologous sequence. RNAI, produced from plasmid ori (ColE1 derived), binds to this homologous RNA, inhibits the repressor translation and restores the essential gene expression. (B): sacB gene is inserted into the genome, fused to RNA-IN sequence. RNA-OUT, encoded by the plasmid, binds to RNA-IN, inhibiting sacB translation, restoring ability of the strain to grow on sucrose containing medium. GoI: Gene of Interest; ori: Origin of replication.
The counterselectable marker sacB is also described in a patented but unpublished system (patent WO2010/135742). In this system, sacB is inserted into E. coli chromosome, under control of the λ Pr promoter. Plasmids expressing the λ cI 857, temperature sensitive repressor lead to the controlled inhibition of sacB which allows bacteria growth in presence of sucrose. The major advantage of this system is the reduction of vector size and plasmid produced can be used for protein expression as well as for DNA vector production for gene therapy.
To produce a recombinant protein, a way to avoid the antibiotic resistance marker is to integrate the gene of interest into bacterial chromosome [83]. With the development of seamless genomic integration by lambda red integrase [84], this kind of system is more and more accessible to easy establishment but cannot be considered for plasmid-based applications.
Striedner et al. [62] developed a plasmid-free, T7-based E. coli expression system in which the target gene is stably integrated into the chromosome of the host at a specific site (Scheme 5). The system, validated in real fermentation conditions, might suffer from some weaknesses due to relatively complex cloning procedure and it is also, important to consider that single copy integration might be a disadvantage for protein over-expression. One possibility to overcome the single-copy drawback might be to take advantage of a position effect, Bryant et al. [85] have shown that expression of gfp may vary by around 300-fold depending on its precise position on the chromosome of E. coli K12.
In contrast, chromosome integration is a technology of choice for some applications, such as the one described by Baumgartner et al. [64]. Using a chromosomal integration of recombinant genes and altering the copy number and expression of 2'-FL and intracellular GDP-L-fucose levels, the authors were able to construct and improve the first selection marker-free E. coli strain producing 2'-FL without the use of expression plasmids.
Another example is provided by Lemuth et al. [63], using a plasmid-free Escherichia coli strain to produce the xanthophyll astaxanthin, a high-value compound with applications in the nutraceutical, cosmetic, food, and animal feed industries.
Scheme 5.
Plasmid-free expression after genomic integration. The gene of interest (GoI) is inserted into the genome to avoid antibiotic resistance gene in the production strain.
It is also important to consider that single copy integration might be a disadvantage for protein over-expression.
For plasmid DNA production, development of minicircle is more and more considered as an obviously interesting option. Minicircle consists of a circular DNA sequence devoid of bacterial backbone. The major advantage of these vectors is their safety due to absence of antibiotic resistance gene, bacterial replication origin and un-methylated immunogenic CpG sequence. To produce minicircle, a producer plasmid has first to be constructed. This plasmid contains a transgene expression cassette flanked by integrase or recombinase recognition sequences and all the bacterial sequences needed for selection and amplification. To propagate the producer plasmid, antibiotic selection is still used but the bioprocess is generally conducted in absence of any antibiotic to avoid minute amounts of antibiotics in the final product and to lower the process cost. After amplification of the producer plasmid, recombination at specific recognition sites lead to the production of minicircles and plasmid backbone (Scheme 6). Minicircles are more difficult to produce than conventional plasmids due to the recombination and purification steps. Minicircle generation can be done using lambda integrase [86], Cre recombinase [87] or ΦC31 integrase [65,68]. In the first production system using ΦC31 integrase, the produced plasmid contained a gene encoding homing endonuclease I-SceI and two copies of the gene encoding ΦC31 integrase and an I-SceI recognition site. ΦC31 integrase expression allows recombination of producer plasmid into minicircles and backbone plasmids. I-SceI, located in the backbone plasmid after recombination, allows the cleavage and the degradation of this plasmid. To optimize the minicircle production process, multiple copies of I-SceI and ΦC31 integrase have been integrated into the bacterial chromosome. This strategy allows for the improvement of production yields by increasing recombination efficiency and reducing contaminant backbone plasmid, facilitating minicircles’ purification.
The development of a totally antibiotic-free minicircle generation process could be considered by implementing a producer plasmid selection system using auxotrophy complementation.
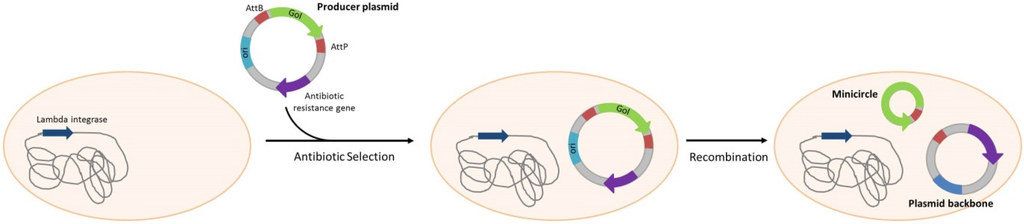
Scheme 6.
Minicircle production. The producer plasmid is selected using an antibiotic resistance gene. Cleavage and recombination leads to the formation of circular plasmid backbones and minicircles. Minicircles do not contain any antibiotic resistance gene. Strategies of in vivo degradation of plasmid backbone are developed to facilitate minicircle purification.
Another way to avoid the antibiotic resistance is to use resistance to other chemicals. Based on this strategy, resistance to Triclosan can be provided by the expression of the plasmid-born Fabl gene Goh and Good [53]. Even if the chemical used is not considered as an antibiotic, this kind of antibiotic-free system presents the same limitations than antibiotic-based systems and same regulatory issues.
