Outbreak Caused by VIM-1- and VIM-4-Positive Proteus mirabilis in a Hospital in Zagreb
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Bacterial Isolates
2.2. Antimicrobial Susceptibility Testing
2.3. Phenotypic Detection of β-Lactamases
2.4. Molecular Detection of Resistance Genes
2.5. Inter-Array Genotyping Kit CarbaResist
2.6. Whole-Genome Sequencing (WGS)
2.7. Characterization of Plasmids
2.8. Detection of Virulence Determinants
3. Results
3.1. Patients and Bacterial Isolates
3.2. Antibiotic Susceptibility
3.3. Phenotypic Detection of β-lactamases
3.4. Molecular Detection of Resistance Genes
3.5. Genotyping by Inter-Array Genotyping Kit CarbaResist
3.6. WGS and Plasmid Analysis
3.7. Detection of Virulence Determinants
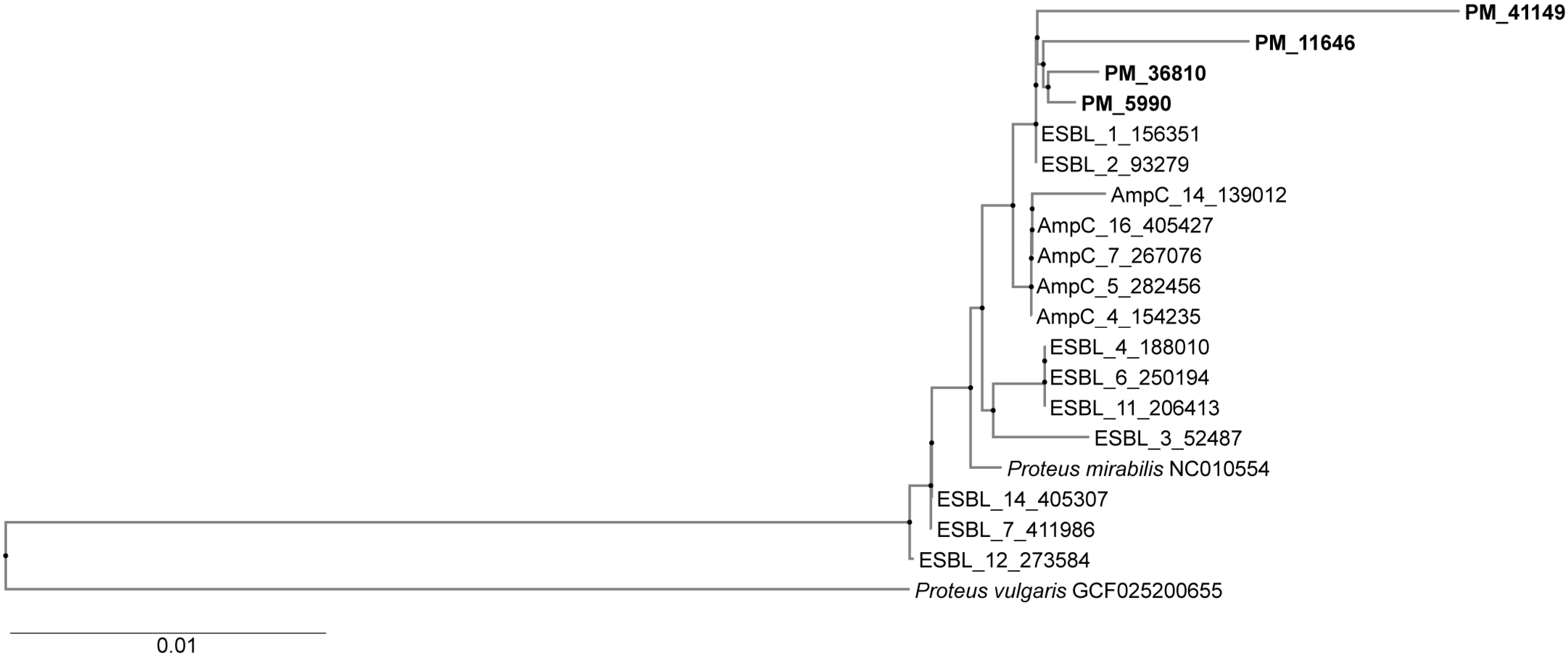
3.8. Genotyping (Phylogenetic Tree)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sanches, M.S.; Silva, L.C.; Silva, C.R.D.; Montini, V.H.; Oliva, B.H.D.; Guidone, G.H.M.; Nogueira, M.C.L.; Menck-Costa, M.F.; Kobayashi, R.K.T.; Vespero, E.C.; et al. Prevalence of Antimicrobial Resistance and Clonal Relationship in ESBL/AmpC-Producing Proteus mirabilis Isolated from Meat Products and Community-Acquired Urinary Tract Infection (UTI-CA) in Southern Brazil. Antibiotics 2023, 10, 370. [Google Scholar] [CrossRef]
- Li, Y.; Yin, M.; Fang, C.; Fu, Y.; Dai, X.; Zeng, W.; Zhang, L. Genetic analysis of resistance and virulence characteristics of clinical multidrug-resistant Proteus mirabilis isolates. Front. Cell. Infect. Microbiol. 2023, 11, 1229194. [Google Scholar] [CrossRef]
- Girlich, D.; Bonnin, R.A.; Dortet, L.; Naas, T. Genetics of Acquired Antibiotic Resistance Genes in Proteus spp. Front. Microbiol. 2020, 21, 256. [Google Scholar] [CrossRef]
