Molecular and Clinical Characterization of Crimean–Congo Hemorrhagic Fever in Bulgaria, 2015–2024
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Data and Sample Collection
2.2. Case Definition
2.3. Serologic Testing
2.4. RNA Extraction
2.5. Real-Time Reverse Transcriptase Polymerase Chain Reaction (RT-qPCR)
2.6. Whole-Genome Sequencing
2.7. Bioinformatic Analysis
2.8. Data Availability
3. Results
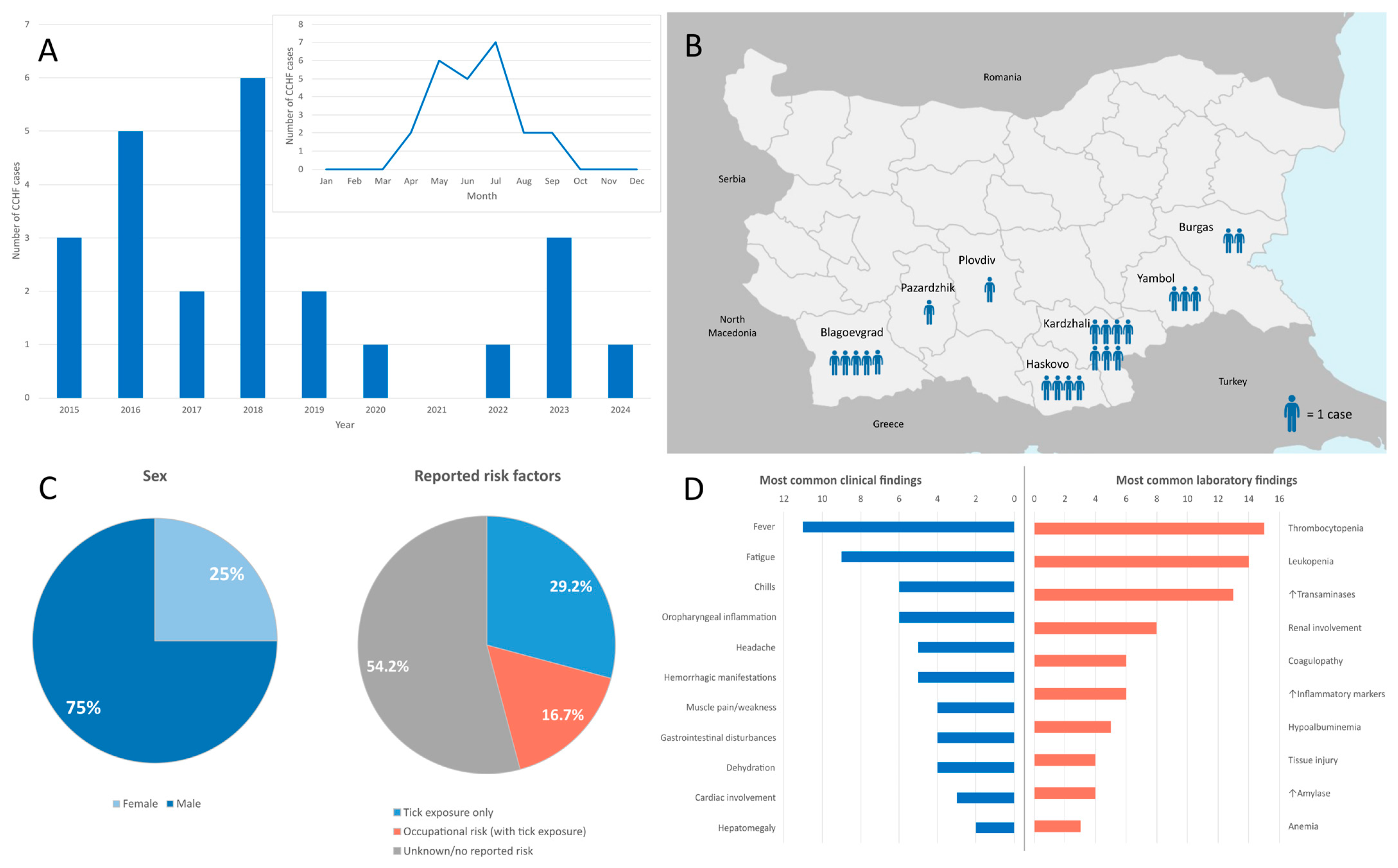
3.1. Epidemiological Characteristics
3.2. Clinical Findings
3.3. Fatal Cases
3.4. Laboratory Findings
3.5. CCHF Laboratory Results
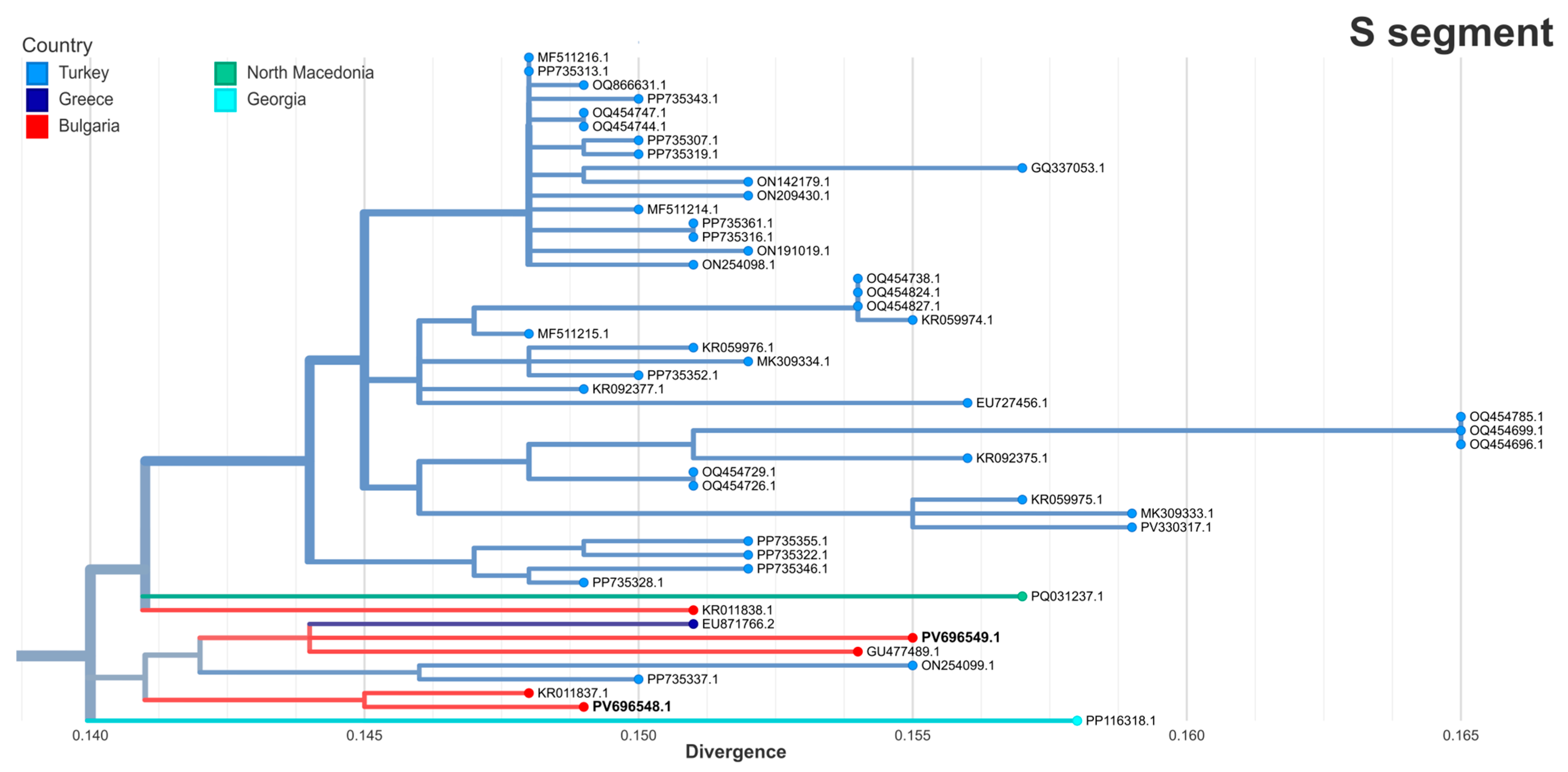
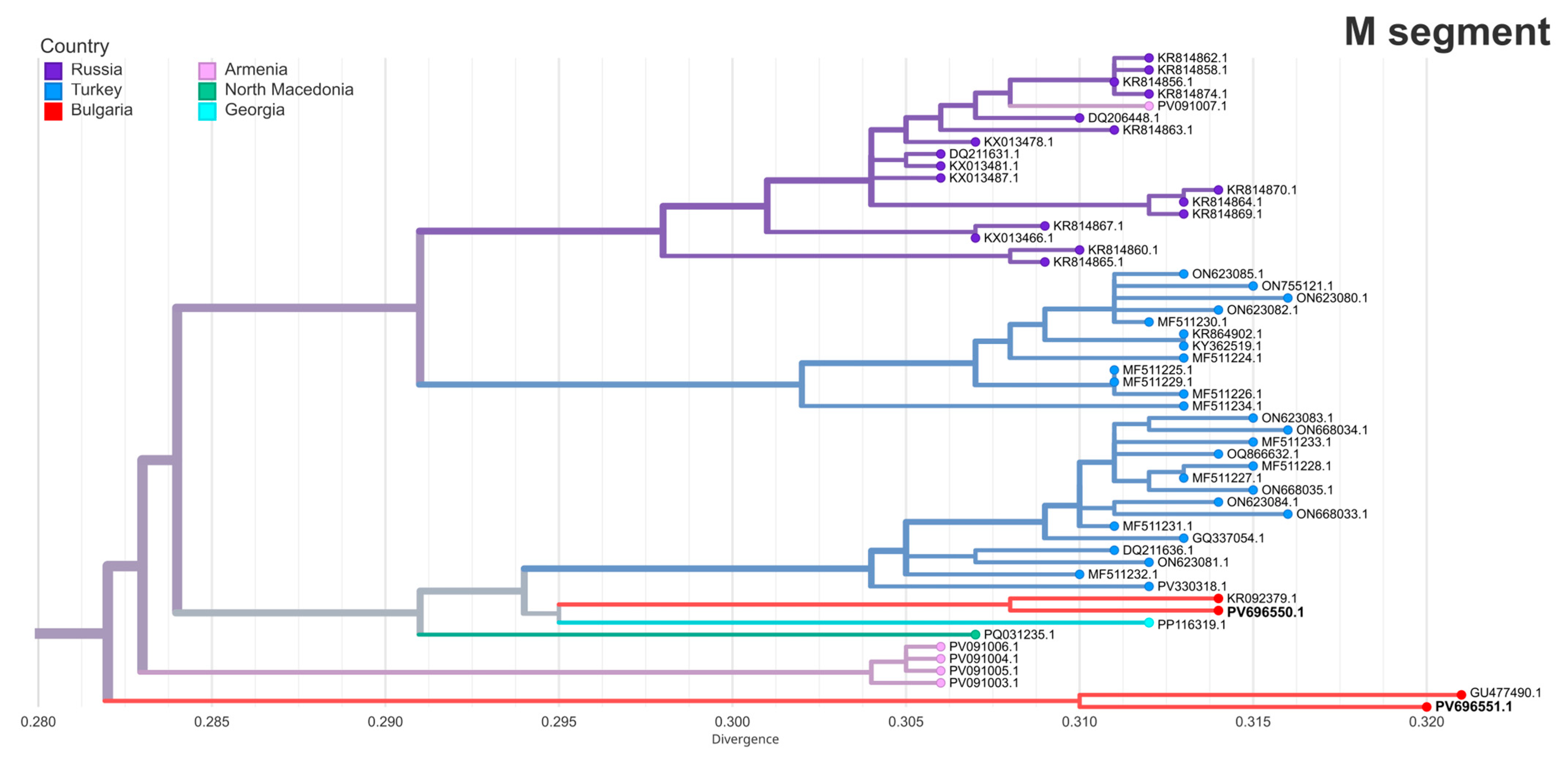
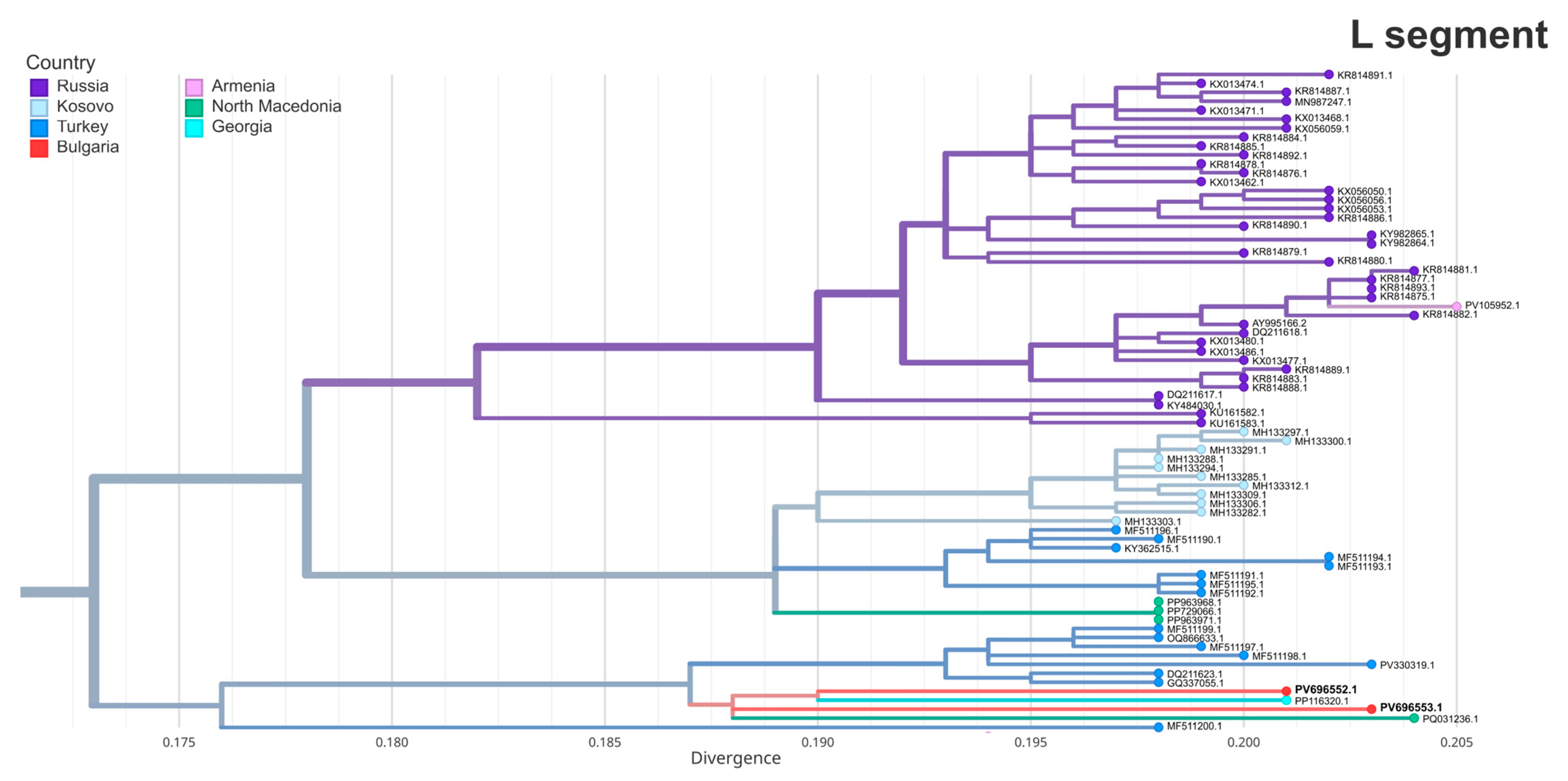
3.6. Phylogenetic Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CCHF | Crimean–Congo hemorrhagic fever |
| CCHFV | Crimean–Congo hemorrhagic fever virus |
