Exploring the Virome of Nile Tilapia (Oreochromis niloticus) Using Metagenomic Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
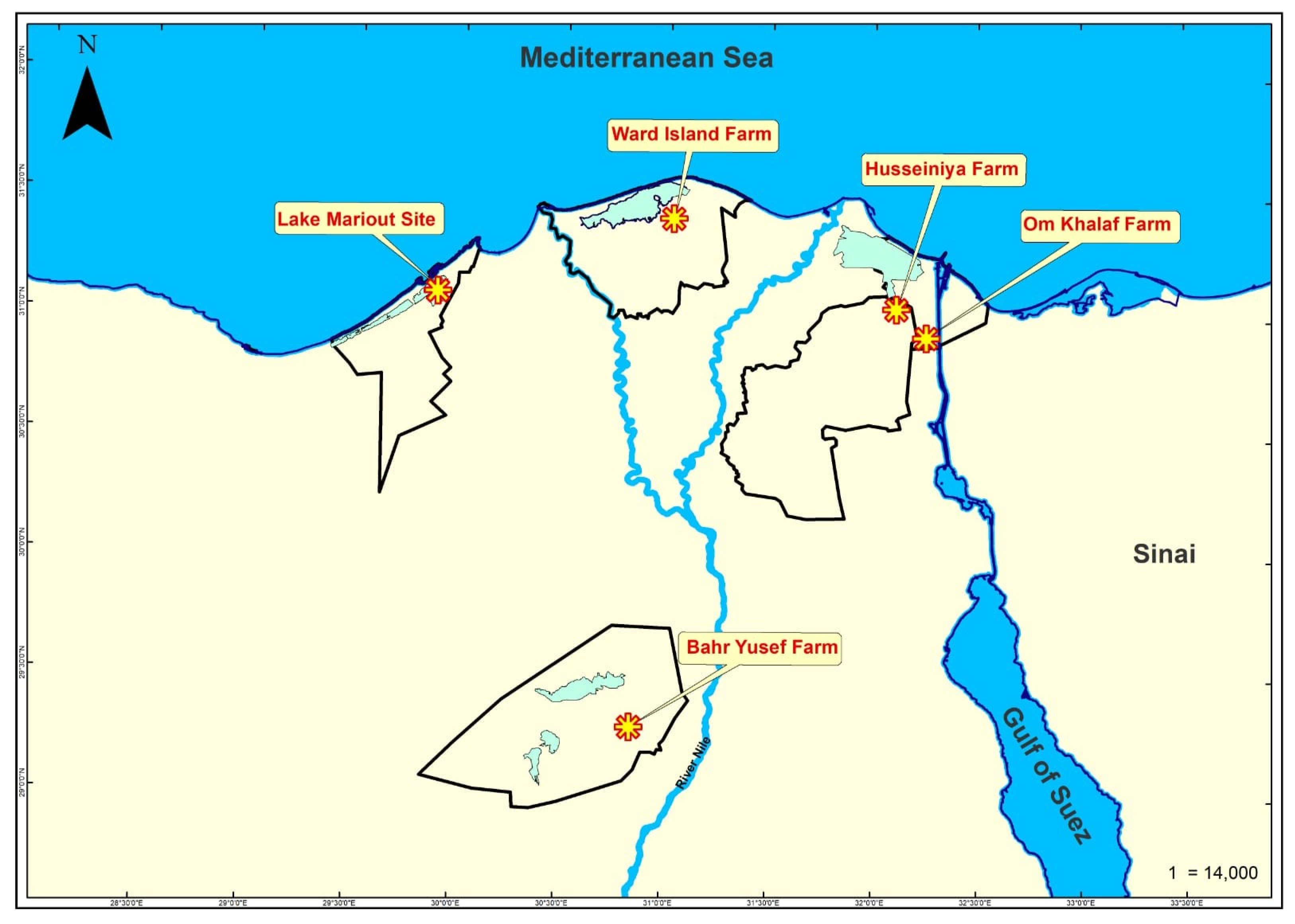
2.2. Fish Sample Collection
2.3. Nucleic Acid Extraction and Reverse Transcription
2.4. Oxford Nanopore Technology (ONT) Sequencing and Data Analysis
2.5. Virome Diversity and Statistical Analysis
(length of the viral sequence in kilobases).
2.6. Further Characterization of Detected Amnoonvirus
| Current ICTV Classification Family | Virus Name | Geographical Location of Occurrence or Sample Collection | Target Host (and Reservoir Host Where Applicable | References, GenBank Accession Number |
|---|---|---|---|---|
| Amnoonviridae | AmnoonvirusEGY1 | Egypt | Tilapia | This Study |
| Amnoonviridae/ Tilapinevirus | Tilapia Lake Virus (TiLV) | Israel | Tilapia | KU751814.1 [34] |
| Amnoonviridae | Flavolineata virus | Bass Strait, Tasmania, Australia | Yellow-striped leatherjacket, Meuschenia flavolineata | MW198700.1 [35] |
| Amnoonviridae | Dolomieu virus | Laurentian Great Lakes, USA | Smallmouth bass, Micropterus dolomieu | GDQU01066121.1 [35] |
| Amnoonviridae | Asotus virus 1 | Japan | Amur catfish, Silurus asotus | GHGF01033499.1 [35] |
| Amnoonviridae | Asotus virus 2 | Japan | Amur catfish, Silurus asotus | GHGF01016319.1 [35] |
| Amnoonviridae | Przewalskii virus | China | Lake Qinghai scaleless carp, Gymnocypris przewalskii | GHYJ01002273.1 [35] |
| Amnoonviridae | Stewartii virus | China | Cyprinid fish, Oxygymnocypris stewartii | GIBO01031171.1 [35] |
| Amnoonviridae | Namensis virus | China | Cyprinid fish, Gymnocypris namensis | GHYH01080462.1 [35] |
| Amnoonviridae | Tilapia lake virus-like virus | Caribbean | Guppy, Poecilia reticulata | BK063200.1 [31] |
| Amnoonviridae | Fancy-tailed guppy virus | USA | Guppy, Poecilia reticulata | PP409995.1 [32] |
| Unclassified Articulavirales | Lauta virus | Australia | Gulf tree gehyra, Gehyra lauta | MT386081.1 [36] |
| Orthomyxoviridae | Infectious salmon anemia virus (ISAV) | Canada | Atlantic salmon, Salmo salar | NC_006503.1 [37] |
| Orthomyxoviridae | Yancheng orthomyxo-like virus | China | Branded goby, Chaeturichthys stigmatias | MG600035.1 [38] |
| Orthomyxoviridae | Rainbow trout orthomyxovirus-1 | USA | Rainbow trout, Oncorhynchus mykiss | KX882062.1 [39] |
| Orthomyxoviridae | Pilchard orthomyxovirus | Australia | Atlantic salmon, Salmo salar | NC_078611.1 [40] |
| Orthomyxoviridae | Wuhan carp Isavirus 1 | China | Goldfish, Carassius auratus | MG600055.1 [38] |
| Orthomyxoviridae | Wuhan spiny eel influenza virus | China | Lesser spiny eel, Macrognathus aculeatus | MG600038.1 [38] |
| Orthomyxoviridae | Wenling hagfish influenza virus | China | Inshore hagfish, Eptatretus burgeri | MG600051.1 [38] |
| Orthomyxoviridae | Wenling orthomyxo-like virus 2 | China | Red spikefish Triacanthodes anomalus | MG600036.1 [38] |
| Orthomyxoviridae | Wuhan asiatic toad influenza virus | China | Asiatic toad, Bufo gargarizans | MG600045.1 [38] |
| Orthomyxoviridae | Xibalbanus thogotovirus 1 | Australia | Cave swimmer, Xibalbanus tulumensis | BK067651.1 [41] |
| Orthomyxoviridae | Influenza A virus | USA | Blue-winged teal, Anas discors | KJ413483.1 [42] |
| Orthomyxoviridae | Influenza B virus | Canada | Egg-grown virus | NC_002204.1 [43] |
| Orthomyxoviridae | Influenza C virus | Japan | Homo sapiens | LC123402.1 [44] |
| Orthomyxoviridae | Influenza D virus | France | Bovine | LN559121.1 [45] |
2.7. Concatenated Phylogenic Analysis Using Partial Nucleotide Sequences of Segments 1, 5, and 7 of Tilapinevirus spp. and AmnoonvirusEGY1
