Bacterial Puppeteering: How the Stealth Bacterium Coxiella Pulls the Cellular Strings
Abstract
1. Introduction
2. Discovery and Characterization of Coxiella burnetii Effector Proteins
2.1. Early Indications and Genomic Insights
2.2. Effector Identification via Bioinformatics and Surrogate Systems
2.3. High-Throughput Screening and Genetic Manipulation
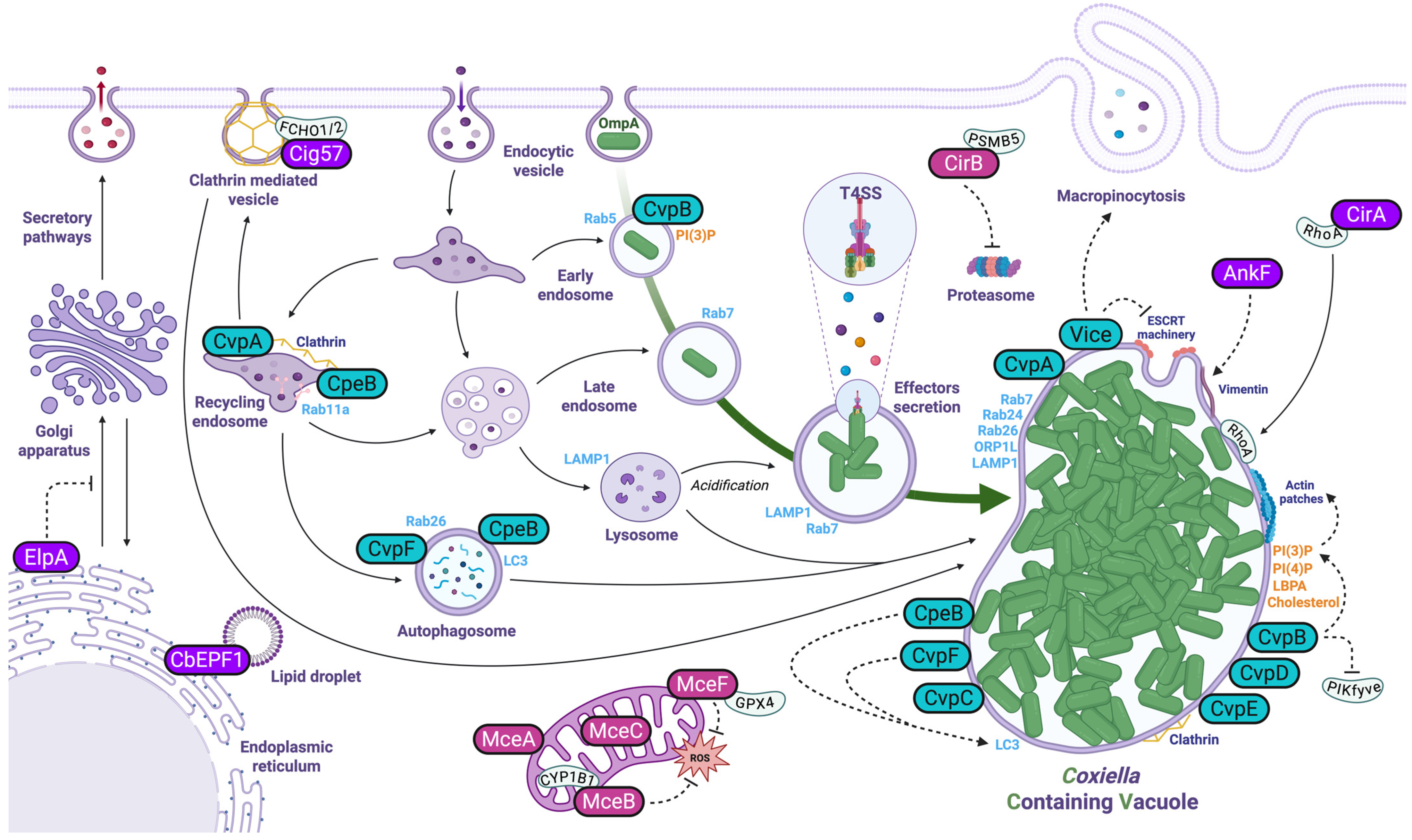
3. Manipulation of Host Cell Endosomal Trafficking
3.1. Effector Proteins Targeting Membrane Trafficking and Endosomal Sorting
3.2. Effector Proteins Manipulating Lipid Metabolism
3.3. Effector Proteins Influencing Autophagy and Host Vesicle Fusion
3.4. Eukaryotic-like Effectors
3.5. Coxiella Effectors and the Manipulation of the Host Cell Cytoskeleton
4. Manipulation of Cell Metabolism
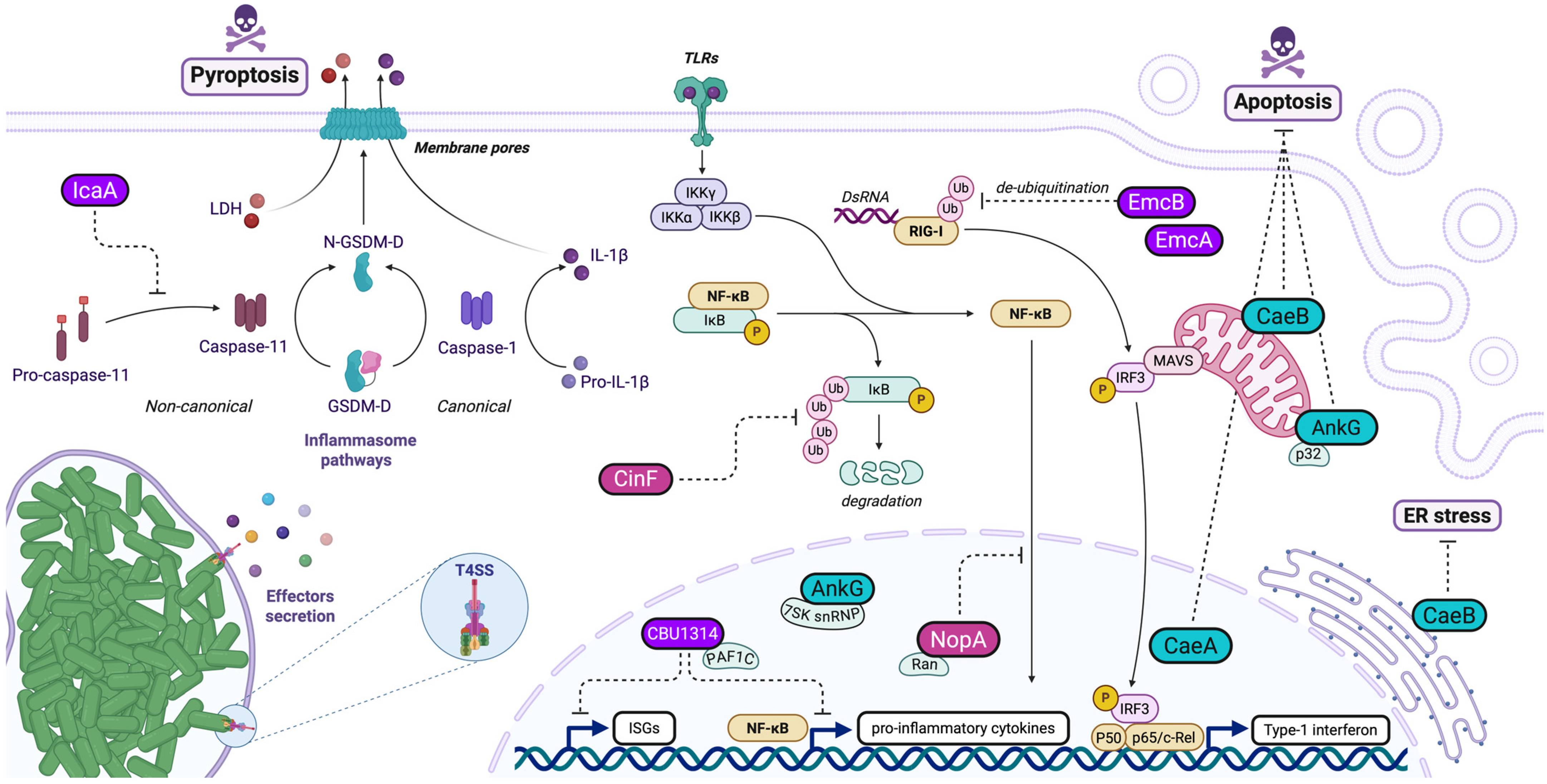
5. Subversion of Host Innate Immunity/Signaling
5.1. Effector Proteins Modulating RIG-I Pathway
5.2. Effector Proteins Interfering with NF-KB Pathway
6. Manipulation of Host Cell Death
6.1. Anti-Apoptotic Effectors in Coxiella burnetii Pathogenesis
6.2. Anti-Pyroptotic Mechanisms in Coxiella burnetii Pathogenesis
7. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Eldin, C.; Mélenotte, C.; Mediannikov, O.; Ghigo, E.; Million, M.; Edouard, S.; Mege, J.-L.; Maurin, M.; Raoult, D. From Q Fever to Coxiella Burnetii Infection: A Paradigm Change. Clin. Microbiol. Rev. 2017, 30, 115–190. [Google Scholar] [CrossRef]
- Burette, M.; Bonazzi, M. From Neglected to Dissected: How Technological Advances Are Leading the Way to the Study of Coxiella Burnetii Pathogenesis. Cell. Microbiol. 2020, 22, e13180. [Google Scholar] [CrossRef] [PubMed]
- Newton, H.J.; Kohler, L.J.; McDonough, J.A.; Temoche-Diaz, M.; Crabill, E.; Hartland, E.L.; Roy, C.R. A Screen of Coxiella Burnetii Mutants Reveals Important Roles for Dot/Icm Effectors and Host Autophagy in Vacuole Biogenesis. PLoS Pathog. 2014, 10, e1004286. [Google Scholar] [CrossRef]
- Larson, C.L.; Beare, P.A.; Voth, D.E.; Howe, D.; Cockrell, D.C.; Bastidas, R.J.; Valdivia, R.H.; Heinzen, R.A. Coxiella Burnetii Effector Proteins That Localize to the Parasitophorous Vacuole Membrane Promote Intracellular Replication. Infect. Immun. 2015, 83, 661–670. [Google Scholar] [CrossRef]
- Martinez, E.; Allombert, J.; Cantet, F.; Lakhani, A.; Yandrapalli, N.; Neyret, A.; Norville, I.H.; Favard, C.; Muriaux, D.; Bonazzi, M. Coxiella Burnetii Effector CvpB Modulates Phosphoinositide Metabolism for Optimal Vacuole Development. Proc. Natl. Acad. Sci. USA 2016, 113, E3260–E3269. [Google Scholar] [CrossRef]
- Kohler, L.J.; Reed, S.R.; Sarraf, S.A.; Arteaga, D.D.; Newton, H.J.; Roy, C.R. Effector Protein Cig2 Decreases Host Tolerance of Infection by Directing Constitutive Fusion of Autophagosomes with the Coxiella-Containing Vacuole. mBio 2016, 7, e01127-16. [Google Scholar] [CrossRef] [PubMed]
- Bird, L.E.; Xu, B.; Hobbs, A.D.; Ziegler, A.R.; Scott, N.E.; Newton, P.; Thomas, D.R.; Edgington-Mitchell, L.E.; Newton, H.J. Coxiella Burnetii Manipulates the Lysosomal Protease Cathepsin B to Facilitate Intracellular Success. Nat. Commun. 2025, 16, 3844. [Google Scholar] [CrossRef] [PubMed]
- Weber, M.M.; Faris, R.; van Schaik, E.J.; McLachlan, J.T.; Wright, W.U.; Tellez, A.; Roman, V.A.; Rowin, K.; Case, E.D.R.; Luo, Z.-Q.; et al. The Type IV Secretion System Effector Protein CirA Stimulates the GTPase Activity of RhoA and Is Required for Virulence in a Mouse Model of Coxiella Burnetii Infection. Infect. Immun. 2016, 84, 2524–2533. [Google Scholar] [CrossRef]
- Weber, M.M.; Chen, C.; Rowin, K.; Mertens, K.; Galvan, G.; Zhi, H.; Dealing, C.M.; Roman, V.A.; Banga, S.; Tan, Y.; et al. Identification of Coxiella Burnetii Type IV Secretion Substrates Required for Intracellular Replication and Coxiella-Containing Vacuole Formation. J. Bacteriol. 2013, 195, 3914–3924. [Google Scholar] [CrossRef]
- Fielden, L.F.; Moffatt, J.H.; Kang, Y.; Baker, M.J.; Khoo, C.A.; Roy, C.R.; Stojanovski, D.; Newton, H.J. A Farnesylated Coxiella Burnetii Effector Forms a Multimeric Complex at the Mitochondrial Outer Membrane during Infection. Infect. Immun. 2017, 85, e01046-16. [Google Scholar] [CrossRef]
