The Molecular Epidemiology of HIV-1 in Russia, 1987–2023: Subtypes, Transmission Networks and Phylogenetic Story
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Overview of Data
2.3. Sequence Alignment and Subtype Assignment
2.4. Definition of HIV Cluster
2.5. Phylodynamic and Phylogeographic Reconstructions
2.6. Statistical Analysis
3. Results
3.1. Demographic Characteristics
3.2. HIV Subtype Diversity
3.3. The Spatial and Temporal Distribution of HIV-1 Subtype
3.4. Molecular Network Analysis and Transmission Clusters
3.5. Correlates of Clustering
3.6. Origin and Migration Pathway
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| HIV | Human immunodeficiency virus |
| MTC | Molecular transmission cluster |
| CRF | Circulating recombinant form |
| AIDS | Acquired immunodeficiency syndrome |
| OR | Odds ratio |
| MSM | Men who have sex with men |
| HET | Heterosexual transmission |
| IDUs | Injecting drug users |
| RuHIV | Russian HIV Antiviral Drug Resistance |
| WHO | World health organization |
| FD | Federal District |
| PR | Protease |
| RT | Reverse transcriptase |
| ML | Maximum likelihood |
| GTR | General time reversible |
| FSU | Former Soviet Union |
| PC | Phylogenetic cluster |
| SHaLRT | Shimodaira–Hasegawa-like approximate likelihood ratio test |
| TN93 | Tamura–Nei |
| ART | Antiretroviral therapy |
| tMRCA | Time to the most recent common ancestor |
| IQR | Interquartile ranges |
| CI | Confidence interval |
| ST | Sexual transmission |
| URF | Unique recombinant form |
| MTCT | Mother-to-child transmission |
| NSC | Nosocomial transmission |
| UNK | Unknown |
References
- Reference Center for Monitoring HIV and HIV-Associated Infections of the Federal Budget Institute of Science; Central Research Institute of Epidemiology. HIV Infection. Newsletter № 48. Available online: http://www.hivrussia.info/wp-content/uploads/2024/11/hiv-infection-info-bulletin-48.pdf (accessed on 20 May 2024).
- Kazennova, E.; Laga, V.; Lapovok, I.; Glushchenko, N.; Neshumaev, D.; Vasilyev, A.; Bobkova, M. HIV-1 Genetic Variants in the Russian Far East. AIDS Res. Hum. Retroviruses 2014, 30, 742–752. [Google Scholar] [CrossRef]
- Dukhovlinova, E.; Masharsky, A.; Toussova, O.; Verevochkin, S.; Solovyeva, T.; Meringof, M.; Paintsil, E.; White, E.; Barbour, R.; Heimer, R.; et al. Two Independent HIV Epidemics in Saint Petersburg, Russia Revealed by Molecular Epidemiology. AIDS Res. Hum. Retroviruses 2015, 31, 608–614. [Google Scholar] [CrossRef]
- Gashnikova, N.M.; Astakhova, E.M.; Gashnikova, M.P.; Bocharov, E.F.; Petrova, S.V.; Pun’kO, O.A.; Popkov, A.V.; Totmenin, A.V. HIV-1 Epidemiology, Genetic Diversity, and Primary Drug Resistance in the Tyumen Oblast, Russia. BioMed Res. Int. 2016, 2016, 1–13. [Google Scholar] [CrossRef]
- Maksimenko, L.V.; Totmenin, A.V.; Gashnikova, M.P.; Astakhova, E.M.; Skudarnov, S.E.; Ostapova, T.S.; Yaschenko, S.V.; Meshkov, I.O.; Bocharov, E.F.; Maksyutov, R.A.; et al. Genetic Diversity of HIV-1 in Krasnoyarsk Krai: Area with High Levels of HIV-1 Recombination in Russia. BioMed Res. Int. 2020, 2020, e9057541. [Google Scholar] [CrossRef]
- Safina, K.R.; Sidorina, Y.; Efendieva, N.; Belonosova, E.; Saleeva, D.; Kirichenko, A.; Kireev, D.; Pokrovsky, V.; A Bazykin, G. Molecular epidemiology of HIV-1 in Oryol Oblast, Russia. Virus Evol. 2022, 8, veac044. [Google Scholar] [CrossRef]