The toxicity of triclosan was evaluated in a rat model through exposure to aerosol inhalation [88] and its potential effect on DNA damage and reproduction was also demonstrated [89]. The results showed that the reproduction of earthworms were significantly reduced (p < 0.05) after exposure to triclosan concentrations ranges from 50 to 300 mg·kg−1. Finally, triclosan, widely used as a biocide, was also shown to induce resistance in Aeromonas salmonicida, a pathogen bacteria for fish [90].
Among all the systems classified in Table 1, some of them can fit two classes. For example, pFAR and pCOR plasmids, based on auxotrophic marker are categorized in Table 1 as “complementation of auxotrophic markers” but could also belong to RNA-based systems as they use t-RNA suppressor. To better understand the mechanism of action of all the antibiotic-free selection and maintenance systems, reviews including schematic have been recently published [28,76,91,92].
4. Gene Therapy; DNA Immunization
For different reasons, including efficiency, easier access to proof of concept and validation in the target species, DNA immunization has proven to be more successful in the domain of animal health. Various DNA vaccines for animal health have been approved.
The Fort Dodge’s West Nile DNA vaccine was approved in 2005 by the US department of agriculture. This plasmid DNA-based vaccine contains gene sequences coding WNV surface antigens prM and E within a vector background pCBWN containing an ampicillin resistance gene. The Novartis licensed DNA vaccine Apex-IHN was approved in 2005 for use in salmon in Canada to prevent from infection of hematopoetic necrosis virus (HNV). The plasmid vector expressing viral glycoprotein of HNV contains an ampicillin resistance gene. A canine melanoma vaccine (Merial, Duluth, GA, USA) was approved in the US in 2007 for the treatment of canine malignant melanoma. This vector background encodes human xenogenoic tyrosinase and contains a kanamycin resistance gene. LifeTide® SW 5 is the first DNA therapy for food animals approved in Australia in 2008 (VGX animal Health, TX, USA). A swine injectable plasmid encoding porcine Growth Hormone Release Hormone carrying a kanamycin resistance gene has been approved in sows of breeding age to increase the number of piglets weaned.
Although these animal DNA vaccines are still based on antibiotic marker selection, very recently the first antibiotic free produced vaccine was approved by USDA in December 2013. The Merial’s Porcine Rotavirus vaccine based on expression of NSP4, VP4 and VP6 rotavirus antigens that prevents pigs from gastroenteritis (US 2013/0209507A1). This antibiotic free selection system uses the stabilization system StabyExpress based on the use of toxin-antitoxin genes [28,93] and anticipates future regulatory requirements.
4.1. DNA Vaccines for Human Application
For human applications, 365 clinical assays were initiated or completed using naked or plasmid DNA [94]. Approaches to develop antibiotic-free plasmid DNA are recommended by regulatory agencies (FDA, USDA, EMEA) for more than 15 years in order to reduce risk of allergic reactions and decrease risk of selection of antibiotic resistant pathogenic bacteria.
Although kanamycin selection marker remains allowed for gene therapy clinical trials [95,96] the first steps to increase biosafety have been performed using antibiotic-free plasmids in clinical models [76]. However, evaluation of a DNA dengue-1 vaccine in a phase I trial by an naval medical research center was still based on a VR1012 plasmid backbone containing kanamycin resistance [97].
First human phase I and II clinical trials using plasmid pCOR encoding fibroblast growth factor 1 have suggested an improvement of perfusion and an increase in amputation free survival in patients with critical limb ischemia [98]. This pCOR plasmid is an antibiotic free selection DNA vector based on complementation of amber mutation tRNA suppressor [39]. The pORT vector was chosen to develop a HIV-1 DNA vaccine, and its delivery induced a specific immune response in patients [99]. Moreover, proof of concept of gene therapy using antibiotic-free pORT plasmid AMEP coding for the recombinant desintergrin domain of ADAM 15 in disseminated melanoma has demonstrated safety and efficacy results of a phase I first-in-man study (European Society for Medical Oncology Meeting in 2012). Approval for clinical trial Phase I/II was obtained by regulatory authorities. This pORT plasmid strategy is based on “operator repressor titration” (see Table 1) [100].
A possible breakthrough technology that was not so far considered as sufficiently come to maturity, is the use of RNA-based nucleic acid vaccination [101,102]. After long and disappointing results, this technique recently entered a new era with the advent of self-replicating RNA viruses [103]. This platform has successfully entered clinical trials after encouraging pre-clinical evaluation.
4.2. Use of Viral Vectors
Since no DNA based vaccines for human applications received FDA approval today there is renewed interest on viral vector based vaccines. Although viral vectors have the same safety concern than nucleic DNA concerning integration capacity, they have the advantage not to contain selection markers. In animal health, ten viral vectors based vaccines are already on the market. Most of them are based on recombinant poxvirus as avian fowlpox, attenuated canarypox (Alvac), and vaccinia. The leader of the viral vector based vaccines is Merial with eight approvals based on recombinant Alvac vector, fowlpox and vaccinia. Avimex and Biomune developed fowlpox virus based and Newcastle disease virus based viral vectors respectively.
For human health, in the past years two products, Advexin (p53 based treatment of squamous cell carcinoma of the head and neck) and Cerepro (use of thymidine kinase for malignant glioma) failed the FDA and EMA approval, respectively. However, Amsterdam-based UniQure’s Glybera (alipogene tiparvovec) was approved for the treatment of Lipoprotein Lipase Deficiency (LPLD) by the European commission on September 2012. Glybera is a replication-deficient, adenovirus associated virus type 1 (AAV1) vector, containing a CMV promoter and expressing Ser447X variant of the human LPL gene [104].