- Larson, C.L.; Pullman, W.; Beare, P.A.; Heinzen, R.A. Identification of Type 4B Secretion System Substrates That Are Conserved among Coxiella Burnetii Genomes and Promote Intracellular Growth. Microbiol. Spectr. 2023, 11, e00696-23. [Google Scholar] [CrossRef]
- Martinez, E.; Huc-Brandt, S.; Brelle, S.; Allombert, J.; Cantet, F.; Gannoun-Zaki, L.; Burette, M.; Martin, M.; Letourneur, F.; Bonazzi, M.; et al. The Secreted Protein Kinase CstK from Coxiella Burnetii Influences Vacuole Development and Interacts with the GTPase-Activating Host Protein TBC1D5 CstK Affects C. Burnetii Vacuole Biogenesis. J. Biol. Chem. 2020, 295, 7391–7403. [Google Scholar] [CrossRef]
- Lifshitz, Z.; Burstein, D.; Schwartz, K.; Shuman, H.A.; Pupko, T.; Segal, G. Identification of Novel Coxiella Burnetii Icm/Dot Effectors and Genetic Analysis of Their Involvement in Modulating a Mitogen-Activated Protein Kinase Pathway. Infect. Immun. 2014, 82, 3740–3752. [Google Scholar] [CrossRef]
- Fu, M.; Liu, Y.; Wang, G.; Wang, P.; Zhang, J.; Chen, C.; Zhao, M.; Zhang, S.; Jiao, J.; Ouyang, X.; et al. A Protein–Protein Interaction Map Reveals That the Coxiella Burnetii Effector CirB Inhibits Host Proteasome Activity. PLoS Pathog. 2022, 18, e1010660. [Google Scholar] [CrossRef] [PubMed]
- Pechstein, J.; Schulze-Luehrmann, J.; Bisle, S.; Cantet, F.; Beare, P.A.; Ölke, M.; Bonazzi, M.; Berens, C.; Lührmann, A. The Coxiella Burnetii T4SS Effector AnkF Is Important for Intracellular Replication. Front. Cell. Infect. Microbiol. 2020, 10, 559915. [Google Scholar] [CrossRef] [PubMed]
- Larson, C.L.; Beare, P.A.; Heinzen, R.A. Dependency of Coxiella Burnetii Type 4B Secretion on the Chaperone IcmS. J. Bacteriol. 2019, 201, e00431-19. [Google Scholar] [CrossRef]
- Zhang, Y.; Fu, J.; Liu, S.; Wang, L.; Qiu, J.; van Schaik, E.J.; Samuel, J.E.; Song, L.; Luo, Z.-Q. Coxiella Burnetii Inhibits Host Immunity by a Protein Phosphatase Adapted from Glycolysis. Proc. Natl. Acad. Sci. USA 2022, 119, e2110877119. [Google Scholar] [CrossRef] [PubMed]
- Crabill, E.; Schofield, W.B.; Newton, H.J.; Goodman, A.L.; Roy, C.R. Dot/Icm-Translocated Proteins Important for Biogenesis of the Coxiella Burnetii-Containing Vacuole Identified by Screening of an Effector Mutant Sublibrary. Infect. Immun. 2018, 86, e00758-17. [Google Scholar] [CrossRef]
- Siadous, F.A.; Cantet, F.; Schaik, E.V.; Burette, M.; Allombert, J.; Lakhani, A.; Bonaventure, B.; Goujon, C.; Samuel, J.; Bonazzi, M.; et al. Coxiella Effector Protein CvpF Subverts RAB26-Dependent Autophagy to Promote Vacuole Biogenesis and Virulence. Autophagy 2020, 17, 706–722. [Google Scholar] [CrossRef]
- Carey, K.L.; Newton, H.J.; Lührmann, A.; Roy, C.R. The Coxiella Burnetii Dot/Icm System Delivers a Unique Repertoire of Type IV Effectors into Host Cells and Is Required for Intracellular Replication. PLoS Pathog. 2011, 7, e1002056. [Google Scholar] [CrossRef]
- Larson, C.L.; Beare, P.A.; Howe, D.; Heinzen, R.A. Coxiella Burnetii Effector Protein Subverts Clathrin-Mediated Vesicular Trafficking for Pathogen Vacuole Biogenesis. Proc. Natl. Acad. Sci. USA 2013, 110, E4770–E4779. [Google Scholar] [CrossRef]
- Lührmann, A.; Nogueira, C.V.; Carey, K.L.; Roy, C.R. Inhibition of Pathogen-Induced Apoptosis by a Coxiella Burnetii Type IV Effector Protein. Proc. Natl. Acad. Sci. USA 2010, 107, 18997–19001. [Google Scholar] [CrossRef]
- Cordsmeier, A.; Rinkel, S.; Jeninga, M.; Schulze-Luehrmann, J.; Ölke, M.; Schmid, B.; Hasler, D.; Meister, G.; Häcker, G.; Petter, M.; et al. The Coxiella Burnetii T4SS Effector Protein AnkG Hijacks the 7SK Small Nuclear Ribonucleoprotein Complex for Reprogramming Host Cell Transcription. PLoS Pathog. 2022, 18, e1010266. [Google Scholar] [CrossRef]
- Eckart, R.A.; Bisle, S.; Schulze-Luehrmann, J.; Wittmann, I.; Jantsch, J.; Schmid, B.; Berens, C.; Lührmann, A. Antiapoptotic Activity of Coxiella Burnetii Effector Protein AnkG Is Controlled by P32-Dependent Trafficking. Infect. Immun. 2014, 82, 2763–2771. [Google Scholar] [CrossRef]
- Fielden, L.F.; Scott, N.E.; Palmer, C.S.; Khoo, C.A.; Newton, H.J.; Stojanovski, D. Proteomic Identification of Coxiella Burnetii Effector Proteins Targeted to the Host Cell Mitochondria During Infection. Mol. Cell. Proteom. 2021, 20, 100005. [Google Scholar] [CrossRef] [PubMed]