| ELISA | Enzyme-linked immunosorbent assay |
| RT-qPCR | Real-time reverse transcriptase polymerase chain reaction |
| GPC | Glycoprotein precursor |
References
- Hawman, D.W.; Feldmann, H. Crimean–Congo Haemorrhagic Fever Virus. Nat. Rev. Microbiol. 2023, 21, 463–477. [Google Scholar] [CrossRef]
- Juanes, H.M.L.; Carbonell, C.; Sendra, B.F.; López-Bernus, A.; Bahamonde, A.; Orfao, A.; Lista, C.V.; Ledesma, M.S.; Negredo, A.I.; Rodríguez-Alonso, B.; et al. Crimean-Congo Hemorrhagic Fever, Spain, 2013–2021. Emerg. Infect. Dis. 2023, 29, 252–259. [Google Scholar] [CrossRef]
- Choubdar, N.; Karimian, F.; Koosha, M.; Oshaghi, M.A. An Integrated Overview of the Bacterial Flora Composition of Hyalomma Anatolicum, the Main Vector of CCHF. PLoS Negl. Trop. Dis. 2021, 15, e0009480. [Google Scholar] [CrossRef]
- Bhowmick, S.; Kasi, K.K.; Gethmann, J.; Fischer, S.; Conraths, F.J.; Sokolov, I.M.; Lentz, H.H.K. Ticks on the Run: A Mathematical Model of Crimean-Congo Haemorrhagic Fever (CCHF)—Key Factors for Transmission. Epidemiologia 2022, 3, 116–134. [Google Scholar] [CrossRef]
- Celina, S.S.; Italiya, J.; Tekkara, A.O.; Černý, J. Crimean-Congo Haemorrhagic Fever Virus in Ticks, Domestic, and Wild Animals. Front. Vet. Sci. 2024, 11, 1513123. [Google Scholar] [CrossRef]
- Serretiello, E.; Astorri, R.; Chianese, A.; Stelitano, D.; Zannella, C.; Folliero, V.; Santella, B.; Galdiero, M.; Franci, G.; Galdiero, M. The Emerging Tick-Borne Crimean-Congo Haemorrhagic Fever Virus: A Narrative Review. Travel. Med. Infect. Dis. 2020, 37, 10187. [Google Scholar] [CrossRef]