In spite of a decreasing attractiveness of DNA based vaccination in human health, some developments are nevertheless in progress. One can mention the work done by Nelson et al. [105] on herpes simplex virus 2 with a DNA construct incorporating the regulatory agency compliant, minimal, antibiotic-free (AF) NTC8485 mammalian expression vector.
5. Live Vaccine; Delivery Systems
Recombinant live vaccine and delivery vectors are a specific category of bio-therapeutic agents that combine the most sophisticated technologies and complex safety/regulatory constraints. This group of biological agents is clearly distinct from old live attenuated vaccines and contains diverse technologies such as genetically attenuated pathogenic strains, GRAS [106] microorganisms (Generally Recognized As Safe) used as delivery vehicles or viral strains carrying foreign antigens.
Neglected tropical diseases, such as Chagas caused by the protozoan parasite Trypanosoma cruzi is a good illustration of the relevant strategy of genetically engineer a complex target for which the identification of protective antigens has proven to be difficult. The protective capacity of genetically attenuated parasite is explained by a better exposition to the different components of the immune response [107].
Lactic acid bacteria and other commensal hosts are candidates of choice for the design of novel oral vaccines and possible alternatives to attenuated pathogens [108]. A recombinant version of the extensively used BCG strain is the basis of a new strategy to improve long-lasting memory response and overcome the relative lack of efficiency of the original vaccine formula [109].
Other bacteria, such as Samonella, capable of activating the innate, humoral and cellular immune responses at both mucosal and systemic compartments, are therefore good vehicles to present antigens from difficult targets like HIV [110].
For all these categories of recombinant microorganisms, the requirement for antibiotic-free selection mean is absolutely essential, as they are by definition live agents prone to propagation of genetic elements.
There are numerous examples of live vaccines, for instance in the domain of animal health the MM-3 Escherichia coli strain a candidate for the protection of neonatal piglets against diarrhea that was originally carrying the cat (chloramphenicol acetyltransferase) gene. The lambda-Red technology was used to replace the first cassette by the aspartate-semialdehyde dehydrogenase gene (asd) and the expression levels of the two antigens K88ac fimbriae and heat-labile enterotoxin B subunit (LTB) in cell lysate were proven equivalent to MM-3 [111].
In another work based on the same kind of antibiotic-free selection, Liang et al. [6] evaluated the efficacy, biodistribution and safety of plasmid delivered by the strain C501. The model is based on a balanced-lethal system using an Asd(+) expression plasmid pVGS/2SS-asd encoding two copies of somatostatin (SS) genes carried by Δasd/Δcrp double mutant Salmonella enterica serovar Choleraesuis.
6. Conclusions
Genetic engineering technologies and regulatory requirements are continuously improving and moving in the same direction. Better techniques are creating new challenges and it is likely that in the near future the regulatory status for antibiotic-free selection will progressively move from preferred to highly recommended and mandatory. This is particularly true if we consider that credible alternatives already exist and moreover that some non-antibiotic selection means can even be advantageous options at the industrial level. Antibiotic-free selection is a field of investigation that will benefit from accumulating knowledge of biological pathways and genetic regulation of industrial microorganisms and drive an impossible step backwards.
Acknowledgments
This work has received, through BIOASTER investment, funding from the French Government through the Investissement d’Avenir program (Grant NO. ANR-10-AIRT-03).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Livermore, D.M. Introduction: The challenge of multiresistance. Int. J. Antimicrob. Agents 2007, 29 (Suppl. S3), S1–7. [Google Scholar] [CrossRef] [PubMed]
- Bax, R.; Green, S. Antibiotics: The changing regulatory and pharmaceutical industry paradigm. J. Antimicrobi. Chemother. 2015. [Google Scholar] [CrossRef]
- Finch, R.; Hunter, P.A. Antibiotic resistance—Action to promote new technologies: Report of an EU Intergovernmental Conference held in Birmingham, UK, 12–13 December 2005. J. Antimicrob. Chemother. 2006, 58 (Suppl. S1), i3–i22. [Google Scholar] [CrossRef] [PubMed]
- Bakker, J.M.; Bleeker, W.K.; Parren, P.W. Therapeutic antibody gene transfer: An active approach to passive immunity. Mol Ther. 2004, 10, 411–416. [Google Scholar] [CrossRef] [PubMed]
- Armengol, G.; Ruiz, L.M.; Orduz, S. The injection of plasmid DNA in mouse muscle results in lifelong persistence of DNA, gene expression, and humoral response. Mol. biotechnol. 2004, 27, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Liang, A.; Riaz, H.; Dong, F.; Luo, X.; Yu, X.; Han, Y.; Chong, Z.; Han, L.; Guo, A.; Yang, L. Evaluation of efficacy, biodistribution and safety of antibiotic-free plasmid encoding somatostatin genes delivered by attenuated Salmonella enterica serovar Choleraesuis. Vaccine 2014, 32, 1368–1374. [Google Scholar] [CrossRef] [PubMed]