- Samuel, J.; Arunima, A.; Niyakan, S.; Butler, S.; Clark, S.; Pinson, A.; Kwak, D.; Qian, X.; de Figueiredo, P.; Schaik, E.V. CYP1B1-AS1 Regulates CYP1B1 to Promote Coxiella Burnetii Pathogenesis by Inhibiting ROS and Host Cell Death. Res. Sq. 2024. [Google Scholar] [CrossRef]
- Burette, M.; Allombert, J.; Lambou, K.; Maarifi, G.; Nisole, S.; Case, E.D.R.; Blanchet, F.P.; Hassen-Khodja, C.; Cabantous, S.; Samuel, J.; et al. Modulation of Innate Immune Signaling by a Coxiella Burnetii Eukaryotic-like Effector Protein. Proc. Natl. Acad. Sci. USA 2020, 117, 13708–13718. [Google Scholar] [CrossRef] [PubMed]
- Weber, M.M.; Faris, R.; McLachlan, J.; Tellez, A.; Wright, W.U.; Galvan, G.; Luo, Z.-Q.; Samuel, J.E. Modulation of the Host Transcriptome by Coxiella Burnetii Nuclear Effector Cbu1314. Microbes Infect. 2016, 18, 336–345. [Google Scholar] [CrossRef]
- Fischer, N.L.; Boyer, M.A.; Bradley, W.P.; Spruce, L.A.; Fazelinia, H.; Shin, S. A Coxiella Burnetii Effector Interacts with the Host PAF1 Complex and Suppresses the Innate Immune Response. bioRxiv 2022. [Google Scholar] [CrossRef]
- Angara, R.K.; Sadi, A.; Gilk, S.D. A Novel Bacterial Effector Protein Mediates ER-LD Membrane Contacts to Regulate Host Lipid Droplets. EMBO Rep. 2024, 25, 5331–5351. [Google Scholar] [CrossRef]
- Duncan-Lowey, J.; Crabill, E.; Jarret, A.; Reed, S.C.O.; Roy, C.R. The Coxiella Burnetii Effector EmcB Is a Deubiquitinase That Inhibits RIG-I Signaling. Proc. Natl. Acad. Sci. USA 2023, 120, e2217602120. [Google Scholar] [CrossRef] [PubMed]
- Klingenbeck, L.; Eckart, R.A.; Berens, C.; Lührmann, A. The Coxiella Burnetii Type IV Secretion System Substrate CaeB Inhibits Intrinsic Apoptosis at the Mitochondrial Level. Cell. Microbiol. 2013, 15, 675–687. [Google Scholar] [CrossRef] [PubMed]
- Bisle, S.; Klingenbeck, L.; Borges, V.; Sobotta, K.; Schulze-Luehrmann, J.; Menge, C.; Heydel, C.; Gomes, J.P.; Lührmann, A. The Inhibition of the Apoptosis Pathway by the Coxiella Burnetii Effector Protein CaeA Requires the EK Repetition Motif, but Is Independent of Survivin. Virulence 2016, 7, 400–412. [Google Scholar] [CrossRef] [PubMed]
- Latomanski, E.A.; Newton, H.J. Interaction between Autophagic Vesicles and the Coxiella-Containing Vacuole Requires CLTC (Clathrin Heavy Chain). Autophagy 2018, 14, 1710–1725. [Google Scholar] [CrossRef]
- Latomanski, E.A.; Newton, P.; Khoo, C.A.; Newton, H.J. The Effector Cig57 Hijacks FCHO-Mediated Vesicular Trafficking to Facilitate Intracellular Replication of Coxiella Burnetii. PLoS Pathog. 2016, 12, e1006101. [Google Scholar] [CrossRef]
- Cunha, L.D.; Ribeiro, J.M.; Fernandes, T.D.; Massis, L.M.; Khoo, C.A.; Moffatt, J.H.; Newton, H.J.; Roy, C.R.; Zamboni, D.S. Inhibition of Inflammasome Activation by Coxiella Burnetii Type IV Secretion System Effector IcaA. Nat. Commun. 2015, 6, 10205. [Google Scholar] [CrossRef]
- Zhao, M.; Zhang, S.; Wan, W.; Zhou, C.; Li, N.; Cheng, R.; Yu, Y.; Ouyang, X.; Zhou, D.; Jiao, J.; et al. Coxiella Burnetii Effector CvpE Maintains Biogenesis of Coxiella-Containing Vacuoles by Suppressing Lysosome Tubulation through Binding PI(3)P and Perturbing PIKfyve Activity on Lysosomes. Virulence 2024, 15, 2350893. [Google Scholar] [CrossRef]
- Bienvenu, A.; Burette, M.; Cantet, F.; Gourdelier, M.; Swain, J.; Cazevieille, C.; Clemente, T.; Sadi, A.; Dupont, C.; Fe, M.L.; et al. The Multifunction Coxiella Effector Vice Stimulates Macropinocytosis and Interferes with the ESCRT Machinery. Proc. Natl. Acad. Sci. USA 2024, 121, e2315481121. [Google Scholar] [CrossRef]
- Voth, D.E.; Beare, P.A.; Howe, D.; Sharma, U.M.; Samoilis, G.; Cockrell, D.C.; Omsland, A.; Heinzen, R.A. The Coxiella Burnetii Cryptic Plasmid Is Enriched in Genes Encoding Type IV Secretion System Substrates. J. Bacteriol. 2011, 193, 1493–1503. [Google Scholar] [CrossRef]
- Beare, P.A.; Sandoz, K.M.; Larson, C.L.; Howe, D.; Kronmiller, B.; Heinzen, R.A. Essential Role for the Response Regulator PmrA in Coxiella Burnetii Type 4B Secretion and Colonization of Mammalian Host Cells. J. Bacteriol. 2014, 196, 1925–1940. [Google Scholar] [CrossRef]