3. Results
3.1. Overview of Nile Tilapia Virome
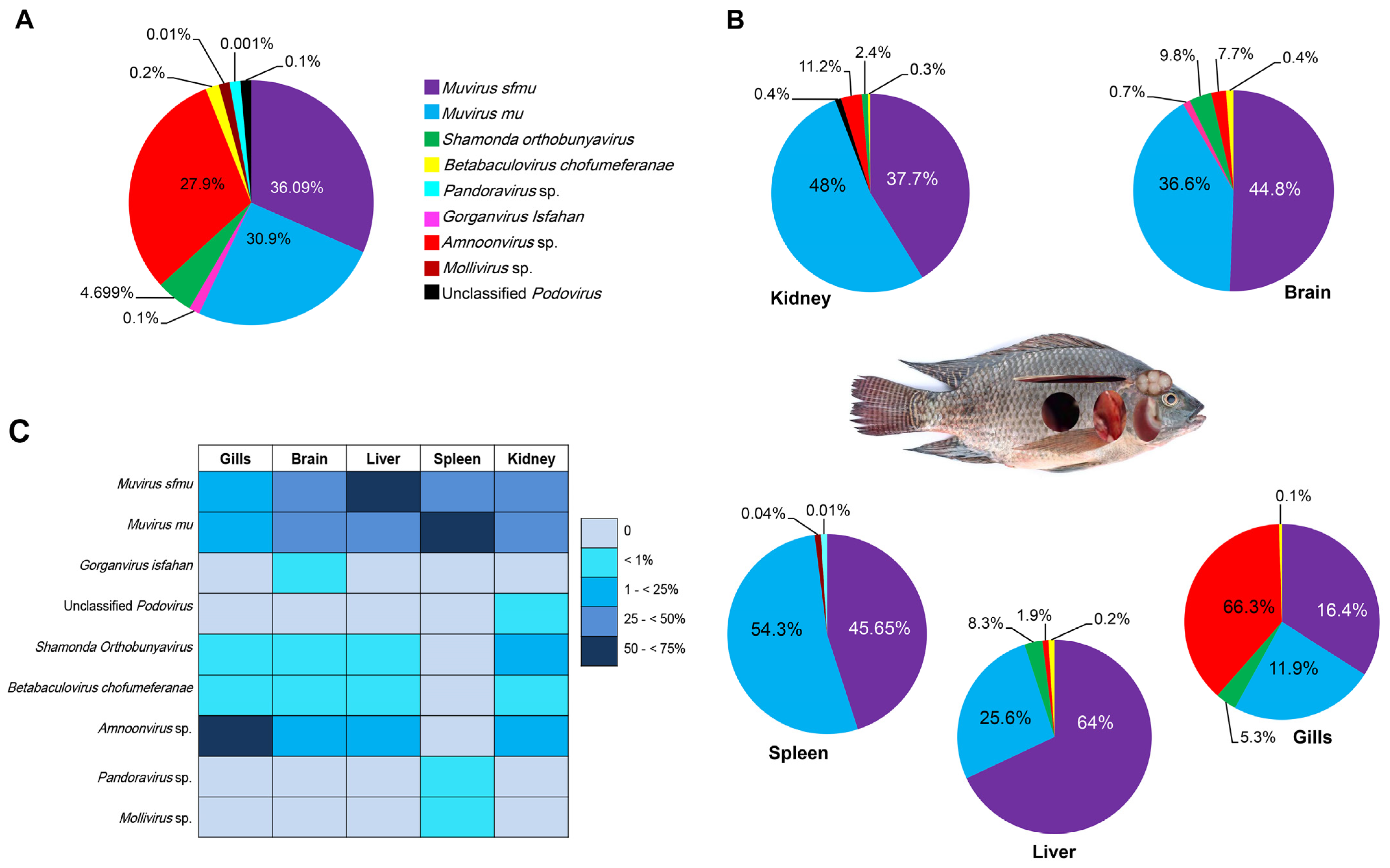
3.2. Distribution of Viruses in Husseiniya Farm Nile Tilapia Organs
3.3. Comparison of Virome Compositions in Organs of Fish Collected from Five Sampling Sites
3.4. Virome Richness in Nile Tilapia Virome
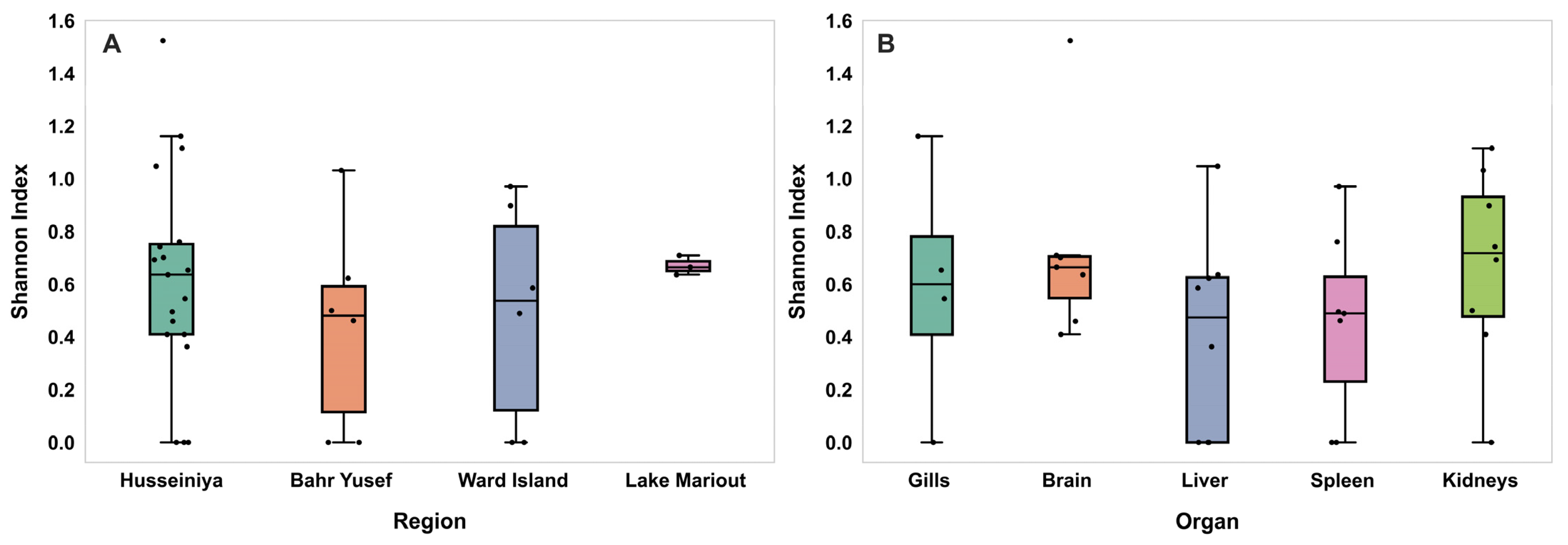
3.5. Shannon Diversity Index of Nile Tilapia Virome
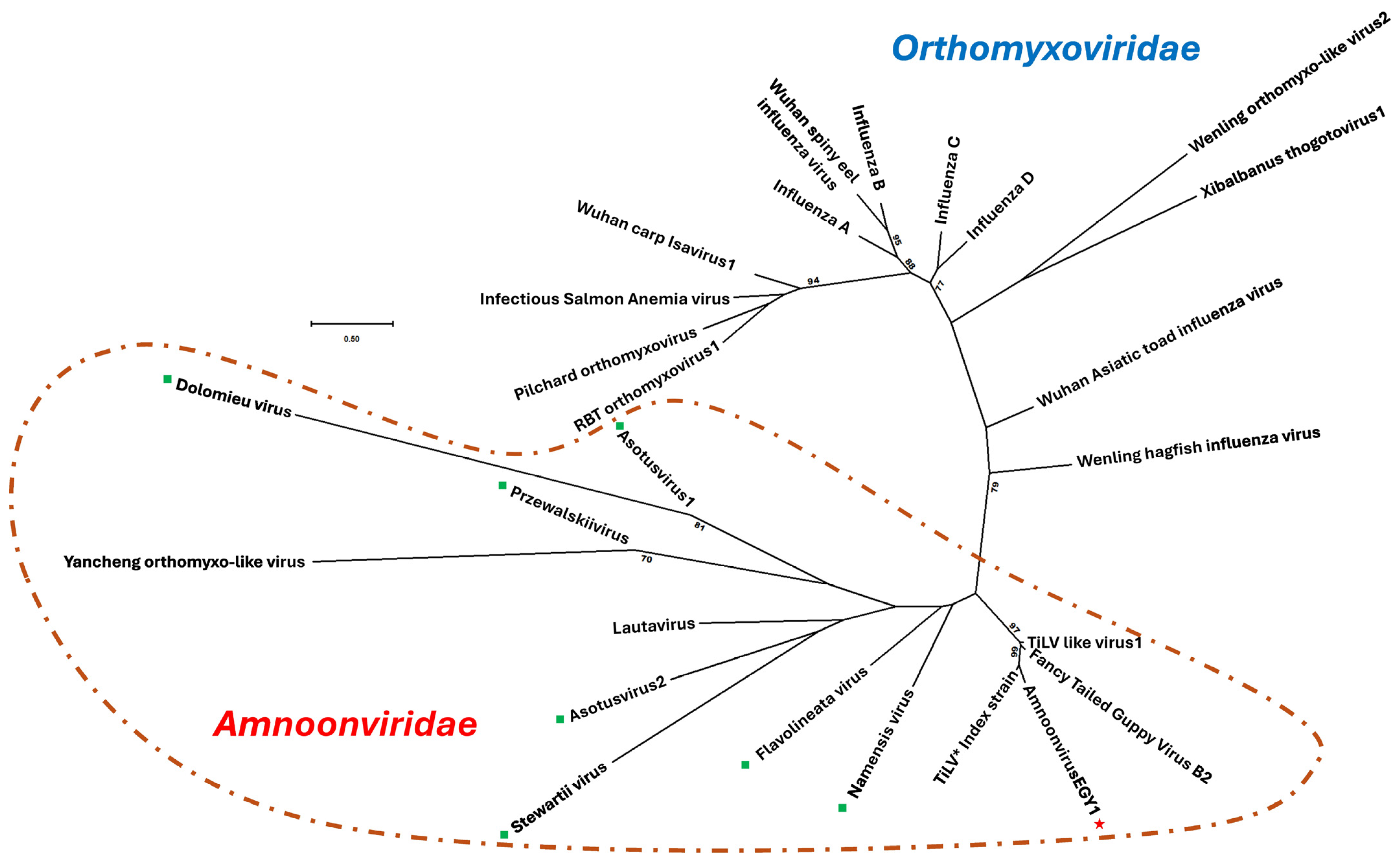
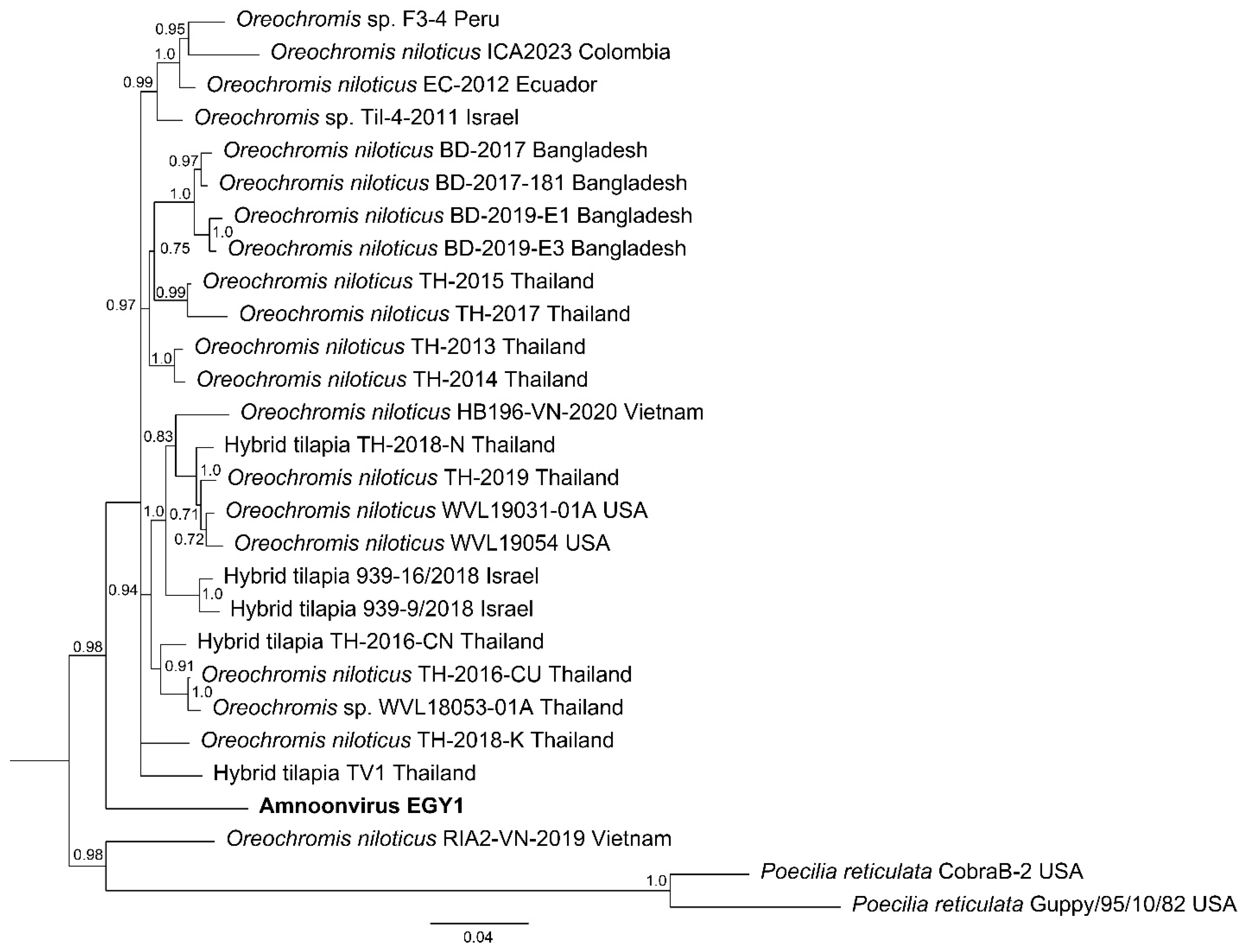
3.6. Phylogenic Analysis of the Amnoonvirus
4. Discussion
4.1. Nile Tilapia Virome Overview
4.2. Phylogenetic Studies on Sequences of AmnoonvirusEGY1
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Prabu, E.; Rajagopalsamy, C.B.T.; Ahilan, B.; Jeevagan, I.J.M.A.; Renuhadevi, M. Tilapia—An excellent candidate species for world aquaculture: A Review. Annu. Res. Rev. Biol. 2019, 31, 1–14. [Google Scholar] [CrossRef]
- FAO. The State of World Fisheries and Aquaculture 2024—Blue Transformation in Action Report; Food and Agriculture Organization of the United Nations: Rome, Italy, 2024. [Google Scholar] [CrossRef]
- Lowe-McConnell, R.H. Tilapias: Biology and Exploitation; Springer: Dordrecht, The Netherlands, 2000; pp. 129–162. [Google Scholar]
- Machimbirike, V.I.; Jansen, M.D.; Senapin, S.; Khunrae, P.; Rattanarojpong, T.; Dong, H.T. Viral infections in tilapines: More than just tilapia lake virus. Aquaculture 2019, 503, 508–518. [Google Scholar] [CrossRef]
- Clyde, C.W.; Tan, J.P.; Yeap, S.K.; Yong, C.Y. Current updates on viral infections affecting tilapia. Aquac. Fish. 2025, 10, 355–371. [Google Scholar] [CrossRef]
- Eyngor, M.; Zamostiano, R.; Kembou Tsofack, J.E.; Berkowitz, A.; Bercovier, H.; Tinman, S.; Lev, M.; Hurvitz, A.; Galeotti, M.; Bacharach, E.; et al. Identification of a novel RNA virus lethal to tilapia. J. Clin. Microbiol. 2014, 52, 4137–4146. [Google Scholar] [CrossRef] [PubMed]
- Aich, N.; Paul, A.; Choudhury, T.G.; Saha, H. Tilapia lake virus (TiLV) disease: Current status of understanding. Aquac. Fish. 2022, 7, 7–17. [Google Scholar] [CrossRef]
- Surachetpong, W.; Roy, S.R.K.; Nicholson, P. Tilapia lake virus: The story so far. J. Fish Dis. 2020, 43, 1115–1132. [Google Scholar] [CrossRef] [PubMed]
- International Committee on Taxonomy of Viruses. Orthomyxoviridae. ICTV 9th Report. 2011. Available online: https://ictv.global/report_9th/RNAneg/Orthomyxoviridae (accessed on 30 May 2025).
- Chengula, A.A.; Mutoloki, S.; Evensen, Ø.; Munang’andu, H.M. Tilapia lake virus does not hemagglutinate avian and piscine erythrocytes and NH4Cl does not inhibit viral replication in vitro. Viruses 2019, 11, 1152. [Google Scholar] [CrossRef]
- Acharya, V.; Chakraborty, H.J.; Rout, A.K.; Balabantaray, S.; Behera, B.K.; Das, B.K. Structural characterization of open reading frame-encoded functional genes from tilapia lake virus (TiLV). Mol. Biotechnol. 2019, 61, 945–957. [Google Scholar] [CrossRef]
- Abu Rass, R.; Kustin, T.; Zamostiano, R.; Smorodinsky, N.; Meir, D.B.; Feder, D.; Mishra, N.; Lipkin, W.I.; Eldar, A.; Ehrlich, M.; et al. Inferring protein function in an emerging virus: Detection of the nucleoprotein in tilapia lake virus. J. Virol. 2022, 96, e01757-21. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Krupovic, M.; Surachetpong, W.; Wolf, Y.I.; Kuhn, J.H. ICTV Virus Taxonomy Profile: Amnoonviridae 2023. J. Gen. Virol. 2023, 104, 001903. [Google Scholar] [CrossRef]
- Kuhn, J.H.; Adkins, S.; Alioto, D.; Alkhovsky, S.V.; Amarasinghe, G.K.; Anthony, S.J.; Avšič-Županc, T.; Ayllón, M.A.; Bahl, J.; Balkema-Buschmann, A.; et al. Taxonomic update for Phylum Negarnaviricota (Riboviria: Orthornavirae), including the large orders Bunyavirales and Mononegavirales. Arch. Virol. 2020, 165, 3023–3072. [Google Scholar] [CrossRef]