- Ambler, R.P. The structure of β-lactamases. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1980, 289, 321–331. [Google Scholar] [CrossRef]
- Paterson, D.L.; Bonomo, R.A. Extended-spectrum β-lactamases: A clinical update. Clin. Microbiol. Rev. 2005, 18, 657–686. [Google Scholar] [CrossRef] [PubMed]
- Cantón, R.; González-Alba, J.M.; Galán, J.C. CTX-M Enzymes: Origin and Diffusion. Front. Microbiol. 2012, 2, 110. [Google Scholar] [CrossRef] [PubMed]
- Potter, R.F.; D’Souza, A.W.; Dantas, G. The rapid spread of carbapenem-resistant Enterobacteriaceae. Drug Resist. Updat. 2016, 9, 30–46. [Google Scholar] [CrossRef] [PubMed]
- Tonkić, M.; Mohar, B.; Šiško-Kraljević, K.; Meško-Meglič, K.; Goić-Barišić, I.; Novak, A.; Kovačić, A.; Punda-Polić, V. High prevalence and molecular characterization of extended-spectrum β-lactamase-producing Proteus mirabilis strains in southern Croatia. J. Med. Microbiol. 2010, 59, 1185–1190. [Google Scholar] [CrossRef]
- Bedenić, B.; Firis, N.; Elveđi-Gašparović, V.; Krilanović, M.; Matanović, K.; Štimac, I.; Luxner, J.; Vraneš, J.; Meštrović, T.; Zarfel, G.; et al. Emergence of multidrug-resistant Proteus mirabilis in a long-term care facility in Croatia. Wien. Klin. Wochenschr. 2016, 128, 404–413. [Google Scholar] [CrossRef]
- Rubić, Z.; Soprek, S.; Jelić, M.; Novak, A.; Goić-Barisić, I.; Radić, M.; Tambić-Andrasević, A.; Tonkić, M. Molecular Characterization of β-Lactam Resistance and Antimicrobial Susceptibility to Possible Therapeutic Options of AmpC-Producing Multidrug-Resistant Proteus mirabilis in a University Hospital of Split, Croatia. Microb. Drug Resist. 2021, 27, 162–169. [Google Scholar] [CrossRef]
- European Committee for Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 12. 2022. Available online: http://www.eucast.org (accessed on 1 October 2024).