- Williams, K.J.; Bax, R.P. Challenges in developing new antibacterial drugs. Curr. Opin. Investig. Drugs 2009, 10, 157–163. [Google Scholar] [PubMed]
- Blair, J.M.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Tuller, T. Codon bias, tRNA pools and horizontal gene transfer. Mob. Genet. elements. 2011, 1, 75–77. [Google Scholar] [CrossRef] [PubMed]
- Juhas, M. Horizontal gene transfer in human pathogens. Crit. rev. Microbiol. 2015, 41, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Datta, N.; Kontomichalou, P. Penicillinase synthesis controlled by infectious R factors in Enterobacteriaceae. Nature 1965, 208, 239–241. [Google Scholar] [CrossRef] [PubMed]
- Garber, N.; Friedman, J. Beta-lactamase and the resistance of Pseudomonas aeruginosa to various penicillins and cephalosporins. J. Gen. Microbiol. 1970, 64, 343–352. [Google Scholar] [CrossRef]
- Wiedenbeck, J.; Cohan, F.M. Origins of bacterial diversity through horizontal genetic transfer and adaptation to new ecological niches. FEMS Microbiol. Rev. 2011, 35, 957–976. [Google Scholar] [CrossRef] [PubMed]
- Hamady, M.; Betterton, M.D.; Knight, R. Using the nucleotide substitution rate matrix to detect horizontal gene transfer. BMC bioinform. 2006, 7, 476. [Google Scholar] [CrossRef]
- Roberts, A.P.; Kreth, J. The impact of horizontal gene transfer on the adaptive ability of the human oral microbiome. Front. Cell. Infect. Microbiol. 2014, 4, 124. [Google Scholar] [CrossRef] [PubMed]
- Sodoyer Regis, C.V.; Peubez, I.; Mignon, C. Antibiotic-free selection for: Moving towards a new “gold standard”. In Antibiotic Resistant Bacteria—A Continuous Challenge in the New Millennium; Marina, P., Ed.; InTech: Rijeka, Croatia, 2012; pp. 531–548. [Google Scholar]
- Georghiou, S.B.; Magana, M.; Garfein, R.S.; Catanzaro, D.G.; Catanzaro, A.; Rodwell, T.C. Evaluation of genetic mutations associated with Mycobacterium tuberculosis resistance to amikacin, kanamycin and capreomycin: A systematic review. PLoS ONE 2012, 7, e33275. [Google Scholar] [CrossRef] [PubMed]
- Ogura, T.; Hiraga, S. Mini-F plasmid genes that couple host cell division to plasmid proliferation. Proc. Natl. Acad. Sci. USA 1983, 80, 4784–4788. [Google Scholar] [CrossRef] [PubMed]
- Jaffe, A.; Ogura, T.; Hiraga, S. Effects of the ccd function of the F plasmid on bacterial growth. J. Bacteriol. 1985, 163, 841–849. [Google Scholar] [PubMed]
- Gerdes, K.; Larsen, J.E.; Molin, S. Stable inheritance of plasmid R1 requires two different loci. J Bacteriol. 1985, 161, 292–298. [Google Scholar] [PubMed]
- Gerdes, K.; Poulsen, L.K.; Thisted, T.; Nielsen, A.K.; Martinussen, J.; Andreasen, P.H. The hok killer gene family in gram-negative bacteria. New Biol. 1990, 2, 946–956. [Google Scholar] [PubMed]
- Pedersen, K.; Gerdes, K. Multiple hok genes on the chromosome of Escherichia coli. Mol. Microbiol. 1999, 32, 1090–1102. [Google Scholar] [CrossRef] [PubMed]
- Steif, A.; Meyer, I.M. he hok mRNA family. RNA biol. 2012, 9, 1399–404. [Google Scholar] [CrossRef] [PubMed]
- Summers, D.K.; Sherratt, D.J. Multimerization of high copy number plasmids causes instability: CoIE1 encodes a determinant essential for plasmid monomerization and stability. Cell 1984, 36, 1097–1103. [Google Scholar] [CrossRef] [PubMed]
- Summers, D.K.; Sherratt, D.J. Resolution of ColE1 dimers requires a DNA sequence implicated in the three-dimensional organization of the cer site. EMBO J. 1988, 7, 851–858. [Google Scholar] [PubMed]
- Balding, C.; Blaby, I.; Summers, D. A mutational analysis of the ColE1-encoded cell cycle regulator Rcd confirms its role in plasmid stability. Plasmid 2006, 56, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Blaby, I.K.; Summers, D.K. The role of FIS in the Rcd checkpoint and stable maintenance of plasmid ColE1. Microbiology (Reading, England) 2009, 155, 2676–82. [Google Scholar]
- Peubez, I.; Chaudet, N.; Mignon, C.; Hild, G.; Husson, S.; Courtois, V.; de Luca, K.; Speck, D.; Sodoyer, R. Antibiotic-free selection in E. coli: New considerations for optimal design and improved production. Microb. Cell Fact. 2010, 9, 65. [Google Scholar] [CrossRef] [PubMed]
- Silva, F.; Queiroz, J.A.; Domingues, F.C. Evaluating metabolic stress and plasmid stability in plasmid DNA production by Escherichia coli. Biotechnol. Adv. 2012, 30, 691–708. [Google Scholar] [CrossRef] [PubMed]
- Bower, D.M.; Prather, K.L. Engineering of bacterial strains and vectors for the production of plasmid DNA. Appl. Microbiol. Biotechnol. 2009, 82, 805–813. [Google Scholar] [CrossRef] [PubMed]
- Stenler, S.; Blomberg, P.; Smith, C.I. Safety and efficacy of DNA vaccines: Plasmids vs. minicircles. Hum. Vaccin. Immunother. 2014, 10, 1306–1308. [Google Scholar] [CrossRef] [PubMed]
- Degryse, E. Stability of a host-vector system based on complementation of an essential gene in Escherichia coli. J. Biotechnol. 1991, 18, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Galan, J.E.; Curtiss, R., 3rd. Distribution of the invA. -B, -C, and -D genes of Salmonella typhimurium among other Salmonella serovars: InvA mutants of Salmonella typhi are deficient for entry into mammalian cells. Infect. Immun. 1991, 59, 2901–2908. [Google Scholar] [PubMed]