- Elsaied, H.E.; Soliman, T.; Abu-Taleb, H.T.; Goto, H.; Jenke-Kodam, H. Phylogenetic characterization of eukaryotic and prokaryotic gut flora of Nile tilapia, Oreochromis niloticus, along niches of Lake Nasser, Egypt, based on rRNA gene high-throughput sequences. Ecol. Genet. Genom. 2019, 11, 100037. [Google Scholar] [CrossRef]
- Bereded, N.K.; Abebe, G.B.; Fanta, S.W.; Curto, M.; Waidbacher, H.; Meimberg, H.; Domig, K.J. The gut bacterial microbiome of Nile tilapia (Oreochromis niloticus) from lakes across an altitudinal gradient. BMC Microbiol. 2022, 22, 87. [Google Scholar] [CrossRef]
- Debnath, S.C.; McMurtrie, J.; Temperton, B.; Delamare-Deboutteville, J.; Mohan, C.V.; Tyler, C.R. Tilapia aquaculture, emerging diseases, and the roles of the skin microbiomes in health and disease. Aquac. Int. 2023, 31, 2945–2976. [Google Scholar] [CrossRef]
- Huavas, J.; Heyse, J.; Props, R.; Delamare-Deboutteville, J.; Shelley, C. Microbiomes of tilapia culture systems: Composition, affecting factors, and future perspectives. Aquac. Res. 2024, 26, 5511461. [Google Scholar] [CrossRef]
- Costa, V.A.; Mifsud, J.C.O.; Gilligan, D.; Williamson, J.E.; Holmes, E.C.; Geoghegan, J.L. Metagenomic sequencing reveals a lack of virus exchange between native and invasive freshwater fish across the Murray–Darling Basin, Australia. Virus Evol. 2021, 7, veab034. [Google Scholar] [CrossRef] [PubMed]
- Gadoin, E.; Desnues, C.; Monteil-Bouchard, S.; Bouvier, T.; Auguet, J.-C.; Roque d’Orbcastel, E.; Bettarel, Y. Fishing for the virome of tropical tuna. Viruses 2021, 13, 1291. [Google Scholar] [CrossRef]
- Ford, C.E.; Dunn, C.D.; Leis, E.M.; Thiel, W.A.; Goldberg, T.L. Five species of wild freshwater sport fish in Wisconsin, USA, reveal highly diverse viromes. Pathogens 2024, 13, 150. [Google Scholar] [CrossRef] [PubMed]
- Alavandi, S.V.; Poornima, M. Viral metagenomics: A tool for virus discovery and diversity in aquaculture. Indian J. Virol. 2012, 23, 88–98. [Google Scholar] [CrossRef]
- Geoghegan, J.L.; Di Giallonardo, F.; Cousins, K.; Shi, M.; Williamson, J.E.; Holmes, E.C. Hidden diversity and evolution of viruses in market fish. Virus Evol. 2018, 4, vey031. [Google Scholar] [CrossRef]
- Perry, B.J.; Darestani, M.M.; Ara, M.G.; Hoste, A.; Jandt, J.M.; Dutoit, L.; Holmes, E.C.; Ingram, T.; Geoghegan, J.L. Viromes of freshwater fish with lacustrine and diadromous life histories differ in composition. Viruses 2022, 14, 257. [Google Scholar] [CrossRef]
- Grimwood, R.M.; Fortune-Kelly, G.; Holmes, E.C.; Ingram, T.; Geoghegan, J.L. Host specificity shapes fish viromes across lakes on an isolated remote island. Virology 2023, 587, 109884. [Google Scholar] [CrossRef]
- Xi, Y.; Jiang, X.; Xie, X.; Zhao, M.; Zhang, H.; Qin, K.; Wang, X.; Liu, Y.; Yang, S.; Shen, Q.; et al. Viromics reveals the high diversity of viruses from fishes of the Tibet Highland. Microbiol. Spectr. 2023, 11, e00946-23. [Google Scholar] [CrossRef]
- Neil, J.A.; Cadwell, K. The Intestinal virome and immunity. J. Immunol. 2018, 201, 1615–1624. [Google Scholar] [CrossRef]
- Smith, S.E.; Huang, W.; Tiamani, K.; Unterer, M.; Mirzaei, M.K.; Deng, L. Emerging technologies in the study of the virome. Curr. Opin. Virol. 2022, 54, 101231. [Google Scholar] [CrossRef] [PubMed]
- Kibenge, F.S.; Kibenge, M.J. Amnoonviruses and non-influenza orthomyxoviruses of fish. In Aquaculture Virology, 2nd ed.; Frederick, S.B.K., Marcos, G.G., Eds.; Academic Press: Cambridge, MA, USA, 2024; pp. 339–353. [Google Scholar] [CrossRef]