- Clinical Laboratory Standard Institution. Performance Standards for Antimicrobial Susceptibility Testing, 28th ed.; Approved Standard M100-S22; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2002, 18, 268–281. [Google Scholar] [CrossRef]
- Jarlier, V.; Nicolas, M.H.; Fournier, G.; Philippon, A. Extended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: Hospital prevalence and susceptibility patterns. Rev. Infect. Dis. 1988, 10, 867–878. [Google Scholar] [CrossRef]
- Jacoby, G.A. AmpC β-lactamases. J. Clin. Microbiol. 2009, 22, 161–182. [Google Scholar] [CrossRef]
- MacDonald, J.W.; Chibabhai, V. Evaluation of the RESIST-4 O.K.N.V Immunochromatographic Lateral Flow Assay for the Rapid Detection of OXA-48, KPC, NDM and VIM Carbapenemases from Cultured Isolates. Access Microbiol. 2019, 1, e000031. [Google Scholar] [CrossRef]
- Lee, K.; Lim, Y.S.; Yong, D.; Yum, J.H.; Chong, Y. Evaluation of the Hodge Test and the Imipenem-EDTA-double-disk Synergy Test for Differentiating Metallo-β-lactamase-producing Isolates of Pseudomonas spp. and Acinetobacter spp. J. Clin. Microbiol. 2003, 41, 4623–4629. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, M100, 31st ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2021. [Google Scholar]
- Arlet, G.; Brami, G.; Decre, D.; Flippo, A.; Gaillot, O.; Lagrange, P.H.; Philippon, A. Molecular characterization by PCR restriction fragment polymorphism of TEM β-lactamases. FEMS Microbiol. Lett. 1995, 134, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Nüesch-Inderbinen, M.T.; Hächler, H.; Kayser, F.H. Detection of genes coding for extended-spectrum SHV β-lactamases in clinical isolates by a molecular genetic method, and comparison with the E test. Eur. J. Clin. Microbiol. Infect. Dis. 1996, 15, 398–402. [Google Scholar] [CrossRef] [PubMed]
- Woodford, N.; Ward, M.E.; Kaufmann, M.E.; Turton, J.; Fagan, E.J.; James, D.; Johnson, A.P.; Pike, R.; Warner, M.; Cheasty, T.; et al. Community and hospital spread of Escherichia coli producing CTX-M extended-spectrum β-lactamases in the UK. J. Antimicrob. Chemother. 2004, 54, 735–743. [Google Scholar] [CrossRef]
- Robicsek, A.; Jacoby, G.A.; Hooper, D.C. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect. Dis. 2006, 6, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Walsh, T.R.; Cuveiller, V.; Nordman, P. Multiplex PCR for Detection of Acquired Carbapenemases Genes. Diagn. Microbiol. Infect. Dis. 2011, 70, 119–123. [Google Scholar] [CrossRef]
- Woodford, N.; Ellington, M.J.; Coelho, J.; Turton, J.; Ward, M.E.; Brown, S.; Amyes, S.G.; Livermore, D.M. Multiplex PCR for genes encoding prevalent OXA carbapenemases. Int. J. Antimicrob. Agents 2006, 27, 351–353. [Google Scholar] [CrossRef] [PubMed]