- Bernard, C.; Kukla, C.J.; Rakotoarivony, I.; Duhayon, M.; Stachurski, F.; Huber, K.; Giupponi, C.; Zortman, I.; Holzmuller, P.; Pollet, T.; et al. Detection of Crimean–Congo Haemorrhagic Fever Virus in Hyalomma Marginatum Ticks, Southern France, May 2022 and April 2023. Eurosurveillance 2024, 29, 2400023. [Google Scholar] [CrossRef]
- Rasikh, A.S.; Aram, M.M.; Noory, A.T. Clinical and Epidemiological Characteristics of 30 Fatal Cases of Crimean-Congo Hemorrhagic Fever in Kabul, Afghanistan: A Retrospective Observational Study. Infect. Drug Resist. 2023, 16, 3469. [Google Scholar] [CrossRef]
- Semper, A.E.; Olver, J.; Warner, J.; Cehovin, A.; Fay, P.C.; Hart, P.J.; Golding, J.P.; Benassi, V.; Preziosi, M.P.; Al-Asadi, K.H.R.; et al. Research and Product Development for Crimean–Congo Haemorrhagic Fever: Priorities for 2024–30. Lancet Infect. Dis. 2024, 25, e223–e234. [Google Scholar] [CrossRef]
- Eslava, M.; Carlos, S.; Reina, G. Crimean-Congo Hemorrhagic Fever Virus: An Emerging Threat in Europe with a Focus on Epidemiology in Spain. Pathogens 2024, 13, 770. [Google Scholar] [CrossRef]
- Shahhosseini, N.; Wong, G.; Babuadze, G.; Camp, J.V.; Ergonul, O.; Kobinger, G.P.; Chinikar, S.; Nowotny, N. Crimean-Congo Hemorrhagic Fever Virus in Asia, Africa and Europe. Microorganisms 2021, 9, 1907. [Google Scholar] [CrossRef]
- Frank, M.G.; Weaver, G.; Raabe, V. Crimean Congo Hemorrhagic Fever Virus for Clinicians-Virology, Pathogenesis, and Pathology. Emerg. Infect. Dis. 2024, 30, 847–853. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control Cases of Crimean–Congo Haemorrhagic Fever Infected in the EU/EEA, 2013–Present. Available online: https://atlas.ecdc.europa.eu/public/index.aspx?Dataset=27&HealthTopic=10 (accessed on 22 April 2025).