- Tacket, C.O.; Kelly, S.M.; Schodel, F.; Losonsky, G.; Nataro, J.P.; Edelman, R.; Levine, M.M.; Curtiss, R., 3rd. Safety and immunogenicity in humans of an attenuated Salmonella typhi vaccine vector strain expressing plasmid-encoded hepatitis B antigens stabilized by the Asd-balanced lethal vector system. Infect. Immun. 1997, 65, 3381–3385. [Google Scholar] [PubMed]
- Morona, R.; Brown, M.H.; Yeadon, J.; Heuzenroeder, M.W.; Manning, P.A. Effect of lipopolysaccharide core synthesis mutations on the production of Vibrio cholerae O-antigen in Escherichia coli K-12. FEMS Microbiol. Lett. 1991, 66, 279–285. [Google Scholar] [CrossRef] [PubMed]
- Ryan, E.T.; Crean, T.I.; Kochi, S.K.; John, M.; Luciano, A.A.; Killeen, K.P.; Klose, K.E.; Calderwood, S.B. Development of a DeltaglnA balanced lethal plasmid system for expression of heterologous antigens by attenuated vaccine vector strains of Vibrio cholerae. Infect. Immun. 2000, 68, 221–226. [Google Scholar] [CrossRef] [PubMed]
- Fiedler, M.; Skerra, A. proBA complementation of an auxotrophic E. coli strain improves plasmid stability and expression yield during fermenter production of a recombinant antibody fragment. Gene 2001, 274, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Velur Selvamani, R.S.; Telaar, M.; Friehs, K.; Flaschel, E. Antibiotic-free segregational plasmid stabilization in Escherichia coli owing to the knockout of triosephosphate isomerase (tpiA). Microb. Cell Fact. 2014, 13, 58. [Google Scholar] [CrossRef] [PubMed]
- Soubrier, F.; Cameron, B.; Manse, B.; Somarriba, S.; Dubertret, C.; Jaslin, G.; Jung, G.; Caer, C.L.; Dang, D.; Mouvault, J.M.; et al. pCOR: A new design of plasmid vectors for nonviral gene therapy. Gene Ther. 1999, 6, 1482–1488. [Google Scholar] [CrossRef] [PubMed]
- Marie, C.; Vandermeulen, G.; Quiviger, M.; Richard, M.; Preat, V.; Scherman, D. pFARs. plasmids free of antibiotic resistance markers, display high-level transgene expression in muscle, skin and tumour cells. J. Gene Med. 2010, 12, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Jawale, C.V.; Lee, J.H. Development of a biosafety enhanced and immunogenic Salmonella enteritidis ghost using an antibiotic resistance gene free plasmid carrying a bacteriophage lysis system. PLoS ONE 2013, 8, e78193. [Google Scholar] [CrossRef] [PubMed]
- Vidal, L.; Pinsach, J.; Striedner, G.; Caminal, G.; Ferrer, P. Development of an antibiotic-free plasmid selection system based on glycine auxotrophy for recombinant protein overproduction in Escherichia coli. J. Biotechnol. 2008, 134, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.R.; Xiang, L.X.; Shao, J.Z. Novel antibiotic-free plasmid selection system based on complementation of host auxotrophy in the NAD de novo synthesis pathway. Appl. Environ. Microbiol. 2010, 76, 2295–2303. [Google Scholar] [CrossRef] [PubMed]
- Gerdes, K.; Bech, F.W.; Jorgensen, S.T.; Lobner-Olesen, A.; Rasmussen, P.B.; Atlung, T.; Boe, L.; Karlstrom, O.; Molin, S.; von Meyenburg, K. Mechanism of postsegregational killing by the hok gene product of the parB system of plasmid R1 and its homology with the relF gene product of the E. coli relB operon. EMBO J. 1986, 5, 2023–2029. [Google Scholar] [PubMed]
- Pecota, D.C.; Kim, C.S.; Wu, K.; Gerdes, K.; Wood, T.K. Combining the hok/sok. parDE, and pnd postsegregational killer loci to enhance plasmid stability. Appl. Environ. Microbiol. 1997, 63, 1917–1924. [Google Scholar] [PubMed]
- Schweder, T.; Schmidt, I.; Herrmann, H.; Neubauer, P.; Hecker, M.; Hofmann, K. An expression vector system providing plasmid stability and conditional suicide of plasmid-containing cells. Appl. Environ. Microbiol. 1992, 38, 91–93. [Google Scholar]
- Galen, J.E.; Nair, J.; Wang, J.Y.; Wasserman, S.S.; Tanner, M.K.; Sztein, M.B.; Levine, M.M. Optimization of plasmid maintenance in the attenuated live vector vaccine strain Salmonella typhi CVD 908-htrA. Infect. Immun. 1999, 67, 6424–6433. [Google Scholar] [PubMed]
- Wegerer, A.; Sun, T.; Altenbuchner, J. Optimization of an E. coli l-rhamnose-inducible expression vector: Test of various genetic module combinations. BMC Biotechnol. 2008, 8, 2. [Google Scholar] [CrossRef] [PubMed]
- Szpirer, C.Y.; Milinkovitch, M.C. Separate-component-stabilization system for protein and DNA production without the use of antibiotics. Biotechniques 2005, 38, 775–381. [Google Scholar] [CrossRef] [PubMed]
- Sevillano, L.; Diaz, M.; Santamaria, R.I. Stable expression plasmids for Streptomyces based on a toxin-antitoxin system. Microb. Cell Fact. 2013, 12, 39. [Google Scholar] [CrossRef] [PubMed]
- Nehlsen, K.; Herrmann, S.; Zauers, J.; Hauser, H.; Wirth, D. Toxin-antitoxin based transgene expression in mammalian cells. Nucleic Acids Res. 2010, 38, e32. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Romero, J.M.; Diaz-Orejas, R.; de Lorenzo, V. Resistance to tellurite as a selection marker for genetic manipulations of Pseudomonas strains. Appl. Environ. Microbiol. 1998, 64, 4040–4046. [Google Scholar] [PubMed]