- Perez-Perez, F.J.; Hanson, N.D. Detection of plasmid-mediated AmpC β-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbiol. 2002, 40, 2153–2162. [Google Scholar] [CrossRef]
- Woodford, N.; Fagan, E.J.; Ellington, M.J. Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum β-lactamases. J. Antimicrob. Chemother. 2006, 57, 154–155. [Google Scholar] [CrossRef] [PubMed]
- Saladin, M.; Cao, V.T.B.; Lambert, T.; Donay, J.L.; Hermann, J.; Ould-Hocine, L. Diversity of CTX-M β-lactamases and Their Promoter Regions from Enterobacteriaceae Isolated in Three Parisian Hospitals. FEMS. Microbiol. Lett. 2002, 209, 161–168. [Google Scholar] [CrossRef]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef]
- Elwell, L.P.; Falkow, S. The characterization of R plasmids and the detection of plasmid-specified genes. In Antibiotics in Laboratory Medicine, 2nd ed.; Lorian, V., Ed.; Williams and Wilkins: Baltimore, MD, USA, 1986; pp. 683–721. [Google Scholar]
- Carattoli, A.; Bertini, A.; Villa, L.; Falbo, V.; Hopkins, K.L.; Threfall, E.J. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 2005, 63, 219–228. [Google Scholar] [CrossRef]
- Carattoli, A.; Seiffert, S.N.; Schwendener, S.; Perreten, V.; Endimiani, A. Differentiation of IncL and IncM Plasmids Associated with the Spread of Clinically Relevant Antimicrobial Resistance. PLoS ONE 2015, 10, e0123063. [Google Scholar] [CrossRef] [PubMed]
- Shaaban, M.; Elshaer, S.L.; Abd El-Rahman, O.A. Prevalence of extended-spectrum β-lactamases, AmpC, and carbapenemases in Proteus mirabilis clinical isolates. BMC Microbiol. 2022, 11, 247. [Google Scholar] [CrossRef]
- Miriagou, V.; Papagiannitsis, C.C.; Tzelepi, E.; Casals, J.B.; Legakis, N.J.; Tzouvelekis, L.S. Detecting VIM-1 production in Proteus mirabilis by an imipenem-dipicolinic acid double disk synergy test. J. Clin. Microbiol. 2010, 48, 667–668. [Google Scholar] [CrossRef]
- Papagiannitsis, C.C.; Miriagou, V.; Kotsakis, S.D.; Tzelepi, E.; Vatopoulos, A.C.; Petinaki, E.; Tzouvelekis, L.S. Characterization of a transmissible plasmid encoding VEB-1 and VIM-1 in Proteus mirabilis. Antimicrob. Agents Chemother. 2012, 56, 4024–4025. [Google Scholar] [CrossRef]
- Protonotariou, E.; Poulou, A.; Politi, L.; Meletis, G.; Chatzopoulou, F.; Malousi, A.; Metallidis, S.; Tsakris, A.; Skoura, L. Clonal outbreak caused by VIM-4-producing Proteus mirabilis in a Greek tertiary-care hospital. Int. J. Antimicrob. Agents 2020, 56, 106060. [Google Scholar] [CrossRef] [PubMed]
