Molecular Surveillance of Plasmodium spp. Infection in Neotropical Primates from Bahia and Minas Gerais, Brazil
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Approval and Permits
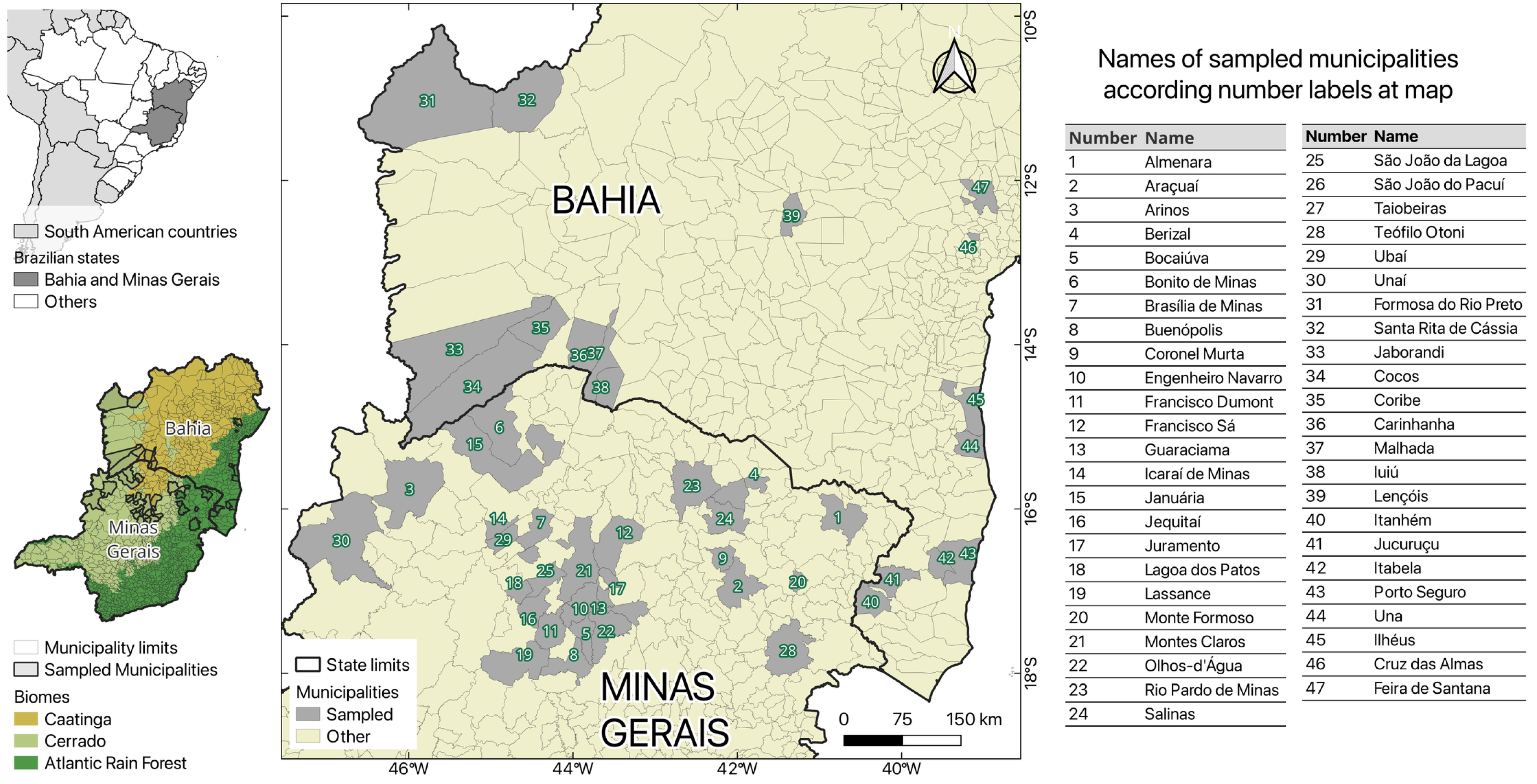
2.2. Study Areas
2.3. Capture of NHPs and Sample Collection
2.4. DNA Extraction
2.5. Mammalian Cytochrome B PCR
2.6. Nested PCR—18S Target
2.7. Nested PCR—Mitochondrial Target and RFLP
2.8. Sequencing
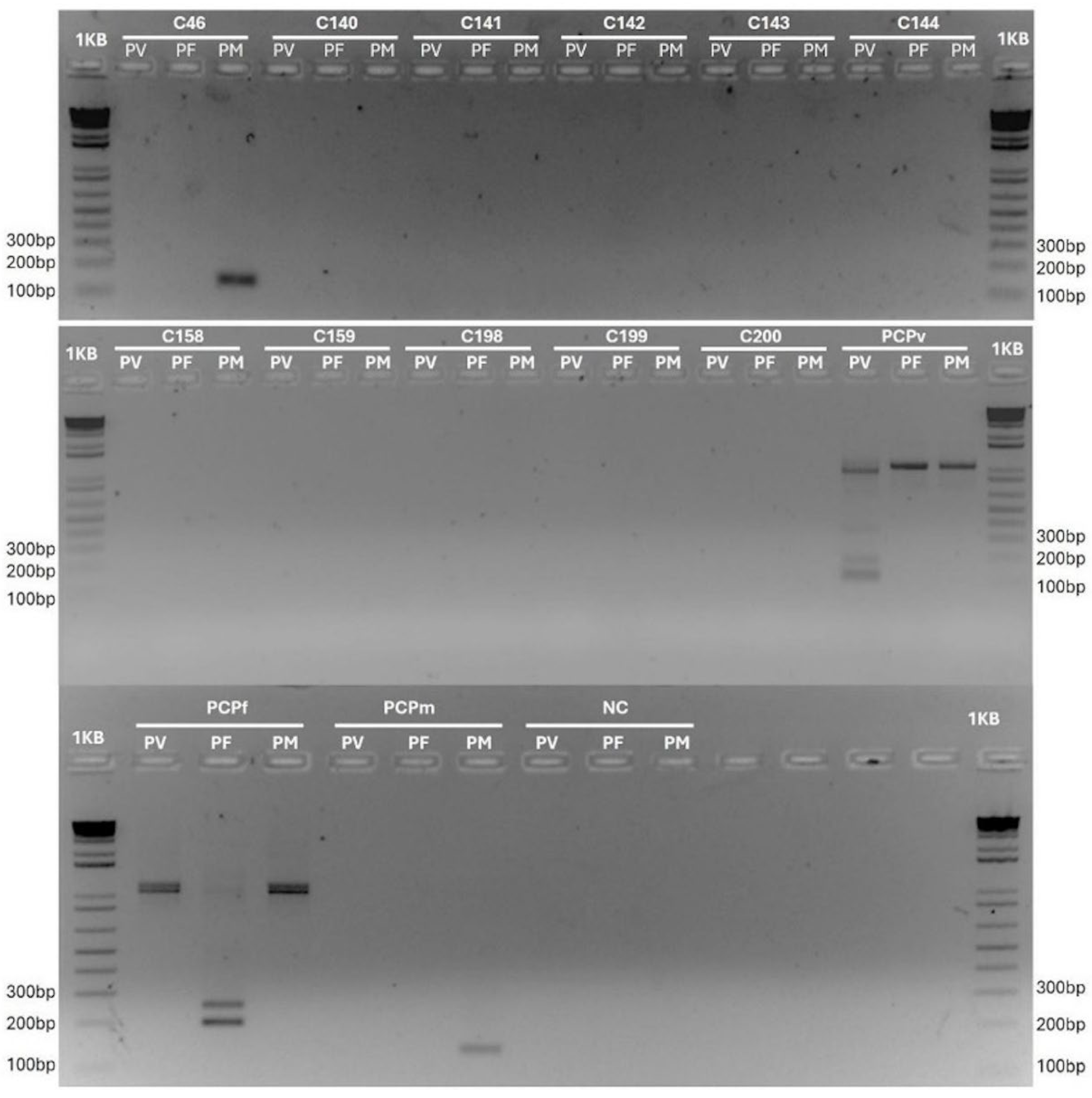
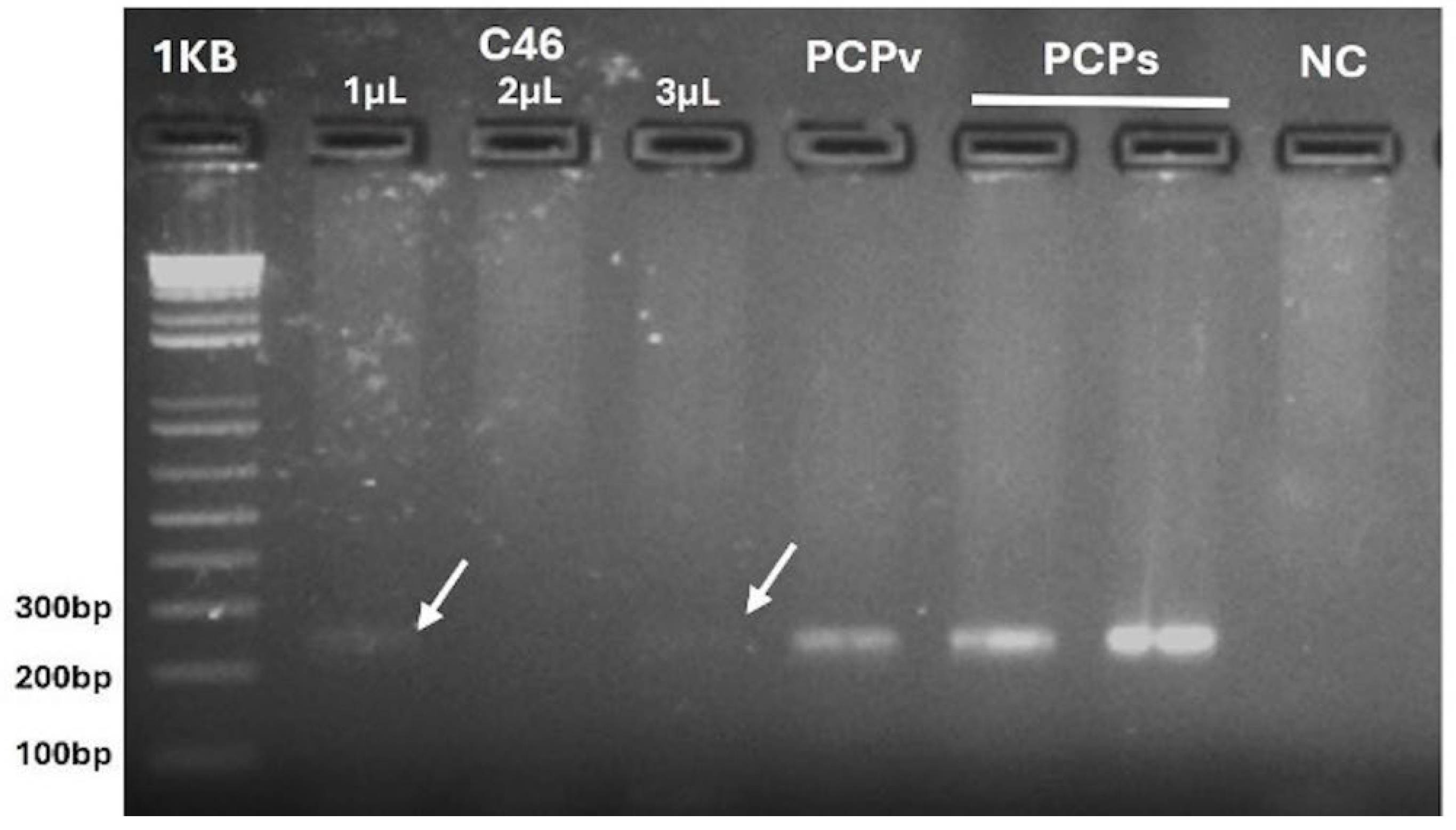
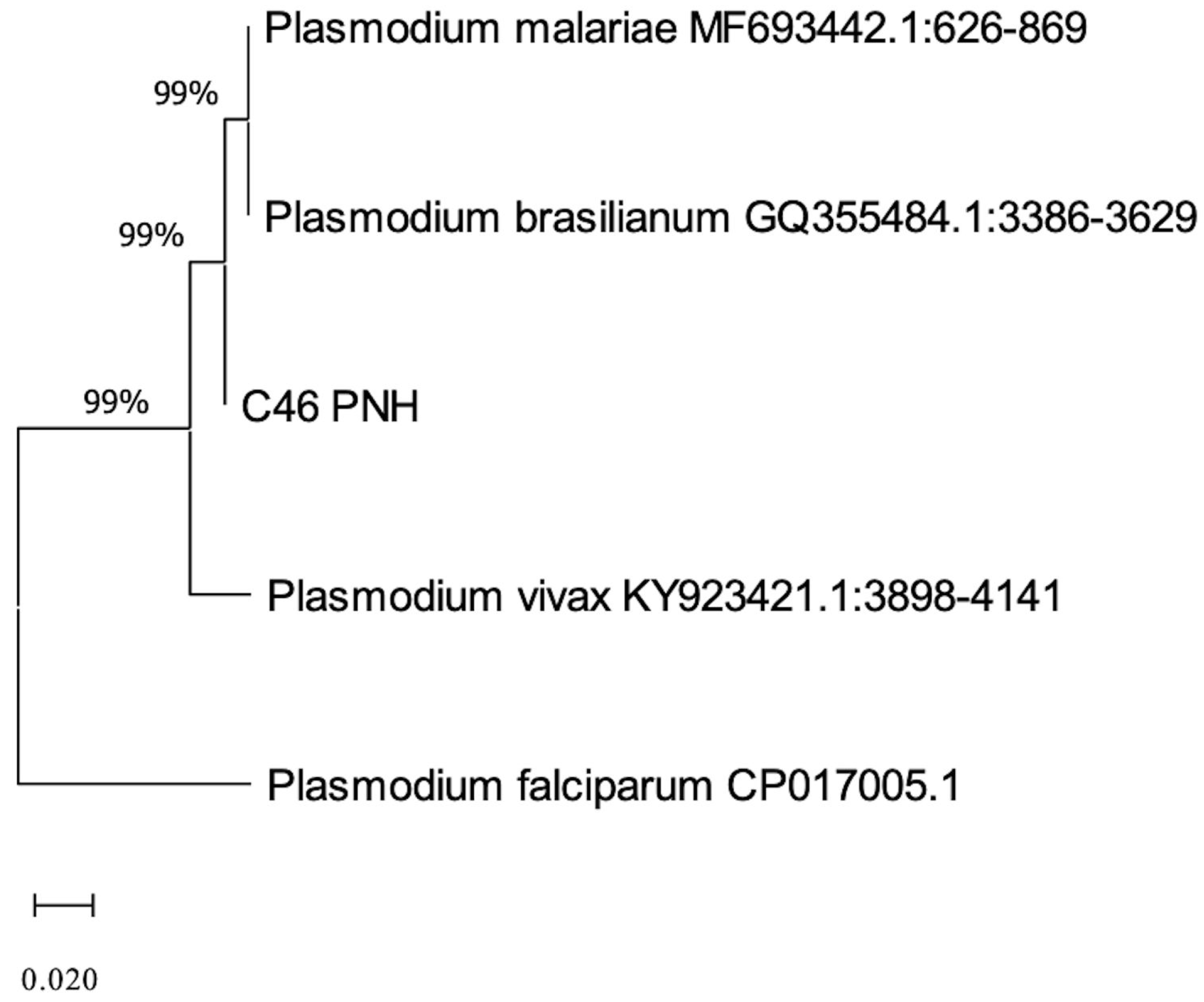
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AF | Atlantic Forest |
| SISBIO | Biodiversity Authorization and Information System |
| ICMBIO | Brazilian Institute of Environment and Natural Resources |
| BA | Bahia |
| MG | Minas Gerais |
References
- Galinski, M.R. Systems Biology of Malaria Explored with Nonhuman Primates. Malar. J. 2022, 21, 177. [Google Scholar] [CrossRef] [PubMed]
- Ministério da Saúde do Brasil. Guia de Vigilância Em Saúde: 6a Edição; Departamento de Ações Estratégicas de Epidemiologia e Vigilância em Saúde e Ambiente: Brasília, Brasil, 2024; Volume 3, ISBN 9786559935031. [Google Scholar]
- Deane, L.M.; Ferreira Neto, J.A.; Lima, M.M. The Vertical Dispersion of Anopheles (Kerteszia) cruzi in a Forest in Southern Brazil Suggests That Human Cases of Malaria of Simian Origin Might Be Expected. Mem. Inst. Oswaldo Cruz 1984, 79, 461–463. [Google Scholar] [CrossRef] [PubMed]
