Nanopore Workflow for Grapevine Viroid Surveillance in Kazakhstan: Bypassing rRNA Depletion Through Non-Canonical Priming
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Total RNA Extraction
2.2. Library Preparation and Nanopore Sequencing
2.3. Bioinformatic Analysis
2.3.1. Basecalling and Barcode Processing
2.3.2. Taxonomic Classification
2.3.3. Alignment and Assembly
2.3.4. Secondary Structure Prediction
2.3.5. Phylogenetic Analysis
3. Results and Discussion
3.1. RNA Quality
3.2. Viroid Detection and Identification Using the Modified Nanopore Protocol
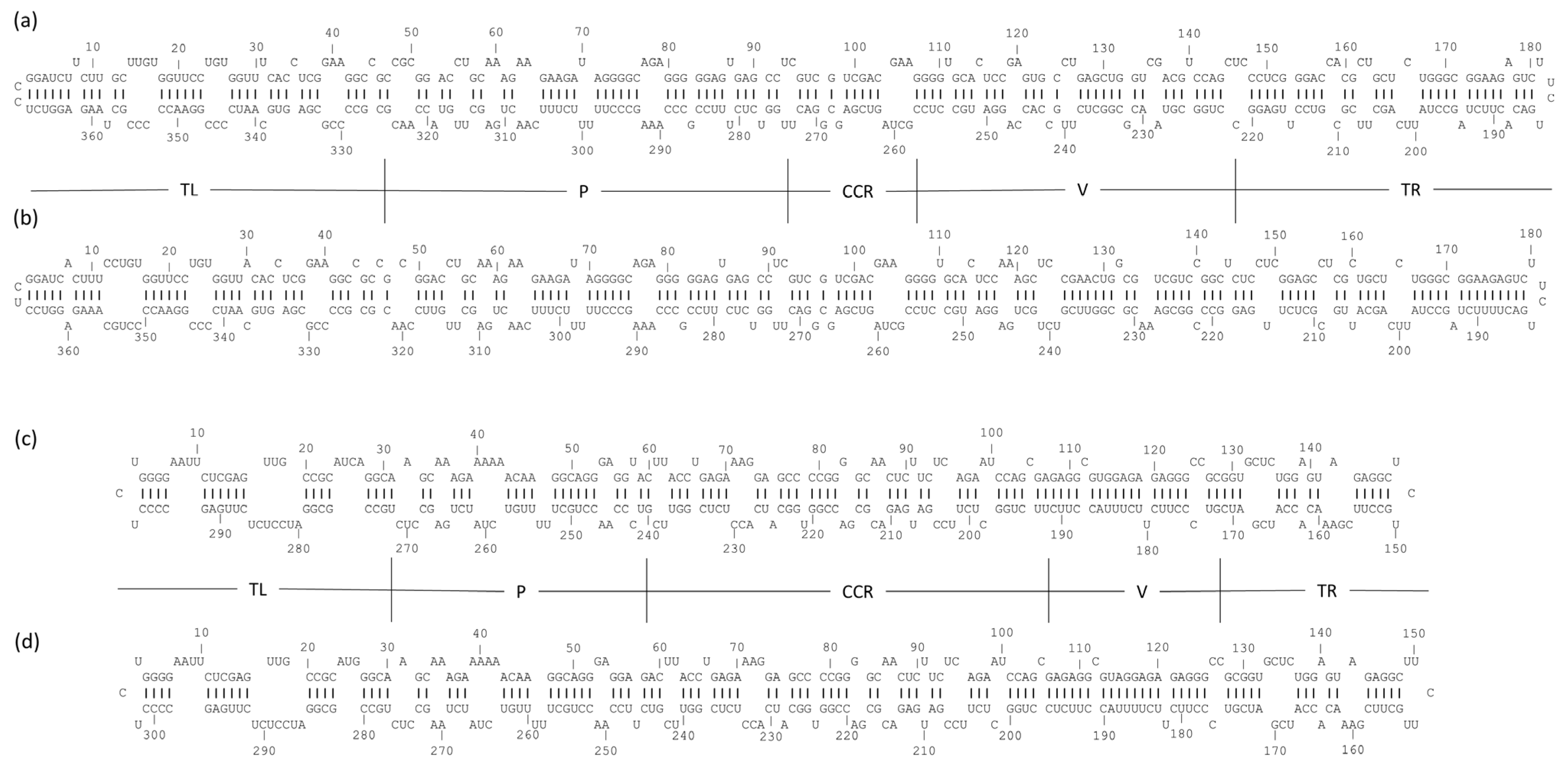
3.3. Predicted Secondary Structure Analysis of Kazakhstani Viroid Isolates
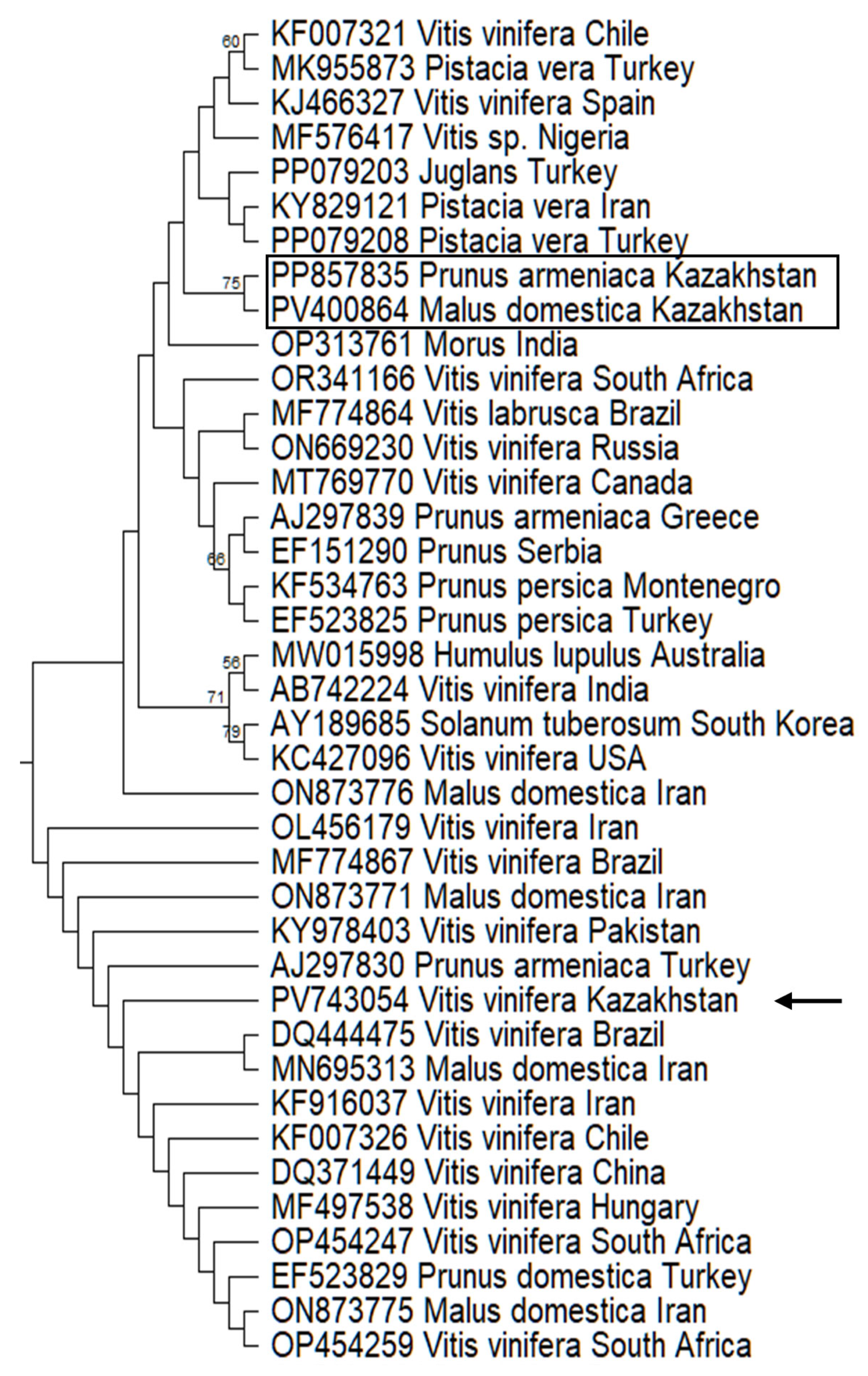
3.4. Phylogenetic Analysis of Kazakhstani Viroid Isolates
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| GYSVd1 | Grapevine yellow speckle viroid 1 |
| HSVd | Hop stunt viroid |
| rRNA | Ribosomal RNA |
| RT | Reverse transcription |
| CRTA | cDNA RT adapter |
| RTP | RT primer |
| HTS | High-throughput sequencing |
| ONT | Oxford Nanopore Technologies |
| CTAB | Cetyltrimethylammonium bromide |
References
- Hadidi, A.; Vidalakis, G.; Sano, T. Chapter 2—Economic Significance of Fruit Tree and Grapevine Viroids. In Viroids and Satellites; Hadidi, A., Flores, R., Randles, J.W., Palukaitis, P., Eds.; Academic Press: Boston, MA, USA, 2017; pp. 15–25. [Google Scholar]
- Basso, M.F.; Fajardo, T.V.M.; Saldarelli, P. Grapevine Virus Diseases:Economic Impact and Current Advances in Viral Prospection and Management. Rev. Bras. Frutic. 2017, 39, 1–22. [Google Scholar] [CrossRef]
- Lee, B.D.; Neri, U.; Oh, C.J.; Simmonds, P.; Koonin, E.V. ViroidDB: A database of viroids and viroid-like circular RNAs. Nucleic Acids Res. 2022, 50, D432–D438. [Google Scholar] [CrossRef]
- Zhang, Y.; Nie, Y.; Wang, L.; Wu, J. Viroid Replication, Movement, and the Host Factors Involved. Microorganisms 2024, 12, 565. [Google Scholar] [CrossRef]