- Markovska, R.; Schneider, I.; Keuleyan, E.; Ivanova, D.; Lesseva, M.; Stoeva, T.; Sredkova, M.; Bauernfeind, A.; Mitov, I. Dissemination of a Multidrug-Resistant VIM-1- and CMY-99-Producing Proteus mirabilis Clone in Bulgaria. Microb. Drug. Resist. 2017, 23, 345–350. [Google Scholar] [CrossRef]
- Fritzenwanker, M.; Falgenhauer, J.; Hain, T.; Imirzalioglu, C.; Chakraborty, T.; Yao, Y. The Detection of Extensively Drug-Resistant Proteus mirabilis Strains Harboring Both VIM-; and VIM-75 Metallo-β-Lactamases from Patients in Germany. Microorganisms 2025, 25, 266. [Google Scholar] [CrossRef]
- Bedenić, B.; Bratić, V.; Mihaljević, S.; Lukić, A.; Vidović, K.; Reiner, K.; Schöenthaler, S.; Barišić, I.; Zarfel, G.; Grisold, A. Multidrug-Resistant Bacteria in a COVID-19 Hospital in Zagreb. Pathogens 2023, 12, 117. [Google Scholar] [CrossRef]
- Sattler, J.; Noster, J.; Stelzer, Y.; Spille, M.; Schäfer, S.; Xanthopoulou, K.; Sommer, J.; Jantsch, J.; Peter, S.; Göttig, S.; et al. OXA-48-like carbapenemases in Proteus mirabilis—Novel genetic environments and a challenge for detection. Emerg. Microbes. Infect. 2024, 13, 2353310. [Google Scholar] [CrossRef] [PubMed]
- Potron, A.; Hocquet, D.; Triponney, P.; Plésiat, P.; Bertrand, X.; Valot, B. Carbapenem-Susceptible OXA-23-Producing Proteus mirabilis in the French Community. Antimicrob. Agents Chemother. 2019, 63, e00191-19. [Google Scholar] [CrossRef] [PubMed]
- Bonnin, R.A.; Girlich, D.; Jousset, A.B.; Gauthier, L.; Cuzon, G.; Bogaerts, P. A single Proteus mirabilis lineage from human and animal sources: A hidden reservoir of OXA-23 or OXA-58 carbapenemases in Enterobacterales. Sci. Rep. 2020, 8, 9160. [Google Scholar] [CrossRef] [PubMed]
- Mendonça, J.; Guedes, C.; Silva, C.; Sá, S.; Oliveira, M.; Accioly, G.; Baylina, P.; Barata, P.; Pereira, C.; Fernandes, R. New CTX-M Group Conferring β-Lactam Resistance: A Compendium of Phylogenetic Insights from Biochemical, Molecular, and Structural Biology. Biology 2022, 11, 256. [Google Scholar] [CrossRef]
| AMX 32 | AMC 32/16 | TZP 32/4 | CXM 32 | CAZ 16 | CTX 4 | CRO 4 | FEP 16 | IMI 4 | MEM 4 | GM 16 | CIP 0.25 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | >128 (R) | 32/16(R) | 8/4(S) | >128(R) | 8(I) | >128(R) | 16(R) | 16(R) | 128(R) | 0.25(S) | >128(R) | >128(R) |
| 2 | >128 (R) | 64/32(R) | 16/4(S) | >128(R) | 8(I) | >128(R) | 16(R) | >128(R) | 128(R) | 0.25(S) | >128(R) | >128(R) |
| 3 | >128 (R) | 64/32(R) | 8/4(S) | >128(R) | 16(R) | >128(R) | 32(R) | 32(R) | 128(R) | 0.25(S) | >128(R) | >128(R) |
| 4 | >128 (R) | 128/64(R) | 8/4(S) | >128(R) | 2(S) | >128(R) | >128(R) | >128(R) | 128(R) | 0.25(S) | >128(R) | >128(R) |