- Papa, A.; Marklewitz, M.; Paraskevopoulou, S.; Garrison, A.R.; Alkhovsky, S.V.; Avsic-Zupanc, T.; Bente, D.A.; Bergeron, E.; Burt, F.; Di Paola, N.; et al. History and Classification of Aigai Virus (Formerly Crimean–Congo Haemorrhagic Fever Virus Genotype VI). J. Gen. Virol. 2022, 103, 001734. [Google Scholar] [CrossRef]
- Bente, D.A.; Forrester, N.L.; Watts, D.M.; McAuley, A.J.; Whitehouse, C.A.; Bray, M. Crimean-Congo Hemorrhagic Fever: History, Epidemiology, Pathogenesis, Clinical Syndrome and Genetic Diversity. Antivir. Res. 2013, 100, 159–189. [Google Scholar] [CrossRef]
- Lukashev, A.N.; Klimentov, A.S.; Smirnova, S.E.; Dzagurova, T.K.; Drexler, J.F.; Gmyl, A.P. Phylogeography of Crimean Congo Hemorrhagic Fever Virus. PLoS ONE 2016, 11, e0166744. [Google Scholar] [CrossRef]
- Panayotova, E.; Papa, A.; Trifonova, I.; Christova, I. Crimean-Congo Hemorrhagic Fever Virus Lineages Europe 1 and Europe 2 in Bulgarian Ticks. Ticks Tick. Borne Dis. 2016, 7, 1024–1028. [Google Scholar] [CrossRef]
- Vladimirova, N.; Minkova, A.; Bogdanov, N.; Petkova, K.; Getsova, Z.; Kamenov, G. Acute Infectious Diseases in Bulgaria in 2023; National center of infectious and parasitic diseases (NCIPD): Sofia, Bulgaria, 2024; Available online: https://ncipd.org/images/Documents/EPI/Analiz_Zarazni_Zabolyavania/Analysis_ZB%20_2023%20FINAL.pdf (accessed on 19 June 2025).
- Norman, F.F.; Arce, O.A.; Díaz-Menéndez, M.; Belhassen-García, M.; González-Sanz, M. Changes in the Epidemiology of Crimean-Congo Hemorrhagic Fever: Impact of Travel and a One Health Approach in the European Region. Travel Med. Infect. Dis. 2025, 64, 102806. [Google Scholar] [CrossRef]
- Baz-Flores, S.; Jiménez-Martín, D.; Peralbo-Moreno, A.; Herraiz, C.; Cano-Terriza, D.; Cuadrado-Matías, R.; García-Bocanegra, I.; Ruiz-Fons, F. Animal Exposure Model for Mapping Crimean-Congo Hemorrhagic Fever Virus Emergence Risk. Emerg. Infect. Dis. 2024, 30, 672–680. [Google Scholar] [CrossRef]
- Fanelli, A.; Christof Schnitzler, J.; De Nardi, M.; Donachie, A.; Capua, I.; Lanave, G.; Buonavoglia, D.; Caceres-Soto, P.; Tizzani, P.; Angela, F.; et al. Epidemic Intelligence Data of Crimean-Congo Haemorrhagic Fever, European Region, 2012 to 2022: A New Opportunity for Risk Mapping of Neglected Diseases. Eurosurveillance 2023, 28, 2200542. [Google Scholar] [CrossRef]
- Ministry of Health Republic of Bulgaria Ordinance 21/18.07.2005 on the Procedure for Registration, Notification, and Reporting of Communicable Diseases in Bulgaria. Available online: https://lex.bg/en/laws/ldoc/2135508238 (accessed on 25 April 2025).
- Garrison, A.R.; Alakbarova, S.; Kulesh, D.A.; Shezmukhamedova, D.; Khodjaev, S.; Endy, T.P.; Paragas, J. Development of a TaqMan®–Minor Groove Binding Protein Assay for the Detection and Quantification of Crimean-Congo Hemorrhagic Fever Virus. Am. J. Trop. Med. Hyg. 2007, 77, 514–520. [Google Scholar] [CrossRef][Green Version]
- Atkinson, B.; Chamberlain, J.; Logue, C.H.; Cook, N.; Bruce, C.; Dowall, S.D.; Hewson, R. Development of a Real-Time RT-PCR Assay for the Detection of Crimean-Congo Hemorrhagic Fever Virus. Vector-Borne Zoonotic Dis. 2012, 12, 786–793. [Google Scholar] [CrossRef]