- Bonenfant, Q.; Noé, L.; Touzet, H. Porechop_ABI: Discovering unknown adapters in Oxford nanopore technology sequencing reads for downstream trimming. Bioinform. Adv. 2022, 3, vbac085. [Google Scholar] [CrossRef] [PubMed]
- Li, M.-S. Discovery of two novel tilapia lake virus-like virus isolates in the transcriptomic data of guppy fish (Poecilia reticulata). J. Fish Dis. 2023, 46, 1015–1019. [Google Scholar] [CrossRef] [PubMed]
- Soto, E.; LaFrentz, B.R.; Yun, S.; Megarani, D.; Henderson, E.; Piewbang, C.; Johnston, A.E.; Techangamsuwan, S.; Fei, T.; Warg, J.; et al. Diagnosis, isolation and description of a novel amnoonvirus recovered from diseased fancy guppies, Poecilia reticulata. J. Fish Dis. 2024, 47, e13937. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Suleski, M.; Sanderford, M.; Sharma, S.; Tamura, K. MEGA12, molecular evolutionary genetic analysis version 12 for adaptive and green Computing. Mol. Biol. Evol. 2024, 41, msae263. [Google Scholar] [CrossRef]
- Bacharach, E.; Mishra, N.; Briese, T.; Zody, M.C.; Kembou Tsofack, J.E.; Zamostiano, R.; Berkowitz, A.; Ng, J.; Nitido, A.; Corvelo, A.; et al. Characterization of a novel Orthomyxo-like virus causing mass die-offs of tilapia. mBio 2016, 7, e00431-16. [Google Scholar] [CrossRef]
- Turnbull, O.M.H.; Ortiz-Báez, A.S.; Eden, J.; Shī, M.; Williamson, J.E.; Gaston, T.F.; Zhang, Y.; Holmes, E.C.; Geoghegan, J.L. Meta-transcriptomic identification of divergent Amnoonviridae in Fish. Viruses 2020, 12, 1254. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Baez, A.S.; Eden, J.-S.; Moritz, C.; Holmes, E.C. A Divergent Articulavirus in an Australian gecko identified using meta-transcriptomics and protein structure comparisons. Viruses 2020, 12, 613. [Google Scholar] [CrossRef]
- Clouthier, S.C.; Rector, T.; Brown, N.E.C.; Anderson, E.D. Genomic organization of infectious salmon anaemia virus. J. Gen. Virol. 2002, 83, 421–428. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Lin, X.-D.; Chen, X.; Tian, J.-H.; Chen, L.-J.; Li, K.; Wang, W.; Eden, J.-S.; Shen, J.-J.; Liu, L.; et al. The Evolutionary history of vertebrate RNA viruses. Nature 2018, 556, 197–202. [Google Scholar] [CrossRef]
- Batts, W.N.; LaPatra, S.E.; Katona, R.; Leis, E.; Ng, T.F.F.; Brieuc, M.S.; Breyta, R.B.; Purcell, M.K.; Conway, C.M.; Waltzek, T.B.; et al. Molecular characterization of a novel orthomyxovirus from rainbow and steelhead trout (Oncorhynchus mykiss). Virus Res. 2017, 230, 38–49. [Google Scholar] [CrossRef]
- Mohr, P.G.; Crane, M.S.J.; Hoad, J.; Williams, L.M.; Cummins, D.; Neave, M.J.; Shiell, B.; Beddome, G.; Michalski, W.; Peck, G.; et al. Pilchard orthomyxovirus (POMV). I. Characterisation of an emerging virus isolated from pilchards Sardinops sagax and Atlantic salmon Salmo salar. Dis. Aquat. Org. 2020, 139, 35–50. [Google Scholar] [CrossRef]
- Petrone, M.E.; Parry, R.; Mifsud, J.C.O.; Brussel, K.V.; Vorhees, I.; Richards, Z.T.; Holmes, E.C. Evidence for an aquatic origin of influenza virus and the Order Articulavirales. Proc. Natl. Acad. Sci. USA 2023, 120, e2310529120. [Google Scholar] [CrossRef]
- Ramey, A.M.; Reeves, A.B.; TeSlaa, J.L.; Nashold, S.; Donnelly, T.; Bahl, J.; Hall, J.S. Evidence for common ancestry among viruses isolated from wild birds in Beringia and highly pathogenic intercontinental reassortant H5N1 and H5N2 influenza A viruses. Infect. Genet. Evol. 2016, 40, 176–185. [Google Scholar] [CrossRef]