| 5 | >128 (R) | 32/16(R) | 16/4(S) | >128(R) | 2(S) | >128(R) | >128(R) | >128(R) | 4(R) | 0.25(S) | >128(R) | >128(R) |
| 6 | >128 (R) | 32/16(R) | 16/4(S) | >128(R) | 8(I) | >128(R) | >128(R) | >128(R) | 4(R) | 0.25(S) | >128(R) | >128(R) |
| 7 | >128 (R) | 128/64(R) | 8/4(S) | >128(R) | 8(I) | >128(R) | >128(R) | >128(R) | 4(R) | 0.25(S) | >128(R) | >128(R) |
| 8 | >128 (R) | 128/64(R) | 8/4(S) | >128(R) | 16(R) | >128(R) | >128(R) | >128(R) | 8(R) | 0.25(S) | >128(R) | >128(R) |
| 9 | >128 (R) | 128/64(R) | 8/4(S) | >128(R) | 16(R) | >128(R) | 64(R) | >128(R) | 4(R) | 0.25(S) | >128(R) | >128(R) |
| 10 | >128 (R) | 128/64(R) | 8/4(S) | >128(R) | 32(R) | >128(R) | >128(R) | >128(R) | 4(R) | 0.25(S) | >128(R) | >128(R) |
| 11 | >128 (R) | 128/64(R) | 8/4(S) | >128(R) | 128(R) | >128(R) | 32(R) | >128(R) | 4(R) | 0.25(S) | >128(R) | >128(R) |
| 12 | >128 (R) | 32/16(R) | 16/4(S) | >128(R) | 2(S) | >128(R) | >128(R) | >128(R) | 8(R) | 0.25(S) | >128(R) | >128(R) |
| 13 | >128 (R) | 128/64(R) | 8/4(S) | >128(R) | 16(R) | >128(R) | 32(R) | >128(R) | 8(R) | 0.25(S) | >128(R) | >128(R) |
| 14 | >128 (R) | 128/64(R) | 16/4(S) | >128(R) | 32(R) | >128(R) | 64((R) | >128(R) | 4(R) | 0.25(S) | >128(R) | >128(R) |
| 15 | >128 (R) | 128/64(R) | 8/4 (S) | >128(R) | 32(R) | >128(R) | 16(R) | >128(R) | 4(R) | 0,25(S) | >128(R) | >128(R) |
| 16 | >128 (R) | 128/64(R) | 16/4(S) | >128(R) | 128(R) | >128(R) | 128(R) | >128(R) | 4(R) | 0,12 S) | >128(R) | >128(R) |
| 17 | >128 (R) | 64/32(R) | 16/4(S) | >128(R) | 16(R) | >128(R) | 32(R) | >128(R) | 8(R) | 0,06 (S) | >128(R) | >128(R) |
| 18 | >128 (R) | 63/32(R) | 8/4(S) | >128(R) | 64(R) | >128(R) | 64(R) | >128(R) | 4(R) | 0,25(S) | >128(R) | >128(R) |
| 19 | >128 (R) | 128/64(R) | 8/4(S) | >128(R) | 64(R) | >128(R) | 32(R) | >128(R) | 4(R) | 0,25(S) | >128(R) | >128(R) |
| 20 | >128 (R) | 64/32(R) | 16/4(S) | >128(R) | 64(R) | >128(R) | 128(R) | >128(R) | 4(R) | 0,5 (S) | >128(R) | >128(R) |
| Isolate and Protocol Number | Res Phenotype | β-Lactam | Aminoglycoside | Sulphamide | Trimethoprim | Integrase Genes |
|---|---|---|---|---|---|---|
| PM 2 | AMX, AMC, TZP, CAZ, CTX, CRO, FEP, GM, AMI CIP | ISEcpblaCTX-M-15 blaTEM blaVIM | aac(6″)IIc aadA1 aadA2 armA | sul1 | dfrA1 | Intl2 |
| PM 3 | AMX, AMC, TZP, CAZ, CTX, CRO, FEP, GM, AMI CIP | ISEcpblaCTX-M-15 blaTEM blaVIM | aac(6″)IIc aadA1 aadA2 armA | sul1 sul2 | dfrA1 | Intl1 Intl2 |
| PM4 | AMX, AMC, TZP, CAZ, CTX, CRO, FEP, GM, AMI CIP | ISEcpblaCTX-M-15 blaTEM blaVIM | aac(6″)IIc aadA1 aadA2 armA | sul1 sul2 | dfrA1 dfrA15 | Intl1 Intl2 |
| PM11 | AMX, AMC, TZP, CAZ, CTX, CRO, FEP, GM, AMI CIP | ISEcpblaCTX-M-15 blaTEM blaVIM | aac(6″)IIc aadA1 aadA2 armA | sul1 sul2 | dfrA1 | Intl1 Intl2 |