- Goh, S.; Good, L. Plasmid selection in Escherichia coli using an endogenous essential gene marker. BMC Biotechnol. 2008, 8, 61. [Google Scholar] [CrossRef] [PubMed]
- Herrero, M.; de Lorenzo, V.; Timmis, K.N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 1990, 172, 6557–6567. [Google Scholar] [PubMed]
- Pfaffenzeller, I.; Mairhofer, J.; Striedner, G.; Bayer, K.; Grabherr, R. Using ColE1-derived RNA I for suppression of a bacterially encoded gene: Implication for a novel plasmid addiction system. Biotechnol. J. 2006, 1, 675–681. [Google Scholar] [CrossRef] [PubMed]
- Mairhofer, J.; Pfaffenzeller, I.; Merz, D.; Grabherr, R. A novel antibiotic free plasmid selection system: Advances in safe and efficient DNA therapy. Biotechnol. J. 2008, 3, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Luke, J.; Carnes, A.E.; Hodgson, C.P.; Williams, J.A. Improved antibiotic-free DNA vaccine vectors utilizing a novel RNA based plasmid selection system. Vaccine 2009, 27, 6454–6459. [Google Scholar] [CrossRef] [PubMed]
- Nuttall, S.D.; Krishnan, U.V.; Hattarki, M.; de Gori, R.; Irving, R.A.; Hudson, P.J. Isolation of the new antigen receptor from wobbegong sharks, and use as a scaffold for the display of protein loop libraries. Mol. Immunol. 2001, 38, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Cranenburgh, R.M.; Lewis, K.S.; Hanak, J.A. Effect of plasmid copy number and lac operator sequence on antibiotic-free plasmid selection by operator-repressor titration in Escherichia coli. J. Mol. Microbiol. Biotechnol. 2004, 7, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Garmory, H.S.; Leckenby, M.W.; Griffin, K.F.; Elvin, S.J.; Taylor, R.R.; Hartley, M.G.; Hanak, J.A.; Williamson, E.D.; Cranenburgh, R.M. Antibiotic-free plasmid stabilization by operator-repressor titration for vaccine delivery by using live Salmonella enterica Serovar typhimurium. Infect. Immun. 2005, 73, 2005–2011. [Google Scholar] [CrossRef] [PubMed]
- Iwanicki, A.; Piatek, I.; Stasilojc, M.; Grela, A.; Lega, T.; Obuchowski, M.; Hinc, K. A system of vectors for Bacillus subtilis spore surface display. Microb. Cell Fact. 2014, 13, 30. [Google Scholar] [CrossRef] [PubMed]
- Striedner, G.; Pfaffenzeller, I.; Markus, L.; Nemecek, S.; Grabherr, R.; Bayer, K. Plasmid-free T7-based Escherichia coli expression systems. Biotechnol. Bioeng. 2010, 105, 786–794. [Google Scholar] [PubMed]
- Lemuth, K.; Steuer, K.; Albermann, C. Engineering of a plasmid-free Escherichia coli strain for improved in vivo biosynthesis of astaxanthin. Microb. Cell Fact. 2011, 10, 29. [Google Scholar] [CrossRef] [PubMed]
- Baumgartner, F.; Seitz, L.; Sprenger, G.A.; Albermann, C. Construction of Escherichia coli strains with chromosomally integrated expression cassettes for the synthesis of 2'-fucosyllactose. Microb. Cell Fact. 2013, 12, 40. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.Y.; He, C.Y.; Kay, M.A. Improved production and purification of minicircle DNA vector free of plasmid bacterial sequences and capable of persistent transgene expression in vivo. Hum. Gene Ther. 2005, 16, 126–131. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.W.; Christensen, L.V.; Lee, M.; Kim, S.W. Efficient expression of vascular endothelial growth factor using minicircle DNA for angiogenic gene therapy. J. Control. Release 2008, 125, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Chen, Z.; Hu, S.; Jia, F.; Li, Z.; Hoyt, G.; Robbins, R.C.; Kay, M.A.; Wu, J.C. Novel minicircle vector for gene therapy in murine myocardial infarction. Circulation 2009, 120, S230–S237. [Google Scholar] [CrossRef] [PubMed]
- Kay, M.A.; He, C.Y.; Chen, Z.Y. A robust system for production of minicircle DNA vectors. Nat. Biotechnol. 2010, 28, 1287–1289. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.; Kim, Y.; Kim, J.; Jung, H.; Rim, Y.A.; Jung, S.M.; Park, S.H.; Ju, J.H. A new strategy to deliver synthetic protein drugs: Self-reproducible biologics using minicircles. Sci. Rep. 2014, 4, 5961. [Google Scholar] [PubMed]
- Zhang, X.; Epperly, M.W.; Kay, M.A.; Chen, Z.Y.; Dixon, T.; Franicola, D.; Greenberger, B.A.; Komanduri, P.; Greenberger, J.S. Radioprotection in vitro and in vivo by minicircle plasmid carrying the human manganese superoxide dismutase transgene. Hum. Gene Ther. 2008, 19, 820–826. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Kong, Q.; Curtiss, R., 3rd. New technologies in developing recombinant attenuated Salmonella vaccine vectors. Microb. Pathog. 2013, 58, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.M.; Sali, M.; Leckenby, M.W.; Radford, D.S.; Huynh, H.A.; Delogu, G.; Cranenburgh, R.M.; Cutting, S.M. Oral delivery of a DNA vaccine against tuberculosis using operator-repressor titration in a Salmonella enterica vector. Vaccine 2010, 28, 7523–7528. [Google Scholar] [CrossRef] [PubMed]
- Bloor, A.E.; Cranenburgh, R.M. An efficient method of selectable marker gene excision by Xer recombination for gene replacement in bacterial chromosomes. Appl. Environ. Microbiol. 2006, 72, 2520–2525. [Google Scholar] [CrossRef] [PubMed]
- Raiford, D.W.; Heizer, E.M., Jr.; Miller, R.V.; Doom, T.E.; Raymer, M.L.; Krane, D.E. Metabolic and translational efficiency in microbial organisms. J. Mol. Evol. 2012, 74, 206–216. [Google Scholar] [CrossRef] [PubMed]