- Kemdirim, S.; Palefsky, J.M.; Briedis, D.J. Influenza B virus PB1 protein; nucleotide sequence of the genome RNA segment predicts a high degree of structural homology with the corresponding influenza A virus polymerase protein. Virology 1986, 152, 126–135. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaki, Y.; Sugawara, K.; Furuse, Y.; Shimotai, Y.; Hongo, S.; Oshitani, H.; Mizuta, K.; Nishimura, H. Genetic lineage and reassortment of Influenza C viruses circulating between 1947 and 2014. J. Virol. 2016, 90, 8251–8265. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ducatez, M.; Pelletier, C.; Meyer, G.G. Influenza D virus in cattle, France, 2011–2014. Emerg. Infect. Dis. 2015, 21, 368–371. [Google Scholar] [CrossRef]
- Zhu, Y.; Shang, J.; Peng, C.; Sun, Y. Phage family classification under Caudoviricetes: A review of current tools using the latest ICTV classification framework. Front. Microbiol. 2022, 13, 1032186. [Google Scholar] [CrossRef]
- International Committee on Taxonomy of Viruses. Family Baculoviridae. Available online: https://ictv.global/report/chapter/baculoviridae/baculoviridae (accessed on 22 March 2025).
- Filipa-Silva, A.; Parreira, R.; Martínez-Puchol, S.; Bofill-Mas, S.; Barreto Crespo, M.T.; Nunes, M. The unexplored virome of two Atlantic coast fish: Contribution of next-generation sequencing to fish virology. Foods 2020, 9, 1634. [Google Scholar] [CrossRef]
- Suttle, C.A. Marine viruses—Major players in the global ecosystem. Nat. Rev. Microbiol. 2007, 5, 801–812. [Google Scholar] [CrossRef]
- Chow, C.-E.T.; Suttle, C.A. Biogeography of viruses in the sea. Annu. Rev. Virol. 2015, 2, 41–66. [Google Scholar] [CrossRef]
- Weitz, J.; Wilhelm, S. Ocean viruses and their effects on microbial communities and biogeochemical cycles. F1000 Biol. Rep. 2012, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Van Regenmortel, M.H.V. Solving the species problem in viral taxonomy: Recommendations on non-latinized binomial species names and on abandoning attempts to assign metagenomic viral sequences to species taxa. Arch. Virol. 2019, 164, 2223–2229. [Google Scholar] [CrossRef] [PubMed]
- Turner, D.; Shkoporov, A.N.; Lood, C.; Millard, A.D.; Dutilh, B.E.; Alfenas-Zerbini, P.; van Zyl, L.J.; Aziz, R.K.; Oksanen, H.M.; Poranen, M.M.; et al. Abolishment of morphology-based taxa and change to binomial species names: 2022 Taxonomy Update of the ICTV Bacterial Viruses Subcommittee. Arch. Virol. 2023, 168, 74. [Google Scholar] [CrossRef]
- Comeau, A.M.; Tremblay, D.; Moineau, S.; Rattei, T.; Kushkina, A.I.; Tovkach, F.I.; Krisch, H.M.; Ackermann, H.-W. Phage morphology recapitulates phylogeny: The comparative genomics of a new group of myoviruses. PLoS ONE 2012, 7, e40102. [Google Scholar] [CrossRef] [PubMed]
- Suresh, J.I.; Sri Janani, M.S.; Sowndharya, R. Bacterial diseases in fish with relation to pollution and their consequences—A Global scenario. In Bacterial Fish Diseases; Dar, G.H., Bhat, R.A., Qadri, H., Al-Ghamdy, K.M., Hakeem, K.R., Eds.; Academic Press: Cambridge, UK, 2022; pp. 113–131. [Google Scholar] [CrossRef]
- Dubos, R.J.; Straus, J.H.; Pierce, C. The Multiplication of bacteriophage in vivo and its protective effect against an experimental infection with Shigella dysenteriae. J. Exp. Med. 1943, 78, 161–168. [Google Scholar] [CrossRef]
- Podlacha, M.; Grabowski, Ł.; Kosznik-Kawśnicka, K.; Zdrojewska, K.; Stasiłojć, M.; Węgrzyn, G.; Węgrzyn, A. Interactions of bacteriophages with animal and human organisms-Safety issues in the light of phage therapy. Int. J. Mol. Sci. 2021, 22, 8937. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Arroyo-Olarte, R.D.; Bravo Rodríguez, R.; Morales-Ríos, E. Genome editing in bacteria: CRISPR-Cas and beyond. Microorganisms 2021, 9, 844. [Google Scholar] [CrossRef]