- Seshadri, R.; Paulsen, I.T.; Eisen, J.A.; Read, T.D.; Nelson, K.E.; Nelson, W.C.; Ward, N.L.; Tettelin, H.; Davidsen, T.M.; Beanan, M.J.; et al. Complete Genome Sequence of the Q-Fever Pathogen Coxiella Burnetii. Proc. Natl. Acad. Sci. USA 2003, 100, 5455–5460. [Google Scholar] [CrossRef]
- Zusman, T.; Aloni, G.; Halperin, E.; Kotzer, H.; Degtyar, E.; Feldman, M.; Segal, G. The Response Regulator PmrA Is a Major Regulator of the Icm/Dot Type IV Secretion System in Legionella Pneumophila and Coxiella Burnetii. Mol. Microbiol. 2007, 63, 1508–1523. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Banga, S.; Mertens, K.; Weber, M.M.; Gorbaslieva, I.; Tan, Y.; Luo, Z.-Q.; Samuel, J.E. Large-Scale Identification and Translocation of Type IV Secretion Substrates by Coxiella Burnetii. Proc. Natl. Acad. Sci. USA 2010, 107, 21755–21760. [Google Scholar] [CrossRef]
- Samoilis, G.; Aivaliotis, M.; Vranakis, I.; Papadioti, A.; Tselentis, Y.; Tsiotis, G.; Psaroulaki, A. Proteomic Screening for Possible Effector Molecules Secreted by the Obligate Intracellular Pathogen Coxiella Burnetii. J. Proteome Res. 2010, 9, 1619–1626. [Google Scholar] [CrossRef] [PubMed]
- Yadav, A.; Brewer, M.N.; Elshahed, M.S.; Shaw, E.I. Comparative Transcriptomics and Genomics from Continuous Axenic Media Growth Identifies Coxiella Burnetii Intracellular Survival Strategies. bioRxiv 2023. [Google Scholar] [CrossRef]
- Vogel, J.P. Turning a Tiger into a House Cat: Using Legionella Pneumophila to Study Coxiella Burnetii. Trends Microbiol. 2004, 12, 103–105. [Google Scholar] [CrossRef]
- Noroy, C.; Lefrançois, T.; Meyer, D.F. Searching Algorithm for Type IV Effector Proteins (S4TE) 2.0: Improved Tools for Type IV Effector Prediction, Analysis and Comparison in Proteobacteria. PLoS Comput. Biol. 2019, 15, e1006847. [Google Scholar] [CrossRef]
- Herweg, J.-A.; Hansmeier, N.; Otto, A.; Geffken, A.C.; Subbarayal, P.; Prusty, B.K.; Becher, D.; Hensel, M.; Schaible, U.E.; Rudel, T.; et al. Purification and Proteomics of Pathogen-Modified Vacuoles and Membranes. Front. Cell. Infect. Microbiol. 2015, 5, 48. [Google Scholar] [CrossRef]
- Padmanabhan, B.; Fielden, L.F.; Hachani, A.; Newton, P.; Thomas, D.R.; Cho, H.-J.; Khoo, C.A.; Stojanovski, D.; Roy, C.R.; Scott, N.E.; et al. Biogenesis of the Spacious Coxiella-Containing Vacuole Depends on Host Transcription Factors TFEB and TFE3. Infect. Immun. 2019, 88, e00534-19. [Google Scholar] [CrossRef]
- Sandoz, K.M.; Popham, D.L.; Beare, P.A.; Sturdevant, D.E.; Hansen, B.; Nair, V.; Heinzen, R.A. Transcriptional Profiling of Coxiella Burnetii Reveals Extensive Cell Wall Remodeling in the Small Cell Variant Developmental Form. PLoS ONE 2016, 11, e0149957. [Google Scholar] [CrossRef] [PubMed]
- Kuley, R.; Bossers-deVries, R.; Smith, H.E.; Smits, M.A.; Roest, H.I.J.; Bossers, A. Major Differential Gene Regulation in Coxiella Burnetii between in Vivo and in Vitro Cultivation Models. BMC Genom. 2015, 16, 953. [Google Scholar] [CrossRef] [PubMed]
- Newton, H.J.; McDonough, J.A.; Roy, C.R. Effector Protein Translocation by the Coxiella Burnetii Dot/Icm Type IV Secretion System Requires Endocytic Maturation of the Pathogen-Occupied Vacuole. PLoS ONE 2013, 8, e54566. [Google Scholar] [CrossRef]
- Martinez, E.; Cantet, F.; Fava, L.; Norville, I.; Bonazzi, M. Identification of OmpA, a Coxiella Burnetii Protein Involved in Host Cell Invasion, by Multi-Phenotypic High-Content Screening. PLoS Pathog. 2014, 10, e1004013. [Google Scholar] [CrossRef]
- Sandoz, K.M.; Beare, P.A.; Cockrell, D.C.; Heinzen, R.A. Complementation of Arginine Auxotrophy for Genetic Transformation of Coxiella Burnetii by Use of a Defined Axenic Medium. Appl. Environ. Microb. 2016, 82, 3042–3051. [Google Scholar] [CrossRef]
- Beare, P.A.; Larson, C.L.; Gilk, S.D.; Heinzen, R.A. Two Systems for Targeted Gene Deletion in Coxiella Burnetii. Appl. Environ. Microb. 2012, 78, 4580–4589. [Google Scholar] [CrossRef]
- Steiner, S.; Roy, C.R. CRISPR-Cas9-Based Approaches for Genetic Analysis and Epistatic Interaction Studies in Coxiella Burnetii. mSphere 2024, 9, e0052324. [Google Scholar] [CrossRef] [PubMed]