- Chen, S. Ultrafast One-Pass FASTQ Data Preprocessing, Quality Control, and Deduplication Using Fastp. iMeta 2023, 2, e107. [Google Scholar] [CrossRef]
- Wang, S.; Sundaram, J.P.; Spiro, D. VIGOR, an Annotation Program for Small Viral Genomes. BMC Bioinform. 2010, 11, 451. [Google Scholar] [CrossRef]
- Hadfield, J.; Megill, C.; Bell, S.M.; Huddleston, J.; Potter, B.; Callender, C.; Sagulenko, P.; Bedford, T.; Neher, R.A. Nextstrain: Real-Time Tracking of Pathogen Evolution. Bioinformatics 2018, 34, 4121–4123. [Google Scholar] [CrossRef]
- Huddleston, J.; Hadfield, J.; Sibley, T.R.; Lee, J.; Fay, K.; Ilcisin, M.; Harkins, E.; Bedford, T.; Neher, R.A.; Hodcroft, E.B. Augur: A Bioinformatics Toolkit for Phylogenetic Analyses of Human Pathogens. J. Open Source Softw. 2021, 6, 2906. [Google Scholar] [CrossRef]
- Papa, A.; Papadimitriou, E.; Christova, I. The Bulgarian Vaccine Crimean-Congo Haemorrhagic Fever Virus Strain. Scand. J. Infect. Dis. 2011, 43, 225–229. [Google Scholar] [CrossRef]
- Papa, A.; Dalla, V.; Papadimitriou, E.; Kartalis, G.N.; Antoniadis, A. Emergence of Crimean-Congo Haemorrhagic Fever in Greece. Clin. Microbiol. Infect. 2010, 16, 843–847. [Google Scholar] [CrossRef]
- Jakimovski, D.; Poposki, K.; Dimzova, M.; Cvetanovska, M.; Cana, F.; Bogdan, I.; Cabezas-Cruz, A.; Zana, B.; Lanszki, Z.; Tauber, Z.; et al. Two Human Infections with Diverse Europe-1 Crimean-Congo Hemorrhagic Fever Virus Strains, North Macedonia, 2024. Emerg. Infect. Dis. 2025, 31, 313–317. [Google Scholar] [CrossRef]
- Ma, X.; Ma, Y.; Wang, Z.; Feng, F.; Han, Y.; Sun, X.; Li, L.; Gao, Z.; Wang, M.; Zhang, R. The Single Amino Acid Change of R516K Enables Efficient Generation of Vesicular Stomatitis Virus-Based Crimean–Congo Hemorrhagic Fever Reporter Virus. J. Med. Virol. 2025, 97, e70457. [Google Scholar] [CrossRef]
- Christova, I.; Panayotova, E.; Trifonova, I.; Taseva, E.; Hristova, T.; Ivanova, V. Country-Wide Seroprevalence Studies on Crimean-Congo Hemorrhagic Fever and Hantavirus Infections in General Population of Bulgaria. J. Med. Virol. 2017, 89, 1720–1725. [Google Scholar] [CrossRef]
- Christova, I.; Panayotova, E.; Groschup, M.H.; Trifonova, I.; Tchakarova, S.; Sas, M.A. High Seroprevalence for Crimean–Congo Haemorrhagic Fever Virus in Ruminants in the Absence of Reported Human Cases in Many Regions of Bulgaria. Exp. Appl. Acarol. 2018, 75, 227–234. [Google Scholar] [CrossRef]
- Mertens, M.; Schuster, I.; Sas, M.A.; Vatansever, Z.; Hubalek, Z.; Güven, E.; Deniz, A.; Georgiev, G.; Peshev, R.; Groschup, M.H. Crimean–Congo Hemorrhagic Fever Virus in Bulgaria and Turkey. Vector-Borne Zoonotic Dis. 2016, 16, 619–623. [Google Scholar] [CrossRef]
- Ministry of Agriculture. Annual Report on the State and Development of Agriculture; Ministry of Agriculture: Sofia, Bulgaria, 2023. Available online: https://www.mzh.government.bg/media/filer_public/2023/01/26/ad_2022_en.pdf (accessed on 19 June 2025).