- Tang, S.; Conte, V.; Zhang, D.J.; Žedaveinytė, R.; Lampe, G.D.; Wiegand, T.; Tang, L.C.; Wang, M.; Walker, M.W.G.; George, J.T.; et al. . De novo gene synthesis by an antiviral reverse transcriptase. Science 2024, 386, eadq0876. [Google Scholar] [CrossRef] [PubMed]
- Mise, K. Isolation and characterization of a new generalized transducing bacteriophage different from Pl in Escherichia coli. J. Virol. 1971, 7, 168–175. [Google Scholar] [CrossRef]
- Yazdi, M.; Bouzari, M.; Ghaemi, E.A. Genomic analyses of a novel bacteriophage (VB_PmiS-Isfahan) within Siphoviridae family Infecting Proteus mirabilis. Genomics 2019, 111, 1283–1291. [Google Scholar] [CrossRef]
- Peeler, E.J.; Taylor, N.G. The Application of epidemiology in aquatic animal health—Opportunities and challenges. Vet. Res. 2011, 42, 94. [Google Scholar] [CrossRef]
- Abeles, S.R.; Pride, D.T. Molecular bases and role of viruses in the human microbiome. J. Mol. Biol. 2014, 426, 3892–3906. [Google Scholar] [CrossRef] [PubMed]
- Dykova, I.; Lom, J.; Schroeder-Diedrich, J.M.; Booton, G.C.; Byers, T.J. Acanthamoeba strains isolated from organs of freshwater fishes. J. Parasitol. 1999, 85, 1106. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Latif, A.M. Gill Parasitic Diseases of Some Freshwater Fishes. Ph.D. Thesis, Faculty of Veterinary Medicine, Benha University, Toukh, Egypt, 2007. [Google Scholar]
- Philippe, N.; Legendre, M.; Doutre, G.; Couté, Y.; Poirot, O.; Lescot, M.; Arslan, D.; Seltzer, V.; Bertaux, L.; Bruley, C.; et al. Pandoraviruses: Amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science 2013, 341, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Morimoto, D.; Tateishi, N.; Takahashi, M.; Nagasaki, K. Isolation of viruses, including mollivirus, with the potential to infect Acanthamoeba from a Japanese warm temperate zone. PLoS ONE 2024, 19, e0301185. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mahmoud, N.E.; Fahmy, M. Investigations on mass mortalities among Oreochromis Niloticus at Mariotteya Stream, Egypt: Parasitic infestation and environmental pollution impacts. Fish. Aquac. J. 2014, 5, 1. [Google Scholar] [CrossRef]
- Taha, E.; Shawky, M.; Ahmed, B.; Moustafa, M.; Yousif, A.; Abdelaziz, M. Emergence of viral nervous necrosis is associated with mass mortality in hatchery-reared tilapia (Oreochromis niloticus) in Egypt. Aquac. Int. 2020, 28, 1811–1823. [Google Scholar] [CrossRef]
- Fathi, M.; Dickson, C.; Dickson, M.; Leschen, W.; Baily, J.; Muir, F.; Ulrich, K.; Weidmann, M. Identification of tilapia lake virus in Egypt in Nile tilapia affected by summer mortality syndrome. Aquaculture 2017, 473, 430–432. [Google Scholar] [CrossRef]
- Nicholson, P.; Fathi, M.A.; Fischer, A.; Mohan, C.; Schieck, E.; Mishra, N.; Heinimann, A.; Frey, J.; Wieland, B.; Jores, J. Detection of tilapia lake virus in Egyptian fish farms experiencing high mortalities in 2015. J. Fish Dis. 2017, 40, 1925–1928. [Google Scholar] [CrossRef]
- Borrego, J.J.; Valverde, E.J.; Labella, A.M.; Castro, D. Lymphocystis disease virus: Its importance in aquaculture. Rev. Aquac. 2015, 9, 179–193. [Google Scholar] [CrossRef]
- Mulei, I.R.; Nyaga, P.N.; Mbuthia, P.G.; Waruiru, R.M.; Njagi, L.W.; Mwihia, E.W.; Gamil, A.A.A.; Evensen, Ø.; Mutoloki, S. Infectious pancreatic necrosis virus isolated from farmed rainbow trout and tilapia in Kenya is identical to European isolates. J. Fish Dis. 2018, 41, 1191–1200. [Google Scholar] [CrossRef]