| PM14 | AMX, AMC, TZP, CAZ, CTX, CRO, FEP, GM, AMI CIP | ISEcpblaCTX-M-15 blaTEM blaVIM | aac(6″)IIc aadA1 aadA2 armA | sul1 sul2 | dfrA1 | Intl1 Intl2 |
| PM 19 | AMX, AMC, TZP, CAZ, CTX, CRO, FEP, GM, AMI CIP | ISEcpblaCTX-M-15 blaTEM blaVIM | aac(6″)IIc aadA1 aadA2 armA | sul1 sul2 | dfrA1 | Intl1 Intl2 |
| PM 20 | AMX, AMC, TZP, CAZ, CTX, CRO, FEP, GM, AMI, CIP | ISEcpblaCTX-M-15 blaTEM blaVIM | aac(6″)IIc aadA1 aadA2 armA | sul1 sul2 | dfrA1 | Intl1 Intl2 |
| Isolate and Protocol Number | β-Lactam | Aminoglycosides | Sulphonamide | Trimethoprim | Chloramphenicol | Tetracycline | Plasmid Replicon |
|---|---|---|---|---|---|---|---|
| PM 3 | blaCTX-M-202, blaTEM-156, blaTEM-1A, blaTEM-2, blaVIM-4, blaVIM-1, | aac(3)-IId, aph(6)-Id, aph(3″)-Ib, aadA1, armA, aac(6′)-IIc, | sul1 sul2 | dfrA1 | cat | tet(J) | IncC |
| PM 5 | blaCTX-M-202, blaTEM-2, blaTEM-1A, blaVIM-4 | aac(3)-IId, aph(6)-Id, aph(3″)-Ib, aadA1, armA, aac(6′)-IIc, | sul1 sul2 | dfrA1 | cat | tet(J) | IncC |
| PM 6 | blaCTX-M-202, blaTEM-2, blaVIM-4 | aph(6)-Id, aph(3″)-Ib, armA, aac(6′)-IIc, aac(3)-IId, aac(6′)-IIc, aadA1 | sul1 sul2 | dfrA1 | cat | tet(J) | IncC |
| PM 8 | blaCTX-M-202, blaTEM-2, blaTEM-1A, blaVIM-1, blaVIM-4 | aac(3)-IId, aph(6)-Id, aph(3″)-Ib, aadA1, armA, aac(6′)-IIc, | sul1 sul2 | dfrA1 | cat | tet(J) | IncC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bedenić, B.; Zarfel, G.; Luxner, J.; Grisold, A.; Nađ, M.; Anušić, M.; Tičić, V.; Dobretzberger, V.; Barišić, I.; Vraneš, J. Outbreak Caused by VIM-1- and VIM-4-Positive Proteus mirabilis in a Hospital in Zagreb. Pathogens 2025, 14, 737. https://doi.org/10.3390/pathogens14080737
Bedenić B, Zarfel G, Luxner J, Grisold A, Nađ M, Anušić M, Tičić V, Dobretzberger V, Barišić I, Vraneš J. Outbreak Caused by VIM-1- and VIM-4-Positive Proteus mirabilis in a Hospital in Zagreb. Pathogens. 2025; 14(8):737. https://doi.org/10.3390/pathogens14080737
Chicago/Turabian StyleBedenić, Branka, Gernot Zarfel, Josefa Luxner, Andrea Grisold, Marina Nađ, Maja Anušić, Vladimira Tičić, Verena Dobretzberger, Ivan Barišić, and Jasmina Vraneš. 2025. "Outbreak Caused by VIM-1- and VIM-4-Positive Proteus mirabilis in a Hospital in Zagreb" Pathogens 14, no. 8: 737. https://doi.org/10.3390/pathogens14080737
APA StyleBedenić, B., Zarfel, G., Luxner, J., Grisold, A., Nađ, M., Anušić, M., Tičić, V., Dobretzberger, V., Barišić, I., & Vraneš, J. (2025). Outbreak Caused by VIM-1- and VIM-4-Positive Proteus mirabilis in a Hospital in Zagreb. Pathogens, 14(8), 737. https://doi.org/10.3390/pathogens14080737