- Soubrier, F.; Laborderie, B.; Cameron, B. Improvement of pCOR plasmid copy number for pharmaceutical applications. Appl. Microbiol. Biotechnol. 2005, 66, 683–688. [Google Scholar] [CrossRef] [PubMed]
- Vandermeulen, G.; Marie, C.; Scherman, D.; Preat, V. New generation of plasmid backbones devoid of antibiotic resistance marker for gene therapy trials. Mol. Ther. 2011, 19, 1942–1949. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Harrison, E.M.; Bi, D.; Tai, C.; He, X.; Ou, H.Y.; Rajakumar, K.; Deng, Z. TADB: A web-based resource for Type 2 toxin-antitoxin loci in bacteria and archaea. Nucleic Acids Res. 2011, 39, D606–D611. [Google Scholar] [CrossRef]
- Galen, J.E.; Wang, J.Y.; Chinchilla, M.; Vindurampulle, C.; Vogel, J.E.; Levy, H.; Blackwelder, W.C.; Pasetti, M.F.; Levine, M.M. A new generation of stable. nonantibiotic, low-copy-number plasmids improves immune responses to foreign antigens in Salmonella enterica serovar Typhi live vectors. Infect. Immun. 2010, 78, 337–347. [Google Scholar]
- Stieber, D.; Gabant, P.; Szpirer, C. The art of selective killing: Plasmid toxin/antitoxin systems and their technological applications. Biotechniques 2008, 45, 344–346. [Google Scholar] [CrossRef] [PubMed]
- Diago-Navarro, E.; Hernandez-Arriaga, A.M.; Lopez-Villarejo, J.; Munoz-Gomez, A.J.; Kamphuis, M.B.; Boelens, R.; Lemonnier, M.; Diaz-Orejas, R. parD toxin-antitoxin system of plasmid R1—Basic contributions, biotechnological applications and relationships with closely-related toxin-antitoxin systems. FEBS J. 2010, 277, 3097–3117. [Google Scholar] [CrossRef] [PubMed]
- De la Cueva-Mendez, G.; Mills, A.D.; Clay-Farrace, L.; Diaz-Orejas, R.; Laskey, R.A. Regulatable killing of eukaryotic cells by the prokaryotic proteins Kid and Kis. EMBO J. 2003, 22, 246–251. [Google Scholar] [CrossRef] [PubMed]
- Luke, J.M.; Carnes, A.E.; Williams, J.A. Development of antibiotic-free selection system for safer DNA vaccination. Methods Mol. Biol. 2014, 1143, 91–111. [Google Scholar] [PubMed]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef] [PubMed]
- Tuntufye, H.N.; Goddeeris, B.M. Use of lambda Red-mediated recombineering and Cre/lox for generation of markerless chromosomal deletions in avian pathogenic Escherichia coli. FEMS Microbiol. Lett. 2011, 325, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Bryant, J.A.; Sellars, L.E.; Busby, S.J.; Lee, D.J. Chromosome position effects on gene expression in Escherichia coli K-12. Nucleic Acids Res. 2014, 42, 11383–11392. [Google Scholar] [CrossRef] [PubMed]
- Darquet, A.M.; Cameron, B.; Wils, P.; Scherman, D.; Crouzet, J. A new DNA vehicle for nonviral gene delivery: Supercoiled minicircle. Gene Ther. 1997, 4, 1341–1349. [Google Scholar] [CrossRef] [PubMed]
- Bigger, B.W.; Tolmachov, O.; Collombet, J.M.; Fragkos, M.; Palaszewski, I.; Coutelle, C. An araC-controlled bacterial cre expression system to produce DNA minicircle vectors for nuclear and mitochondrial gene therapy. J. Biol. Chem. 2001, 276, 23018–23027. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.S.; Kwon, J.T.; Shim, I.; Kim, H.M.; Kim, P.; Kim, J.C.; Lee, K. Evaluation of toxicity to triclosan in rats following 28days of exposure to aerosol inhalation. Regul. Toxicol. Pharmacol. 2015, 71, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Li, Y.; Zhou, Q.; Xu, Y.; Wang, D. Effect of triclosan on reproduction. DNA damage and heat shock protein gene expression of the earthworm Eisenia fetida. Ecotoxicology 2014, 23, 1826–1832. [Google Scholar] [CrossRef] [PubMed]
- Khan, R.; Lee, M.H.; Joo, H.; Jung, Y.H.; Ahmad, S.; Choi, J.; Lee, S.W. Triclosan resistance in a bacterial fish pathogen aeromonas salmonicida subsp. salmonicida is mediated by an enoyl reductase FabV. J. Microbiol. Biotechnol. 2014. [Google Scholar] [PubMed]
- Oliveira, P.H.; Mairhofer, J. Marker-free plasmids for biotechnological applications—Implications and perspectives. Trends Biotechnol. 2013, 31, 539–547. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.A. Improving DNA vaccine performance through vector design. Curr.Gene Ther. 2014, 14, 170–189. [Google Scholar] [CrossRef] [PubMed]
- Reschner, A.; Scohy, S.; Vandermeulen, G.; Daukandt, M.; Jacques, C.; Michel, B.; Nauwynck, H.; Xhonneux, F.; Preat, V.; Vanderplasschen, A.; et al. Use of Staby((R)) technology for development and production of DNA vaccines free of antibiotic resistance gene. Hum. vaccin. Immunother. 2013, 9, 2203–2210. [Google Scholar] [CrossRef] [PubMed]
- Ginn, S.L.; Alexander, I.E.; Edelstein, M.L.; Abedi, M.R.; Wixon, J. Gene Therapy Clinical Trials Worldwide to 2012. J. Gene Med. 2013, 15, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Note for guidance on the quality, preclinical and clinical aspects of gene transfer medicinal products. Annex on non-clinical testing for indadvertent germline transmission of gene transfer vectors. (2001:648)
- WHO. Guidelines for assuring the quality of non clinical safety evaluation of DNA vaccines. In Proceedings of the 56th Meeting of the WHO Expert Committee on Biological Standardization, Geneva, Switzerland, 24–28 October 2005.