- Ramírez-Paredes, J.G.; Paley, R.K.; Hunt, W.; Feist, S.W.; Stone, D.M.; Field, T.R.; Haydon, D.J.; Ziddah, P.A.; Nkansa, M.; Guilder, J.; et al. First detection of infectious spleen and kidney necrosis virus (ISKNV) associated with massive mortalities in farmed tilapia in Africa. Transbound. Emerg. Dis. 2020, 68, 1550–1563. [Google Scholar] [CrossRef]
- Shlapobersky, M.; Sinyakov, M.S.; Katzenellenbogen, M.; Sarid, R.; Don, J.; Avtalion, R.R. Viral encephalitis of tilapia larvae: Primary characterization of a novel herpes-like Virus. Virology 2010, 399, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Schoch, C.L.; Ciufo, S.; Domrachev, M.; Hotton, C.L.; Kannan, S.; Khovanskaya, R.; Leipe, D.; Mcveigh, R.; O’Neill, K.; Robbertse, B.; et al. NCBI Taxonomy: A Comprehensive Update on Curation, Resources and Tools. Database 2020, 2020, baaa062. [Google Scholar] [CrossRef]
- Chaput, D.L.; Bass, D.; Alam, M.M.; Al Hasan, N.; Stentiford, G.D.; van Aerle, R.; Moore, K.; Bignell, J.P.; Haque, M.M.; Tyler, C.R. The segment matters: Probable reassortment of tilapia lake virus (TiLV) complicates phylogenetic analysis and inference of geographical origin of new isolate from Bangladesh. Viruses 2020, 12, 258. [Google Scholar] [CrossRef] [PubMed]
- Tran, T.H.; Nguyen, V.T.H.; Bui, H.C.N.; Tran, Y.B.T.; Tran, H.T.T.; Le, T.T.T.; Vu, H.T.T.; Ngo, T.P.H. Tilapia lake virus (TiLV) from Vietnam Is genetically distantly related to TiLV Strains from other countries. J. Fish Dis. 2022, 45, 1389–1401. [Google Scholar] [CrossRef]
- Verma, D.K.; Sood, N.; Paria, A.; Swaminathan, T.R.; Mohan, C.V.; Rajendran, K.V.; Pradhan, P.K. Reassortment and evolutionary dynamics of tilapia lake virus genomic segments. Virus Res. 2021, 308, 198625. [Google Scholar] [CrossRef]
- Arragain, B.; Pelosse, M.; Thompson, A.; Cusack, S. Structural and functional analysis of the minimal orthomyxovirus-like polymerase of tilapia lake virus from the highly diverged Amnoonviridae family. Nat. Commun. 2023, 14, 8145. [Google Scholar] [CrossRef]
- Venkataraman, S.; Prasad, B.; Selvarajan, R. RNA dependent RNA polymerases: Insights from structure, function and evolution. Viruses 2018, 10, 76. [Google Scholar] [CrossRef] [PubMed]
- Taengphu, S.; Sangsuriya, P.; Phiwsaiya, K.; Debnath, P.P.; Delamare-Deboutteville, J.; Mohan, C.V.; Dong, H.T.; Senapin, S. Genetic diversity of tilapia lake virus genome segment 1 from 2011 to 2019 and a newly validated semi-nested RT-PCR method. Aquaculture 2020, 526, 735423. [Google Scholar] [CrossRef]
- Lueangyangyuen, A.; Senapin, S.; Dong, H.T.; Unajak, S.; Wangkahart, E.; Khunrae, P. Expression and Purification of S5196-272 and S6200-317 proteins from tilapia lake virus (TiLV) and their potential use as vaccines. Protein Expr. Purif. 2021, 190, 106013. [Google Scholar] [CrossRef] [PubMed]
- Mugimba, K.K.; Chengula, A.A.; Wamala, S.; Mwega, E.D.; Kasanga, C.J.; Byarugaba, D.K.; Mdegela, R.H.; Tal, S.; Bornstein, B.; Dishon, A.; et al. Detection of tilapia lake virus (TiLV) infection by PCR in farmed and wild Nile tilapia (Oreochromis niloticus) from Lake Victoria. J. Fish Dis. 2018, 41, 1181–1189. [Google Scholar] [CrossRef]
- Child, H.T.; Airey, G.; Maloney, D.M.; Parker, A.; Wild, J.; McGinley, S.; Evens, N.; Porter, J.; Templeton, K.; Paterson, S.; et al. Comparison of metagenomic and targeted methods for sequencing human pathogenic viruses from wastewater. mBio 2023, 14, e01468-23. [Google Scholar] [CrossRef] [PubMed]
- Cook, R.; Brown, N.; Rihtman, B.; Michniewski, S.; Redgwell, T.; Clokie, M.; Stekel, D.J.; Chen, Y.; Scanlan, D.J.; Hobman, J.L.; et al. The long and short of it: Benchmarking viromics using Illumina, Nanopore and PacBio sequencing technologies. Microb. Genom. 2024, 10, 001198. [Google Scholar] [CrossRef]