- Navrotskaya, E.; Porotikova, E.; Yurchenko, E.; Galbacs, Z.N.; Varallyay, E.; Vinogradova, S. High-Throughput Sequencing of Small RNAs for Diagnostics of Grapevine Viruses and Viroids in Russia. Viruses 2021, 13, 2432. [Google Scholar] [CrossRef] [PubMed]
- Hadidi, A.; Sun, L.; Randles, J.W. Modes of Viroid Transmission. Cells 2022, 11, 719. [Google Scholar] [CrossRef] [PubMed]
- Chupradit, S.; Huy, D.T.N.; Hachem, K.; Shichiyakh, R.A.; Bokov, D.; Mahmudiono, T.; Al-Rekaby, H.Q.; Kadhim, M.M.; Thangavelu, L. Agrobiological evaluations of newly introduced grapes varieties under climatic conditions of the south of Kazakhstan. Braz. J. Biol. 2022, 84, e258275. [Google Scholar] [CrossRef] [PubMed]
- Pozharskiy, A.S.; Aubakirova, K.P.; Gritsenko, D.A.; Tlevlesov, N.I.; Karimov, N.Z.; Galiakparov, N.N.; Ryabushkina, N.A. Genotyping and morphometric analysis of Kazakhstani grapevine cultivars versus Asian and European cultivars. Genet. Mol. Res. 2020, 19, 18482. [Google Scholar]
- OIV Guidelines for the Harmonization of Requirements for Exchange of Viticultural Plant Material: Phytosanitary and Generic Aspects. Available online: https://www.oiv.int/node/2838 (accessed on 9 July 2025).
- Costa, L.C.; Atha, B.; Hu, X.; Lamour, K.; Yang, Y.; O’Connell, M.; McFarland, C.; Foster, J.A.; Hurtado-Gonzales, O.P. High-throughput detection of a large set of viruses and viroids of pome and stone fruit trees by multiplex PCR-based amplicon sequencing. Front. Plant Sci. 2022, 13, 1072768. [Google Scholar] [CrossRef]
- Boonham, N.; Kreuze, J.; Winter, S.; van der Vlugt, R.; Bergervoet, J.; Tomlinson, J.; Mumford, R. Methods in virus diagnostics: From ELISA to next generation sequencing. Virus Res. 2014, 186, 20–31. [Google Scholar] [CrossRef]
- Gaafar, Y.Z.A.; Ziebell, H. Comparative study on three viral enrichment approaches based on RNA extraction for plant virus/viroid detection using high-throughput sequencing. PLoS ONE 2020, 15, e0237951. [Google Scholar] [CrossRef]
- Pecman, A.; Adams, I.; Gutiérrez-Aguirre, I.; Fox, A.; Boonham, N.; Ravnikar, M.; Kutnjak, D. Systematic Comparison of Nanopore and Illumina Sequencing for the Detection of Plant Viruses and Viroids Using Total RNA Sequencing Approach. Front. Microbiol. 2022, 13, 883921. [Google Scholar] [CrossRef]