- Beckett, C.G.; Tjaden, J.; Burgess, T.; Danko, J.R.; Tamminga, C.; Simmons, M.; Wu, S.J.; Sun, P.; Kochel, T.; Raviprakash, K.; et al. Evaluation of a prototype dengue-1 DNA vaccine in a Phase 1 clinical trial. Vaccine 2011, 29, 960–968. [Google Scholar] [CrossRef]
- Nikol, S.; Baumgartner, I.; van Belle, E.; Diehm, C.; Visona, A.; Capogrossi, M.C.; Ferreira-Maldent, N.; Gallino, A.; Wyatt, M.G.; Wijesinghe, L.D.; et al. Therapeutic angiogenesis with intramuscular NV1FGF improves amputation-free survival in patients with critical limb ischemia. Mol. Ther. 2008, 16, 972–978. [Google Scholar] [CrossRef] [PubMed]
- Mwau, M.; Cebere, I.; Sutton, J.; Chikoti, P.; Winstone, N.; Wee, E.G.; Beattie, T.; Chen, Y.H.; Dorrell, L.; McShane, H.; et al. A human immunodeficiency virus 1 (HIV-1) clade A vaccine in clinical trials: Stimulation of HIV-specific T-cell responses by DNA and recombinant modified vaccinia virus Ankara (MVA) vaccines in humans. J. Gen. Virol. 2004, 85, 911–919. [Google Scholar] [CrossRef] [PubMed]
- Cranenburgh, R.M.; Hanak, J.A.; Williams, S.G.; Sherratt, D.J. Escherichia coli strains that allow antibiotic-free plasmid selection and maintenance by repressor titration. Nucleic Acids Res. 2001, 29, E26. [Google Scholar] [CrossRef] [PubMed]
- Andries, O.; Kitada, T.; Bodner, K.; Sanders, N.N.; Weiss, R. Synthetic biology devices and circuits for RNA-based “smart vaccines”: A propositional review. Expert Rev. Vaccin. 2015, 14, 313–331. [Google Scholar] [CrossRef]
- Ulmer, J.B.; Mason, P.W.; Geall, A.; Mandl, C.W. RNA-based vaccines. Vaccine 2012, 30, 4414–4418. [Google Scholar] [CrossRef] [PubMed]
- Ljungberg, K.; Liljestrom, P. Self-replicating alphavirus RNA vaccines. Expert Rev. Vaccin. 2015, 14, 177–194. [Google Scholar] [CrossRef]
- Melchiorri, D.; Pani, L.; Gasparini, P.; Cossu, G.; Ancans, J.; Borg, J.J.; Drai, C.; Fiedor, P.; Flory, E.; Hudson, I.; et al. Regulatory evaluation of Glybera in Europe—Two committees, one mission. Nat. Rev. Drug Discov. 2013, 12, 719. [Google Scholar] [CrossRef] [PubMed]
- Nelson, J.; Rodriguez, S.; Finlayson, N.; Williams, J.; Carnes, A. Antibiotic-free production of a herpes simplex virus 2 DNA vaccine in a high yield cGMP process. Hum. Vaccin. Immunother. 2013, 9, 2211–2215. [Google Scholar] [CrossRef] [PubMed]
- Mattia, A.; Merker, R. Regulation of probiotic substances as ingredients in foods: Premarket approval or “generally recognized as safe” notification. Clin. Infect. Dis. 2008, 46, S115–118, discussion S144–151. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Valdez, F.J.; Perez Brandan, C.; Ferreira, A.; Basombrio, M.A. Gene-deleted live-attenuated Trypanosoma cruzi parasites as vaccines to protect against Chagas disease. Expert Rev. Vaccin. 2014, 1–17. [Google Scholar] [CrossRef]
- Trombert, A. Recombinant lactic acid bacteria as delivery vectors of heterologous antigens: The future of vaccination? Benef. Microbes 2014, 1–12. [Google Scholar] [CrossRef]
- Da Costa, A.C.; Nogueira, S.V.; Kipnis, A.; Junqueira-Kipnis, A.P. Recombinant BCG: Innovations on an Old Vaccine. Scope of BCG Strains and Strategies to Improve Long-Lasting Memory. Front. Immunol. 2014, 5, 152. [Google Scholar] [CrossRef] [PubMed]
- Chin’ombe, N.; Ruhanya, V. Recombinant salmonella bacteria vectoring HIV/AIDS vaccines. Open Virol. J. 2013, 7, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.L.; Wang, P.; Tao, H.X.; Liu, X.X.; Wang, Y.C.; Zhan, D.W.; Liu, C.J.; Zhang, Z.S. Removal of antibiotic resistance of live vaccine strain Escherichia coli MM-3 and evaluation of the immunogenicity of the new strain. Acta Biochim. Biophys. Sin. 2006, 38, 844–856. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).