- Wachter, S.; Cockrell, D.C.; Miller, H.E.; Virtaneva, K.; Kanakabandi, K.; Darwitz, B.; Heinzen, R.A.; Beare, P.A. The Endogenous Coxiella Burnetii Plasmid Encodes a Functional Toxin–Antitoxin System. Mol. Microbiol. 2022, 118, 744–764. [Google Scholar] [CrossRef]
- Wachter, S.; Bonazzi, M.; Shifflett, K.; Moses, A.S.; Raghavan, R.; Minnick, M.F. A CsrA-Binding, trans-Acting sRNA of Coxiella Burnetii Is Necessary for Optimal Intracellular Growth and Vacuole Formation during Early Infection of Host Cells. J. Bacteriol. 2019, 201, e00524-19. [Google Scholar] [CrossRef]
- Graham, J.G.; Winchell, C.G.; Sharma, U.M.; Voth, D.E. Identification of ElpA, a Coxiella Burnetii Pathotype-Specific Dot/Icm Type IV Secretion System Substrate. Infect. Immun. 2015, 83, 1190–1198. [Google Scholar] [CrossRef]
- Matteis, M.A.D.; Godi, A. PI-Loting Membrane Traffic. Nat. Cell Biol. 2004, 6, 487–492. [Google Scholar] [CrossRef] [PubMed]
- Hilbi, H.; Weber, S.; Finsel, I. Anchors for Effectors: Subversion of Phosphoinositide Lipids by Legionella. Front. Microbiol. 2011, 2, 91. [Google Scholar] [CrossRef]
- Mulye, M.; Samanta, D.; Winfree, S.; Heinzen, R.A.; Gilk, S.D. Elevated Cholesterol in the Coxiella Burnetii Intracellular Niche Is Bacteriolytic. mBio 2017, 8, e02313-16. [Google Scholar] [CrossRef]
- Clemente, T.M.; Ratnayake, R.; Samanta, D.; Augusto, L.; Beare, P.A.; Heinzen, R.A.; Gilk, S.D. Coxiella Burnetii Sterol-Modifying Protein Stmp1 Regulates Cholesterol in the Intracellular Niche. mBio 2022, 13, e03073-21. [Google Scholar] [CrossRef]
- Gilk, S.D.; Beare, P.A.; Heinzen, R.A. Coxiella Burnetii Expresses a Functional Δ24 Sterol Reductase. J. Bacteriol. 2010, 192, 6154–6159. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Weigele, B.A.; Orchard, R.C.; Jimenez, A.; Cox, G.W.; Alto, N.M. A Systematic Exploration of the Interactions between Bacterial Effector Proteins and Host Cell Membranes. Nat. Commun. 2017, 8, 532. [Google Scholar] [CrossRef]
- van Schaik, E.J.; Case, E.D.; Martinez, E.; Bonazzi, M.; Samuel, J.E. The SCID Mouse Model for Identifying Virulence Determinants in Coxiella Burnetii. Front. Cell. Infect. Microbiol. 2017, 7, 25. [Google Scholar] [CrossRef]
- Wang, T.; Wang, C.; Li, C.; Song, L. The Intricate Dance: Host Autophagy and Coxiella Burnetii Infection. Front. Microbiol. 2023, 14, 1281303. [Google Scholar] [CrossRef] [PubMed]
- Berón, W.; Gutierrez, M.G.; Rabinovitch, M.; Colombo, M.I. Coxiella Burnetii Localizes in a Rab7-Labeled Compartment with Autophagic Characteristics. Infect. Immun. 2002, 70, 5816–5821. [Google Scholar] [CrossRef] [PubMed]
- Romano, P.S.; Gutierrez, M.G.; Berón, W.; Rabinovitch, M.; Colombo, M.I. The Autophagic Pathway Is Actively Modulated by Phase II Coxiella Burnetii to Efficiently Replicate in the Host Cell. Cell. Microbiol. 2007, 9, 891–909. [Google Scholar] [CrossRef]
- Gutierrez, M.G.; Vázquez, C.L.; Munafó, D.B.; Zoppino, F.C.M.; Berón, W.; Rabinovitch, M.; Colombo, M.I. Autophagy Induction Favours the Generation and Maturation of the Coxiella-replicative Vacuoles. Cell. Microbiol. 2005, 7, 981–993. [Google Scholar] [CrossRef]
- Winchell, C.G.; Graham, J.G.; Kurten, R.C.; Voth, D.E. Coxiella Burnetii Type IV Secretion-Dependent Recruitment of Macrophage Autophagosomes. Infect. Immun. 2014, 82, 2229–2238. [Google Scholar] [CrossRef]
- Fu, M.; Zhang, J.; Zhao, M.; Zhang, S.; Dai, L.; Ouyang, X.; Yu, Y.; Wen, B.; Zhou, D.; Sun, Y.; et al. Coxiella Burnetii Plasmid Effector B Promotes LC3-II Accumulation and Contributes To Bacterial Virulence in a SCID Mouse Model. Infect. Immun. 2022, 90, e00016-22. [Google Scholar] [CrossRef]
- Martyn, J.E.; Gomez-Valero, L.; Buchrieser, C. The Evolution and Role of Eukaryotic-like Domains in Environmental Intracellular Bacteria: The Battle with a Eukaryotic Cell. FEMS Microbiol. Rev. 2022, 46, fuac012. [Google Scholar] [CrossRef]
- Miller, H.E.; Larson, C.L.; Heinzen, R.A. Actin Polymerization in the Endosomal Pathway, but Not on the Coxiella-Containing Vacuole, Is Essential for Pathogen Growth. PLoS Pathog. 2018, 14, e1007005. [Google Scholar] [CrossRef]
- Flores, R.M.O.; Distel, J.S.; Aguilera, M.O.; Berón, W. The Role of Microtubules and the Dynein/Dynactin Motor Complex of Host Cells in the Biogenesis of the Coxiella Burnetii-Containing Vacuole. PLoS ONE 2019, 14, e0209820. [Google Scholar] [CrossRef]