| Husseiniya | Bahr Yusef | Ward Island | Om Khalaf | Lake Mariout | |
|---|---|---|---|---|---|
| Muvirus sfmu | |||||
| Muvirus mu | |||||
| Muvirus 108 | |||||
| Gorganvirus isfahan | |||||
| Unclassified Podovirus | |||||
| Betabaculovirus chofumeferanae | |||||
| Shamonda orthobunyavirus | |||||
| Amnoonvirus sp. | |||||
| Pandoravirus sp. | |||||
| Mollivirus sp. |
| Organ | Virus | Husseiniya (%) | Bahr Yusef (%) | Ward Island (%) | Om Khalaf (%) | Lake Mariout (%) |
|---|---|---|---|---|---|---|
| Gills | Muvirus sfmu | 16.4 | 70 | |||
| Muvirus mu | 11.9 | 30 | ||||
| Amnoonvirus sp. | 66.3 | 0 | ||||
| Betabaculovirus chofumeferanae | 0.1 | 0 | ||||
| Shamonda orthobunyavirus | 5.3 | 0 | ||||
| Brain | Muvirus sfmu | 44.8 | 26.1 | 46.7 | 63 | 67.5 |
| Muvirus mu | 36.6 | 15 | 17.7 | 37 | 30 | |
| Amnoonvirus sp. | 7.7 | 31.3 | 1.3 | 0 | 0.1 | |
| Betabaculovirus chofumeferanae | 0.4 | 3 | 33.6 | 0 | 2.4 | |
| Shamonda orthobunyavirus | 9.8 | 24 | 0 | 0 | 0 | |
| Gorganvirus isfahan | 0.7 | 0.6 | 0 | 0 | 0 | |
| Muvirus 108 | 0 | 0 | 0.7 | 0 | 0 | |
| Liver | Muvirus sfmu | 64 | 32.5 | 66.4 | 55 | 80 |
| Muvirus mu | 25.6 | 67.5 | 33.6 | 45 | 20 | |
| Amnoonvirus sp. | 1.9 | 0 | 0 | 0 | 0 | |
| Betabaculovirus chofumeferanae | 0.2 | 0 | 0 | 0 | 0 | |
| Shamonda orthobunyavirus | 8.3 | 0 | 0 | 0 | 0 | |
| Spleen | Muvirus sfmu | 45.65 | 66.5 | 33 | 67 | 43 |
| Muvirus mu | 54.3 | 33.5 | 45 | 33 | 57 | |
| Betabaculovirus chofumeferanae | 0 | 0 | 0.6 | 0 | 0 | |
| Shamonda orthobunyavirus | 0 | 0 | 21.4 | 0 | 0 | |
| Mollivirus sp. | 0.04 | 0 | 0 | 0 | 0 | |
| Pandoravirus sp. | 0.01 | 0 | 0 | 0 | 0 | |
| Kidney | Muvirus sfmu | 37.7 | 58 | 27.4 | ||
| Muvirus mu | 48 | 13.3 | 55.6 | |||
| Amnoonvirus sp. | 11.2 | 10 | 0 | |||
| Betabaculovirus chofumeferanae | 0.3 | 0.3 | 17 | |||
| Shamonda orthobunyavirus | 2.4 | 17.4 | 0 | |||
| Unclassified Podovirus | 0.4 | 0 | 0 | |||
| Muvirus 108 | 0 | 1 | 0 |
| Husseiniya | Bahr Yusef | Ward island | Om Khalaf | Lake Mariout | Total | |
|---|---|---|---|---|---|---|
| Gills | 5 | 2 | 0 | 0 | 0 | 5 |
| Liver | 5 | 2 | 2 | 2 | 2 | 5 |
| Spleen | 4 | 2 | 4 | 2 | 2 | 6 |
| Kidneys | 6 | 6 | 3 | 0 | 0 | 7 |
| Brain | 6 | 6 | 5 | 2 | 4 | 7 |
| Total | 9 | 7 | 5 | 2 | 4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ezzat, A.; Abd El Wahed, A.; Ceruti, A.; El Asely, A.M.; Khalifa, M.S.; Winters, A.D.; Truyen, U.; Shaheen, A.A.; Faisal, M. Exploring the Virome of Nile Tilapia (Oreochromis niloticus) Using Metagenomic Analysis. Pathogens 2025, 14, 935. https://doi.org/10.3390/pathogens14090935
Ezzat A, Abd El Wahed A, Ceruti A, El Asely AM, Khalifa MS, Winters AD, Truyen U, Shaheen AA, Faisal M. Exploring the Virome of Nile Tilapia (Oreochromis niloticus) Using Metagenomic Analysis. Pathogens. 2025; 14(9):935. https://doi.org/10.3390/pathogens14090935
Chicago/Turabian StyleEzzat, Amira, Ahmed Abd El Wahed, Arianna Ceruti, Amel M. El Asely, Mohamed Shawky Khalifa, Andrew D. Winters, Uwe Truyen, Adel A. Shaheen, and Mohamed Faisal. 2025. "Exploring the Virome of Nile Tilapia (Oreochromis niloticus) Using Metagenomic Analysis" Pathogens 14, no. 9: 935. https://doi.org/10.3390/pathogens14090935
APA StyleEzzat, A., Abd El Wahed, A., Ceruti, A., El Asely, A. M., Khalifa, M. S., Winters, A. D., Truyen, U., Shaheen, A. A., & Faisal, M. (2025). Exploring the Virome of Nile Tilapia (Oreochromis niloticus) Using Metagenomic Analysis. Pathogens, 14(9), 935. https://doi.org/10.3390/pathogens14090935