- Guimarães, A.E.; Arlé, M.; Machado, R.N.M. Mosquitos No Parque Nacional da Serra dos Órgãos, Estado Do Rio de Janeiro, Brasil. II. Distribuição Vertical. Mem. Inst. Oswaldo Cruz 1985, 80, 171–185. [Google Scholar] [CrossRef]
- Ashley, E.A.; Pyae Phyo, A.; Woodrow, C.J. Malaria. The Lancet 2018, 391, 1608–1621. [Google Scholar] [CrossRef] [PubMed]
- Davidson, G.; Chua, T.H.; Cook, A.; Speldewinde, P.; Weinstein, P. Defining the Ecological and Evolutionary Drivers of Plasmodium knowlesi Transmission Within a Multi-Scale Framework. Malar. J. 2019, 18, 18–66. [Google Scholar] [CrossRef] [PubMed]
- Deane, L.M.; Deane, M.P.; Ferreira Neto, J. Studies on Transmission of Simian Malaria and on a Natural Infection of Man with Plasmodium simium in Brazil. Bull. World Health Organ. 1966, 35, 805–808. [Google Scholar] [PubMed]
- Lalremruata, A.; Magris, M.; Vivas-Martínez, S.; Koehler, M.; Esen, M.; Kempaiah, P.; Jeyaraj, S.; Perkins, D.J.; Mordmüller, B.; Metzger, W.G. Natural Infection of Plasmodium brasilianum in Humans: Man and Monkey Share Quartan Malaria Parasites in the Venezuelan Amazon. EBioMedicine 2015, 2, 1186–1192. [Google Scholar] [CrossRef] [PubMed]
- Brasil, P.; Zalis, M.G.; de Pina-Costa, A.; Siqueira, A.M.; Júnior, C.B.; Silva, S.; Areas, A.L.L.; Pelajo-Machado, M.; de Alvarenga, D.A.M.; da Silva Santelli, A.C.F.; et al. Outbreak of Human Malaria Caused by Plasmodium simium in the Atlantic Forest in Rio de Janeiro: A Molecular Epidemiological Investigation. Lancet Glob. Health 2017, 5, e1038–e1046. [Google Scholar] [CrossRef] [PubMed]