- Salinas, R.P.; Flores, R.M.O.; Distel, J.S.; Aguilera, M.O.; Colombo, M.I.; Berón, W. Coxiella Burnetii Phagocytosis Is Regulated by GTPases of the Rho Family and the RhoA Effectors mDia1 and ROCK. PLoS ONE 2015, 10, e0145211. [Google Scholar] [CrossRef]
- Fahlgren, A.; Westermark, L.; Akopyan, K.; Fällman, M. Cell Type-specific Effects of Yersinia Pseudotuberculosis Virulence Effectors. Cell. Microbiol. 2009, 11, 1750–1767. [Google Scholar] [CrossRef]
- Truong, D.; Boddy, K.C.; Canadien, V.; Brabant, D.; Fairn, G.D.; D’Costa, V.M.; Coyaud, E.; Raught, B.; Pérez-Sala, D.; Park, W.S.; et al. Salmonella Exploits Host Rho GTPase Signalling Pathways through the Phosphatase Activity of SopB. Cell. Microbiol. 2018, 20, e12938. [Google Scholar] [CrossRef]
- Ma, K.; Shu, R.; Liu, H.; Ge, J.; Liu, J.; Lu, Q.; Fu, J.; Liu, X.; Qiu, J. Legionella Effectors SidC/SdcA Ubiquitinate Multiple Small GTPases and SNARE Proteins to Promote Phagosomal Maturation. Cell. Mol. Life Sci. 2024, 81, 249. [Google Scholar] [CrossRef]
- Woida, P.J.; Satchell, K.J.F. The Vibrio Cholerae MARTX Toxin Silences the Inflammatory Response to Cytoskeletal Damage before Inducing Actin Cytoskeleton Collapse. Sci. Signal. 2020, 13, eaaw9447. [Google Scholar] [CrossRef]
- Yang, Z.; Duncan-Lowey, J.K.; Roy, C.R. Identification of a Coxiella Burnetii Outer Membrane Porin Required for Intracellular Replication. Infect. Immun. 2025, 93, e00448-24. [Google Scholar] [CrossRef]
- Loterio, R.K.; Thomas, D.R.; Andrade, W.; Lee, Y.W.; Santos, L.L.; Mascarenhas, D.P.A.; Steiner, T.M.; Chiaratto, J.; Fielden, L.F.; Lopes, L.; et al. Coxiella Co-Opts the Glutathione Peroxidase 4 to Protect the Host Cell from Oxidative Stress–Induced Cell Death. Proc. Natl. Acad. Sci. USA 2023, 120, e2308752120. [Google Scholar] [CrossRef]
- Kapsenberg, M.L. Dendritic-Cell Control of Pathogen-Driven T-Cell Polarization. Nat. Rev. Immunol. 2003, 3, 984–993. [Google Scholar] [CrossRef]
- Medzhitov, R.; Janeway, C., Jr. Innate Immune Recognition: Mechanisms and Pathways. Immunol. Rev. 2000, 173, 89–97. [Google Scholar] [CrossRef]
- Wicherska-Pawłowska, K.; Wróbel, T.; Rybka, J. Toll-Like Receptors (TLRs), NOD-Like Receptors (NLRs), and RIG-I-Like Receptors (RLRs) in Innate Immunity. TLRs, NLRs, and RLRs Ligands as Immunotherapeutic Agents for Hematopoietic Diseases. Int. J. Mol. Sci. 2021, 22, 13397. [Google Scholar] [CrossRef]
- Asrat, S.; Davis, K.M.; Isberg, R.R. Modulation of the Host Innate Immune and Inflammatory Response by Translocated Bacterial Proteins. Cell. Microbiol. 2015, 17, 785–795. [Google Scholar] [CrossRef]
- González, C.M.; Wang, L.; Damania, B. Kaposi’s Sarcoma-Associated Herpesvirus Encodes a Viral Deubiquitinase. J. Virol. 2009, 83, 10224–10233. [Google Scholar] [CrossRef]
- Lamotte, L.-A.; Tafforeau, L. How Influenza A Virus NS1 Deals with the Ubiquitin System to Evade Innate Immunity. Viruses 2021, 13, 2309. [Google Scholar] [CrossRef]
- Johannessen, M.; Askarian, F.; Sangvik, M.; Sollid, J.E. Bacterial Interference with Canonical NFκB Signalling. Microbiology 2013, 159, 2001–2013. [Google Scholar] [CrossRef]
- Nelson, R.H.; Nelson, D.E. Signal Distortion: How Intracellular Pathogens Alter Host Cell Fate by Modulating NF-κB Dynamics. Front. Immunol. 2018, 9, 2962. [Google Scholar] [CrossRef]
- Yang, S.; Deng, Q.; Sun, L.; Zhu, Y.; Dong, K.; Wu, S.; Huang, R.; Li, Y. Salmonella Effector SpvB Inhibits NF-κB Activity via KEAP1-Mediated Downregulation of IKKβ. Front. Cell. Infect. Microbiol. 2021, 11, 641412. [Google Scholar] [CrossRef]
- Kim, D.W.; Lenzen, G.; Page, A.-L.; Legrain, P.; Sansonetti, P.J.; Parsot, C. The Shigella Flexneri Effector OspG Interferes with Innate Immune Responses by Targeting Ubiquitin-Conjugating Enzymes. Proc. Natl. Acad. Sci. USA 2005, 102, 14046–14051. [Google Scholar] [CrossRef]
- de Jong, M.F.; Liu, Z.; Chen, D.; Alto, N.M. Shigella Flexneri Suppresses NF-κB Activation by Inhibiting Linear Ubiquitin Chain Ligation. Nat. Microbiol. 2016, 1, 16084. [Google Scholar] [CrossRef]
- Guan, H.; Fu, J.; Yu, T.; Wang, Z.; Gan, N.; Huang, Y.; Perčulija, V.; Li, Y.; Luo, Z.; Ouyang, S. Molecular Basis of Ubiquitination Catalyzed by the Bacterial Transglutaminase MavC. Adv. Sci. 2020, 7, 2000871. [Google Scholar] [CrossRef]