- Liefting, L.W.; Waite, D.W.; Thompson, J.R. Application of Oxford Nanopore Technology to Plant Virus Detection. Viruses 2021, 13, 1424. [Google Scholar] [CrossRef] [PubMed]
- Japelaghi, R.H.; Haddad, R.; Garoosi, G.-A. Rapid and efficient isolation of high quality nucleic acids from plant tissues rich in polyphenols and polysaccharides. Mol. Biotechnol. 2011, 49, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Rincon, N.; Wood, D.E.; Breitwieser, F.P.; Pockrandt, C.; Langmead, B.; Salzberg, S.L.; Steinegger, M. Metagenome analysis using the Kraken software suite. Nat. Protoc. 2022, 17, 2815–2839. [Google Scholar] [CrossRef]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.S. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Reuter, J.S.; Mathews, D.H. RNAstructure: Software for RNA secondary structure prediction and analysis. BMC Bioinform. 2010, 11, 129. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Suleski, M.; Sanderford, M.; Sharma, S.; Tamura, K.; Battistuzzi, F.U. MEGA12: Molecular Evolutionary Genetic Analysis Version 12 for Adaptive and Green Computing. Mol. Biol. Evol. 2024, 41, msae263. [Google Scholar] [CrossRef]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: Oxford, UK, 2000; p. 333. [Google Scholar]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Amoia, S.S.; Chiumenti, M.; Minafra, A. Application of Nanopore-Based Sequencing to Identify Virus Infections in Woody Plants. Methods Mol. Biol. 2024, 2732, 265–278. [Google Scholar] [CrossRef] [PubMed]
- Flores, R.; Hernández, C.; de Alba, A.E.M.; Daròs, J.A.; Di Serio, F. Viroids and viroid-host interactions. Annu. Rev. Phytopathol. 2005, 43, 117–139. [Google Scholar] [CrossRef]
- Kemenczeiová, J.; Alaxin, P.; Predajňa, L.; Šubr, Z.; Glasa, M.; Achs, A. High prevalence and low genetic diversity of grapevine yellow speckle viroid 1 in Slovakia: Down the rabbit hole of RNA secondary structure and phylogeny. Acta Virol. 2025, 69, 14361. [Google Scholar] [CrossRef]