- Fuehrer, H.P.; Campino, S.; Sutherland, C.J. The Primate Malaria Parasites Plasmodium malariae, Plasmodium brasilianum and Plasmodium ovale Genomic Insights into Distribution, Dispersal and Host Transitions. Malar. J. 2022, 21, 21–138. [Google Scholar] [CrossRef] [PubMed]
- Tazi, L.; Ayala, F.J. Unresolved Direction of Host Transfer of Plasmodium vivax v. P. simium and P. malariae v. P. brasilianum. Infect. Genet. Evol. 2011, 11, 209–221. [Google Scholar] [CrossRef]
- Lal, A.A.; De la Cruz, V.F.; Collins, W.E.; Campbell, G.H.; Procell, P.M.; McCutchan, T.F. Circumsporozoite Protein Gene from Plasmodium brasilianum. Animal Reservoirs for Human Malaria Parasites? J. Biol. Chem. 1988, 263, 5495–5498. [Google Scholar] [CrossRef] [PubMed]
- Guimarães, L.O.; Bajay, M.M.; Wunderlich, G.; Bueno, M.G.; Röhe, F.; Catão-Dias, J.L.; Neves, A.; Malafronte, R.S.; Curado, I.; Kirchgatter, K. The Genetic Diversity of Plasmodium malariae and Plasmodium brasilianum from Human, Simian and Mosquito Hosts in Brazil. Acta Trop. 2012, 124, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Fandeur, T.; Volney, B.; Peneau, C.; De Thoisy, B. Monkeys of the Rainforest in French Guiana Are Natural Reservoirs for P. brasilianum/P. malariae Malaria. Parasitology 2000, 120, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Duarte, A.M.R.d.C.; Malafronte, R.d.S.; Cerutti, C., Jr.; Curado, I.; de Paiva, B.R.; Maeda, A.Y.; Yamasaki, T.; Summa, M.E.L.; Neves, D.D.V.D.d.A.; de