- Vescio, F.M.; Busani, L.; Mughini-Gras, L.; Khoury, C.; Avellis, L.; Taseva, E.; Rezza, G.; Christova, I. Environmental Correlates of Crimean-Congo Haemorrhagic Fever Incidence in Bulgaria. BMC Public. Health 2012, 12, 1116. [Google Scholar] [CrossRef]
- Qaderi, S.; Mardani, M.; Shah, A.; Shah, J.; Bazgir, N.; Sayad, J.; Ghandchi, E.; Samsami, M.; Bagherpour, J.Z. Crimean-Congo Hemorrhagic Fever (CCHF) in Afghanistan: A Retrospective Single Center Study. Int. J. Infect. Dis. 2021, 103, 323–328. [Google Scholar] [CrossRef]
- Balinandi, S.; Whitmer, S.; Mulei, S.; Nyakarahuka, L.; Tumusiime, A.; Kyondo, J.; Baluku, J.; Mutyaba, J.; Mugisha, L.; Malmberg, M.; et al. Clinical and Molecular Epidemiology of Crimean-Congo Hemorrhagic Fever in Humans in Uganda, 2013–2019. Am. J. Trop. Med. Hyg. 2021, 106, 88. [Google Scholar] [CrossRef]
- Bozkurt, I.; Erdeniz, E.H.; Riley, M.J.; Şensoy, L.; Beeching, N.J.; Aydogdu, S.; Leblebicioglu, H.; Korukluoglu, G.; Fletcher, T.E. A Comparison of Clinical and Laboratory Features of Crimean-Congo Hemorrhagic Fever in Children and Adults: A Retrospective Single-Center Cohort Study and Literature Review. Vector Borne Zoonotic Dis. 2025, 25, 81–91. [Google Scholar] [CrossRef]
- Aslani, D.; Salehi-Vaziri, M.; Baniasadi, V.; Jalali, T.; Azad-Manjiri, S.; Mohammadi, T.; Khakifirouz, S.; Fazlalipour, M. Crimean-Congo Hemorrhagic Fever among Children in Iran. Arch. Virol. 2017, 162, 721–725. [Google Scholar] [CrossRef]
- Umair, M.; Rehman, Z.; Whitmer, S.; Mobley, M.; Fahim, A.; Ikram, A.; Salman, M.; Montgomery, J.M.; Klena, J.D. Crimean-Congo Hemorrhagic Fever Virus Diversity and Reassortment, Pakistan, 2017–2020. Emerg. Infect. Dis. 2024, 30, 654–664. [Google Scholar] [CrossRef]
- Khurshid, A.; Hassan, M.; Alam, M.M.; Aamir, U.B.; Rehman, L.; Sharif, S.; Shaukat, S.; Rana, M.S.; Angez, M.; Zaidi, S.S.Z. CCHF Virus Variants in Pakistan and Afghanistan: Emerging Diversity and Epidemiology. J. Clin. Virol. 2015, 67, 25–30. [Google Scholar] [CrossRef]
- Fillâtre, P.; Revest, M.; Tattevin, P. Crimean-Congo Hemorrhagic Fever: An Update. Med. Mal. Infect. 2019, 49, 574–585. [Google Scholar] [CrossRef]
- Raabe, V.N. Diagnostic Testing for Crimean-Congo Hemorrhagic Fever. J. Clin. Microbiol. 2020, 58, 10–1028. [Google Scholar] [CrossRef]
- Frank, M.G.; Weaver, G.; Raabe, V. Crimean-Congo Hemorrhagic Fever Virus for Clinicians—Diagnosis, Clinical Management, and Therapeutics. Emerg. Infect. Dis. 2024, 30, 864–873. [Google Scholar] [CrossRef]
- Muzammil, K.; Rayyani, S.; Abbas Sahib, A.; Gholizadeh, O.; Naji Sameer, H.; Jwad Kazem, T.; Badran Mohammed, H.; Ghafouri Kalajahi, H.; Zainul, R.; Yasamineh, S. Recent Advances in Crimean-Congo Hemorrhagic Fever Virus Detection, Treatment, and Vaccination: Overview of Current Status and Challenges. Biol. Proced. Online 2024, 26, 20. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ngoc, K.; Stoikov, I.; Trifonova, I.; Panayotova, E.; Taseva, E.; Trifonova, I.; Christova, I. Molecular and Clinical Characterization of Crimean–Congo Hemorrhagic Fever in Bulgaria, 2015–2024. Pathogens 2025, 14, 785. https://doi.org/10.3390/pathogens14080785
Ngoc K, Stoikov I, Trifonova I, Panayotova E, Taseva E, Trifonova I, Christova I. Molecular and Clinical Characterization of Crimean–Congo Hemorrhagic Fever in Bulgaria, 2015–2024. Pathogens. 2025; 14(8):785. https://doi.org/10.3390/pathogens14080785
Chicago/Turabian StyleNgoc, Kim, Ivan Stoikov, Ivelina Trifonova, Elitsa Panayotova, Evgenia Taseva, Iva Trifonova, and Iva Christova. 2025. "Molecular and Clinical Characterization of Crimean–Congo Hemorrhagic Fever in Bulgaria, 2015–2024" Pathogens 14, no. 8: 785. https://doi.org/10.3390/pathogens14080785
APA StyleNgoc, K., Stoikov, I., Trifonova, I., Panayotova, E., Taseva, E., Trifonova, I., & Christova, I. (2025). Molecular and Clinical Characterization of Crimean–Congo Hemorrhagic Fever in Bulgaria, 2015–2024. Pathogens, 14(8), 785. https://doi.org/10.3390/pathogens14080785