- Ward, L.I.; Burnip, G.M.; Liefting, L.W.; Harper, S.J.; Clover, G.R.G. First Report of Grapevine yellow speckle viroid 1 and Hop stunt viroid in Grapevine (Vitis vinifera) in New Zealand. Plant Dis. 2011, 95, 617. [Google Scholar] [CrossRef] [PubMed]
- Maddahian, M.; Massumi, H.; Heydarnejad, J.; Hosseinipour, A.; Khezri, A.; Sano, T. Biological and molecular characterization of hop stunt viroid variants from pistachio trees in Iran. J. Phytopathol. 2019, 167, 163–173. [Google Scholar] [CrossRef]
- Röckel, F.; Trapp, O.; Hundemer, M.; Merkel, R.; Moock, C.; Walk, M.; Kecke, S.; Wolck, A.; Ganesch, A. Vitis International Variety Catalogue. Available online: www.vivc.de (accessed on 9 July 2025).


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aubakirova, K.P.; Bakytzhanova, Z.N.; Rakhatkyzy, A.; Yerbolova, L.S.; Malakhova, N.P.; Galiakparov, N.N. Nanopore Workflow for Grapevine Viroid Surveillance in Kazakhstan: Bypassing rRNA Depletion Through Non-Canonical Priming. Pathogens 2025, 14, 782. https://doi.org/10.3390/pathogens14080782
Aubakirova KP, Bakytzhanova ZN, Rakhatkyzy A, Yerbolova LS, Malakhova NP, Galiakparov NN. Nanopore Workflow for Grapevine Viroid Surveillance in Kazakhstan: Bypassing rRNA Depletion Through Non-Canonical Priming. Pathogens. 2025; 14(8):782. https://doi.org/10.3390/pathogens14080782
Chicago/Turabian StyleAubakirova, Karlygash P., Zhibek N. Bakytzhanova, Akbota Rakhatkyzy, Laura S. Yerbolova, Natalya P. Malakhova, and Nurbol N. Galiakparov. 2025. "Nanopore Workflow for Grapevine Viroid Surveillance in Kazakhstan: Bypassing rRNA Depletion Through Non-Canonical Priming" Pathogens 14, no. 8: 782. https://doi.org/10.3390/pathogens14080782
APA StyleAubakirova, K. P., Bakytzhanova, Z. N., Rakhatkyzy, A., Yerbolova, L. S., Malakhova, N. P., & Galiakparov, N. N. (2025). Nanopore Workflow for Grapevine Viroid Surveillance in Kazakhstan: Bypassing rRNA Depletion Through Non-Canonical Priming. Pathogens, 14(8), 782. https://doi.org/10.3390/pathogens14080782