Oliveira, S.G.; et al. Natural Plasmodium Infections in Brazilian Wild Monkeys: Reservoirs for Human Infections? Acta Trop. 2008, 107, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Fuentes-Ramírez, A.; Jiménez-Soto, M.; Castro, R.; Romero-Zuñiga, J.J.; Dolz, G. Molecular Detection of Plasmodium malariae/Plasmodium brasilianum in Non-Human Primates in Captivity in Costa Rica. PLoS One 2017, 12, e0170704. [Google Scholar] [CrossRef] [PubMed]
- Ministério da Saúde do Brasil. Guia de Vigilância de Epizootias em Primatas Não Humanos e Entomologia Aplicada à Vigilância da Febre Amarela, 2nd ed.; Ministério da Saúde do Brasil: São Paulo, Brazil, 2014; ISBN 9788533421028. [Google Scholar]
- Teixeira, D.S.; Bernal-Valle, S.; Ramos, A.V.V.; de Santos, L.K.N.S.S.; de Abreu, F.V.S.; dos Santos, E.; de Bandeira, J.C.; de Almeida, R.M. Capture and Collection of Biological Samples from Free-Living Neotropical Primates. Primate Conserv. 2022, 2022, 91–102. [Google Scholar]
- Gamble, K.C. Primates. In Exotic Animal Formulary; Carpenter, J.W., Marion, C.J., Eds.; Elsevier Saunders: Maryland Heights, MO, USA, 2023; pp. 575–615. ISBN 9780323444507. [Google Scholar]
- De Alvarenga, D.A.M.; Culleton, R.; De Pina-Costa, A.; Rodrigues, D.F.; Bianco, C.; Silva, S.; Nunes, A.J.D.; De Souza, J.C.; Hirano, Z.M.B.; Moreira, S.B.; et al. An Assay for the Identification of Plasmodium simium Infection for Diagnosis of Zoonotic Malaria in the Brazilian Atlantic Forest. Sci. Rep. 2018, 8, 86. [Google Scholar] [CrossRef] [PubMed]