- Evans, S.M.; Rodino, K.G.; Adcox, H.E.; Carlyon, J.A. Orientia Tsutsugamushi Uses Two Ank Effectors to Modulate NF-κB P65 Nuclear Transport and Inhibit NF-κB Transcriptional Activation. PLoS Pathog. 2018, 14, e1007023. [Google Scholar] [CrossRef]
- Rolhion, N.; Furniss, R.C.D.; Grabe, G.; Ryan, A.; Liu, M.; Matthews, S.A.; Holden, D.W. Inhibition of Nuclear Transport of NF-ĸB P65 by the Salmonella Type III Secretion System Effector SpvD. PLoS Pathog. 2016, 12, e1005653. [Google Scholar] [CrossRef]
- Marazzi, I.; Ho, J.S.Y.; Kim, J.; Manicassamy, B.; Dewell, S.; Albrecht, R.A.; Seibert, C.W.; Schaefer, U.; Jeffrey, K.L.; Prinjha, R.K.; et al. Suppression of the Antiviral Response by an Influenza Histone Mimic. Nature 2012, 483, 428–433. [Google Scholar] [CrossRef]
- Petit, M.J.; Kenaston, M.W.; Pham, O.H.; Nagainis, A.A.; Fishburn, A.T.; Shah, P.S. Nuclear Dengue Virus NS5 Antagonizes Expression of PAF1-Dependent Immune Response Genes. PLoS Pathog. 2021, 17, e1010100. [Google Scholar] [CrossRef]
- Clemente, T.M.; Mulye, M.; Justis, A.V.; Nallandhighal, S.; Tran, T.M.; Gilk, S.D. Coxiella Burnetii Blocks Intracellular Interleukin-17 Signaling in Macrophages. Infect. Immun. 2018, 86, e00532-18. [Google Scholar] [CrossRef]
- Hata, K.; Andoh, A.; Shimada, M.; Fujino, S.; Bamba, S.; Araki, Y.; Okuno, T.; Fujiyama, Y.; Bamba, T. IL-17 Stimulates Inflammatory Responses via NF-κB and MAP Kinase Pathways in Human Colonic Myofibroblasts. Am. J. Physiol. Gastrointest. Liver Physiol. 2002, 282, G1035–G1044. [Google Scholar] [CrossRef]
- Elliott, A.; Schoenlaub, L.; Freches, D.; Mitchell, W.; Zhang, G. Neutrophils Play an Important Role in Protective Immunity against Coxiella Burnetii Infection. Infect. Immun. 2015, 83, 3104–3113. [Google Scholar] [CrossRef]
- Schäfer, W.; Eckart, R.A.; Schmid, B.; Cagköylü, H.; Hof, K.; Muller, Y.A.; Amin, B.; Lührmann, A. Nuclear Trafficking of the Anti-apoptotic Coxiella Burnetii Effector Protein AnkG Requires Binding to P32 and Importin-α1. Cell. Microbiol. 2016, 19, e12634. [Google Scholar] [CrossRef]
- Schäfer, W.; Schmidt, T.; Cordsmeier, A.; Borges, V.; Beare, P.A.; Pechstein, J.; Schulze-Luehrmann, J.; Holzinger, J.; Wagner, N.; Berens, C.; et al. The Anti-Apoptotic Coxiella Burnetii Effector Protein AnkG Is a Strain Specific Virulence Factor. Sci. Rep. 2020, 10, 15396. [Google Scholar] [CrossRef]
- Friedrich, A.; Beare, P.A.; Schulze-Luehrmann, J.; Cordsmeier, A.; Pazen, T.; Sonnewald, S.; Lührmann, A. The Coxiella Burnetii Effector Protein CaeB Modulates Endoplasmatic Reticulum (ER) Stress Signalling and Is Required for Efficient Replication in Galleria Mellonella. Cell. Microbiol. 2021, 23, e13305. [Google Scholar] [CrossRef] [PubMed]
- Boudaher, E.; Shaffer, C.L. Inhibiting Bacterial Secretion Systems in the Fight against Antibiotic Resistance. MedChemComm 2019, 10, 682–692. [Google Scholar] [CrossRef]
- Martinez, E.; Siadous, F.A.; Bonazzi, M. Tiny Architects: Biogenesis of Intracellular Replicative Niches by Bacterial Pathogens. FEMS Microbiol. Rev. 2018, 42, 425–447. [Google Scholar] [CrossRef]
| Gene (CDS) | Acronym | Cellular Function and Target (If Known) | Reference |
|---|---|---|---|
| CBU0021 | CvpB/Cig2 | Interacts with phosphatidylinositol 3-phosphate (PI3P), inhibits PIKfyve activity and stimulates PI3P and LC3 recruitment to the CCV, participates in Cathepsin B removal from the CCV | [3,4,5,6,7] |
| CBU0041 | CirA/CoxCC1 | Stimulates RhoA GTPase activity | [8,9] |
| CBU0077 | MceA | Is farnesylated by the host cell and localizes to the mitochondrial outer membrane | [10] |
| CBU0122 | CvpM | Localizes to Mitochondria and CCV | [11] |
| CBU0175 | CstK | Interacts with TBC1D5 and displays threonine and tyrosine kinase activity | [12] |
| CBU0388 | CetCb2 | Enhances MAP kinase pathway in yeast | [13] |
| CBU0425 | CirB | Interacts with proteasomal subunit PSMB5 and inhibits host proteasome activity | [9,14] |
| CBU0447 | AnkF | Interacts with and recruits vimentin to the CCV | [15,16] |
| CBU0513 | CinF | Displays phosphatase activity and dephosphorylates IkBα | [17,18] |