- Kocher, T.D.; Thomas, W.K.; Meyer, A.; Edwards, S.V.; Paabo, S.; Villablanca, F.X.; Wilson, A.C. Dynamics of Mitochondrial DNA Evolution in Animals: Amplification and Sequencing with Conserved Primers. Proc. Natl. Acad. Sci. U S A 1989, 86, 6196–6200. [Google Scholar] [CrossRef] [PubMed]
- Snounou, G.; Viriyakosol, S.; Jarra, W.; Thaithong, S.; Brown, K.N. Identification of the Four Human Malaria Parasite Species in Field Samples by the Polymerase Chain Reaction and Detection of a High Prevalence of Mixed Infections. Mol. Biochem. Parasitol. 1993, 58, 283–292. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M. Estimation of the Number of Nucleotide Substitutions in the Control Region of Mitochondrial DNA in Humans and Chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef] [PubMed]
- Deane, L.M. Simian Malaria in Brazil. Mem. Inst. Oswaldo Cruz 1992, 87, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Araújo, M.S.; Messias, M.R.; Figueiró, M.R.; Gil, L.H.S.; Probst, C.M.; Vidal, N.M.; Katsuragawa, T.H.; Krieger, M.A.; Pereira Da Silva, L.H.; Ozaki, L.S. Natural Plasmodium Infection in Monkeys in the State of Rondônia (Brazilian Western Amazon). Malar. J. 2013, 12, 180. [Google Scholar] [CrossRef] [PubMed]
- Alvarenga, D.A.M.; Pina-Costa, A.; Bianco, C.; Moreira, S.B.; Brasil, P.; Pissinatti, A.; Daniel-Ribeiro, C.T.; Brito, C.F.A. New Potential Plasmodium brasilianum Hosts: Tamarin and Marmoset Monkeys (Family Callitrichidae). Malar. J. 2017, 16, 16–71. [Google Scholar] [CrossRef] [PubMed]
- Lourenço-de-Oliveira, R.; Deane, L.M. Simian Malaria at Two Sites in the Brazilian Amazon. I--The Infection Rates of Plasmodium brasilianum in Non-Human Primates. Mem. Inst. Oswaldo Cruz 1995, 90, 331–339. [Google Scholar] [CrossRef] [PubMed]
- Aitken, E.H.; Bueno, M.G.; Dos Santos Ortolan, L.; Alvaréz, J.M.; Pissinatti, A.; Kierulff, M.C.M.; Catão-Dias, J.L.; Epiphanio, S. Survey of Plasmodium in the Golden-Headed Lion Tamarin (Leontopithecus chrysomelas) Living in Urban Atlantic Forest in Rio de Janeiro, Brazil. Malar. J. 2016, 15, 93. [Google Scholar] [CrossRef] [PubMed]
- Rylands, A.B. Marmosets and Tamarins: Systematics, Behavior, and Ecology.; Oxford University Press: Oxford, UK; New York, NY, USA, 1993. [Google Scholar]
- Kindlovits, L.M.; Kindlovits, A. Febre Amarela. Clínica e Terapêutica Em Primatas Tropicais. Livros, L.F., Ed.; 2a ed.SciELO—Scientific Electronic Library Online: Rio de Janeiro, Brazil, 2009. [Google Scholar]
- Ministério da Saúde do Brasil. Malária. Available online: https://www.gov.br/saude/pt-br/composicao/svsa/cnie/painel-malaria (accessed on 10 December 2024).
- de Abreu, F.V.S.; dos Santos, E.; Gomes, M.Q.; Vargas, W.P.; de Oliveira Passos, P.H.; Nunes e Silva, C.; Araújo, P.C.; Pires, J.R.; Romano, A.P.M.; Teixeira, D.S.; et al. Capture of Alouatta guariba clamitans for the Surveillance of Sylvatic Yellow Fever and Zoonotic Malaria: Which Is the Best Strategy in the Tropical Atlantic Forest? Am. J. Primatol. 2019, 81, e23000. [Google Scholar] [CrossRef] [PubMed]