| CBU0626 | CvpF | Recruits RAB26 to the CCV | [19] |
| CBU0635 | Interferes with host protein secretion | [16,20] | |
| CBU0665 | CvpA | Interacts with clathrin adaptor complex AP2 | [16,21] |
| CBU0781 | AnkG | Interacts with p32 (gClqR), DDX21 and 7SK snRNP to inhibit apoptosis and participate in transcriptional reprogramming | [22,23,24] |
| CBU0937 | MceB/CirC | Interacts with mitochondrial protein CYP1B1 | [9,25,26] |
| CBU1217 | NopA | Interacts with Ran to perturb nucleocytoplasmic trafficking and innate immune signaling | [27] |
| CBU1314 | coxCC6 | Interacts with PAF1C to inhibit NF-κB-, MAPK-, and type I IFN-dependent gene expression | [28,29] |
| CBU1370 | CbEPF1 | Interacts with lipid droplets | [30] |
| CBU1387 | EmcA/Cem6 | Inhibits RIG-I signaling | [31] |
| CBU1425 | MceC | Interacts with mitochondrial protein YME1L | [25] |
| CBU1524 | CaeA | Inhibit intrinsic apoptosis pathway | [32,33] |
| CBU1532 | CaeB | Inhibit intrinsic apoptosis pathway | [32] |
| CBU1751 | Cig57 | Interacts with FCHO2 to co-opt clathrin-mediated trafficking and autophagy | [3,34,35] |
| CBU1823 | IcaA | Inhibits non-canonical inflammasome | [36] |
| CBU1863 | CvpE | Interacts with Phosphatidylinositol 3-phosphate (PI3P), perturbs PIKfyve activity and suppresses lysosomal calcium transient receptor potential channel mucolipin 1 (TRPML1) activity | [4,37] |
| CBU2007 | Vice | Interacts with LBPA/CHMP3/ALIX, stimulates macropinocytosis and inhibits ESCRT machinery | [38] |
| CBU2013 | EmcB | Displays ubiquitin-specific cysteine protease activity and inhibits RIG-I signaling | [31] |
| CBUA0013 | CpeB | Promotes LC3-II accumulation and contributes to virulence in SCID mouse model | [14,16,39] |
| Gene (CDS) | Acronym | Reference |
|---|---|---|
| CBU0072 | AnkA | [16] |
| CBU0080 | [16] | |
| CBU0129 | [16] | |
| CBU0145 | [16] | |
| CBU0201 | AnkC | [27] |
| CBU0295 | [16] | |
| CBU0329 | [16] | |
| CBU0410 | Cig12 | [16] |
| CBU0414 | CoxH1 | [16,18] |
| CBU0505 | Cig14 | [27] |
| CBU0519 | DedA | [27] |
| CBU0534 | [4,16] | |
| CBU0542 | LigA | [27] |
| CBU0547 | [27] | |
| CBU0794 | [16] | |
| CBU0885 | CetCb4 | [4,16] |
| CBU0978 | Cem3 | [18] |
| CBU1024 | [16] | |
| CBU1045 | [16] | |
| CBU1107 | [16] | |
| CBU1108 | [16] | |
| CBU1198 | [11] | |
| CBU1213 | AnkI | [27] |
| CBU1366 | Cig40 | [27] |
| CBU1457 | Cig43 | [27] |
| CBU1460 | [16] | |
| CBU1461 | [16] | |
| CBU1493 | [4,16] | |
| CBU1525 | [16] | |
| CBU1530 | [40] | |
| CBU1543 | [4] | |
| CBU1556 | CvpC | [4,16] |
| CBU1569 | [16] | |
| CBU1594 | MceD | [25] |
| CBU1614 | [16,40] | |
| CBU1676 | [4,16] | |
| CBU1677 | MceE | [25] |
| CBU1685 | [40] | |
| CBU1686 | [40] | |
| CBU1724 | CetCb6 | [27] |
| CBU1752 | [40] | |
| CBU1776 | [16] | |
| CBU1780 | [3] | |
| CBU1790 | [16] | |
| CBU1799 | [27] | |
| CBU1818 | CvpD | [4,16] |
| CBU1819 | [4,16] | |
| CBU1825 | [16] | |
| CBU1863 | [16] | |
| CBU1963 | [16] | |
| CBU2028 | [18] | |
| CBU2052 | CirD | [9,16] |
| CBU2056 | [16] | |
| CBU2059 | CirE | [9] |
| CBUA0006 | CpeA | [39] |
| CBUA0014 | CpeC | [16,39] |
| CBUA0015 | CpeD | [16,39] |
| CBUA0016 | CpeE | [16,39] |
| CBUA0023 | CpeF | [39] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruart, D.; Riedinger, J.; Zitouni, S.; Bienvenu, A.; Bonazzi, M.; Martinez, E. Bacterial Puppeteering: How the Stealth Bacterium Coxiella Pulls the Cellular Strings. Pathogens 2025, 14, 896. https://doi.org/10.3390/pathogens14090896
Ruart D, Riedinger J, Zitouni S, Bienvenu A, Bonazzi M, Martinez E. Bacterial Puppeteering: How the Stealth Bacterium Coxiella Pulls the Cellular Strings. Pathogens. 2025; 14(9):896. https://doi.org/10.3390/pathogens14090896
Chicago/Turabian StyleRuart, Dylan, Juliette Riedinger, Sihem Zitouni, Arthur Bienvenu, Matteo Bonazzi, and Eric Martinez. 2025. "Bacterial Puppeteering: How the Stealth Bacterium Coxiella Pulls the Cellular Strings" Pathogens 14, no. 9: 896. https://doi.org/10.3390/pathogens14090896
APA StyleRuart, D., Riedinger, J., Zitouni, S., Bienvenu, A., Bonazzi, M., & Martinez, E. (2025). Bacterial Puppeteering: How the Stealth Bacterium Coxiella Pulls the Cellular Strings. Pathogens, 14(9), 896. https://doi.org/10.3390/pathogens14090896