- Nunes, A.J.D.; de Alvarenga, D.A.M.; de Souza Junior, J.C.; Peruchi, A.R.; Gonçalves, G.H.P.; Hirano, Z.M.B.; de Brito, C.F.A.; Cremer, M.J. Plasmodium Infection and Its Association with Biochemical and Haematological Parameters in Free-Living Alouatta guariba clamitans (Cabrera, 1940) (Primates: Atelidae) in Southern Brazil. Mem. Inst. Oswaldo Cruz 2019, 114, e190210. [Google Scholar] [CrossRef] [PubMed]
- Neto, S.P.G.d.C.; Da Silva, L.T. Turismo e Desenvolvimento: Transformações No Território da Região Do Extremo Sul Da Bahia. Caminhos Geogr. 2015, 16, 74–88. [Google Scholar] [CrossRef]
- Multini, L.C.; Wilke, A.B.B.; Marrelli, M.T. Neotropical Anopheles (Kerteszia) Mosquitoes Associated with Bromeliad-Malaria Transmission in a Changing World. Acta Trop. 2020, 205, 105413. [Google Scholar] [CrossRef] [PubMed]
- Secretaria da Saúde do Estado da Bahia (SESAB). Carta Anofélica da Bahia, Brasil (Diptera: Culicidae). Boletim Entomológico; Laboratório Central de Saúde Pública, Ed.; Secretaria Estadual de Saúde da Bahia: Salvador-Bahia, 2016; Volume 3. [Google Scholar]
- Secretaria da Saúde do Estado da Bahia. Malária—Confirmed Cases Reported in the Notification Disease Information System (According to Year 1st Symptom(s) and to Municipality of Infection). Available online: https://www.gov.br/saude/pt-br/assuntos/saude-de-a-a-z/m/malaria (accessed on 10 December 2024).
- Bajic, M.; Ravishankar, S.; Sheth, M.; Rowe, L.A.; Pacheco, M.A.; Patel, D.S.; Batra, D.; Loparev, V.; Olsen, C.; Escalante, A.A.; et al. The First Complete Genome of the Simian Malaria Parasite Plasmodium brasilianum. Sci. Rep. 2022, 12, 19802. [Google Scholar] [CrossRef] [PubMed]
| Plasmodium Positives | Plasmodium Negatives | |||||
|---|---|---|---|---|---|---|
| Variable | Category | n | (%) | n | (%) | Total |
| Gender | Female | 0 | 0.0 | 142 | 100.0 | 144 |
| Male | 1 | 0.7 | 155 | 99.3 | 156 | |
| Living condition | Free-living | 1 | 0.3 | 291 | 99.7 | 292 |
| Captive | 0 | 0.0 | 6 | 100.0 | 6 | |
| Age range | Baby | 0 | 0.0 | 12 | 100.0 | 12 |
| Juvenile | 0 | 0.0 | 65 | 100.0 | 65 | |
| Adult | 1 | 0.4 | 220 | 99.6 | 221 | |
| Species | Alouatta caraya | 0 | 0.0 | 25 | 100.0 | 25 |
| Alouatta guariba clamitans | 0 | 0.0 | 1 | 100.0 | 1 | |
| Callicebus melanochir | 0 | 0.0 | 1 | 100.0 | 1 | |
| Callithrix spp. | 0 | 0.0 | 15 | 100.0 | 15 | |
| Callithrix geoffroyi | 1 | 3.6 | 27 | 96.4 | 28 | |
| Callithrix jacchus | 0 | 0.0 | 4 | 100.0 | 4 | |
| Calllithrix khulii | 0 | 0.0 | 31 | 100.0 | 31 | |
| Callithrix penicillata | 0 | 0.0 | 175 | 100.0 | 175 | |
| Leontopithecus chrysomelas | 0 | 0.0 | 16 | 100.0 | 16 | |
| Sapajus robustus | 0 | 0.0 | 1 | 100.0 | 1 | |
| Sapajus xanthosthernos | 0 | 0.0 | 1 | 100.0 | 1 | |
| Total | 10 species | 1 | 0.3 | 297 | 99.7 | 298 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santos, L.K.N.S.S.; Aquino-Teixeira, S.M.; Bernal-Valle, S.; Daltro, B.S.; Noetzold, M.; Silva, A.R.C.; Alvarenga, D.A.M.; Silva, L.B.; Oliveira, R.S.; Oliveira, C.H.; et al. Molecular Surveillance of Plasmodium spp. Infection in Neotropical Primates from Bahia and Minas Gerais, Brazil. Pathogens 2025, 14, 757. https://doi.org/10.3390/pathogens14080757
Santos LKNSS, Aquino-Teixeira SM, Bernal-Valle S, Daltro BS, Noetzold M, Silva ARC, Alvarenga DAM, Silva LB, Oliveira RS, Oliveira CH, et al. Molecular Surveillance of Plasmodium spp. Infection in Neotropical Primates from Bahia and Minas Gerais, Brazil. Pathogens. 2025; 14(8):757. https://doi.org/10.3390/pathogens14080757
Chicago/Turabian StyleSantos, Luana Karla N. S. S., Sandy M. Aquino-Teixeira, Sofía Bernal-Valle, Beatriz S. Daltro, Marina Noetzold, Aloma Roberta C. Silva, Denise Anete M. Alvarenga, Luisa B. Silva, Ramon S. Oliveira, Cirilo H. Oliveira, and et al. 2025. "Molecular Surveillance of Plasmodium spp. Infection in Neotropical Primates from Bahia and Minas Gerais, Brazil" Pathogens 14, no. 8: 757. https://doi.org/10.3390/pathogens14080757
APA StyleSantos, L. K. N. S. S., Aquino-Teixeira, S. M., Bernal-Valle, S., Daltro, B. S., Noetzold, M., Silva, A. R. C., Alvarenga, D. A. M., Silva, L. B., Oliveira, R. S., Oliveira, C. H., Celestino, I. A., Gonçalves-dos-Santos, M. E., Teixeira, T. J., Sevá, A. P., Campos, F. S., Ribeiro, B. M., Roehe, P. M., Simonini-Teixeira, D., Abreu, F. V. S., ... Albuquerque, G. R. (2025). Molecular Surveillance of Plasmodium spp. Infection in Neotropical Primates from Bahia and Minas Gerais, Brazil. Pathogens, 14(8), 757. https://doi.org/10.3390/pathogens14080757