- Kazennova, E.; Laga, V.; Gromov, K.; Lebedeva, N.; Zhukova, E.; Pronin, A.; Grezina, L.; Dement’Eva, N.; Shemshura, A.; Bobkova, M. Genetic Variants of HIV Type 1 in Men Who Have Sex with Men in Russia. AIDS Res. Hum. Retroviruses 2017, 33, 1061–1064. [Google Scholar] [CrossRef]
- Karamov, E.; Epremyan, K.; Siniavin, A.; Zhernov, Y.; Cuevas, M.T.; Delgado, E.; Sánchez-Martínez, M.; Carrera, C.; Kornilaeva, G.; Turgiev, A.; et al. HIV-1 Genetic Diversity in Recently Diagnosed Infections in Moscow: Predominance of AFSU, Frequent Branching in Clusters, and Circulation of the Iberian Subtype G Variant. AIDS Res. Hum. Retroviruses 2018, 34, 629–634. [Google Scholar] [CrossRef]
- Shcherbakova, N.S.; Shalamova, L.A.; Delgado, E.; Fernández-García, A.; Vega, Y.; Karpenko, L.I.; Ilyichev, A.A.; Sokolov, Y.V.; Shcherbakov, D.N.; Pérez-Álvarez, L.; et al. Short Communication: Molecular Epidemiology, Phylogeny, and Phylodynamics of CRF63_02A1, a Recently Originated HIV-1 Circulating Recombinant Form Spreading in Siberia. AIDS Res. Hum. Retroviruses 2014, 30, 912–919. [Google Scholar] [CrossRef]
- Díez-Fuertes, F.; Cabello, M.; Thomson, M.M. Bayesian phylogeographic analyses clarify the origin of the HIV-1 subtype A variant circulating in former Soviet Union’s countries. Infect. Genet. Evol. 2015, 33, 197–205. [Google Scholar] [CrossRef]
- Kostaki, E.-G.; Karamitros, T.; Bobkova, M.; Oikonomopoulou, M.; Magiorkinis, G.; Garcia, F.; Hatzakis, A.; Paraskevis, D. Spatiotemporal Characteristics of the HIV-1 CRF02_AG/CRF63_02A1 Epidemic in Russia and Central Asia. AIDS Res. Hum. Retroviruses 2018, 34, 415–420. [Google Scholar] [CrossRef]
- Aibekova, L.; Foley, B.; Hortelano, G.; Raees, M.; Abdraimov, S.; Toichuev, R.; Ali, S.; Sandstrom, P. Molecular epidemiology of HIV-1 subtype A in former Soviet Union countries. PLoS ONE 2018, 13, e0191891. [Google Scholar] [CrossRef]
- Murzakova, A.; Kireev, D.; Baryshev, P.; Lopatukhin, A.; Serova, E.; Shemshura, A.; Saukhat, S.; Kolpakov, D.; Matuzkova, A.; Suladze, A.; et al. Molecular Epidemiology of HIV-1 Subtype G in the Russian Federation. Viruses 2019, 11, 348. [Google Scholar] [CrossRef]
- Lebedev, A.; Pasechnik, O.; Ozhmegova, E.; Antonova, A.; Blokh, A.; Grezina, L.; Sandyreva, T.; Dementeva, N.; Kazennova, E.; Bobkova, M.; et al. Prevalence and spatiotemporal dynamics of HIV-1 Circulating Recombinant Form 03_AB (CRF03_AB) in the Former Soviet Union countries. PLoS ONE 2020, 15, e0241269, Erratum in PLoS ONE 2021, 16, e0247611. [Google Scholar] [CrossRef]
- Schlösser, M.; Kartashev, V.V.; Mikkola, V.H.; Shemshura, A.; Saukhat, S.; Kolpakov, D.; Suladze, A.; Tverdokhlebova, T.; Hutt, K.; Heger, E.; et al. HIV-1 Sub-Subtype A6: Settings for Normalised Identification and Molecular Epidemiology in the Southern Federal District, Russia. Viruses 2020, 12, 475. [Google Scholar] [CrossRef]
- Abidi, S.H.; Aibekova, L.; Davlidova, S.; Amangeldiyeva, A.; Foley, B.; Ali, S.; Switzer, W.M. Origin and evolution of HIV-1 subtype A6. PLoS ONE 2021, 16, e0260604. [Google Scholar] [CrossRef]
- Mustafa, A.; Akbay, B.; Davlidova, S.; Abidi, S.H.; Ali, S. Origin and evolution of subtype B variants in the former Soviet Union countries. Infect. Genet. Evol. 2023, 108, 105402. [Google Scholar] [CrossRef]
- Sivay, M.V.; Maksimenko, L.V.; Osipova, I.P.; Nefedova, A.A.; Gashnikova, M.P.; Zyryanova, D.P.; Ekushov, V.E.; Totmenin, A.V.; Nalimova, T.M.; Ivlev, V.V.; et al. Spatiotemporal dynamics of HIV-1 CRF63_02A6 sub-epidemic. Front. Microbiol. 2022, 13, 946787. [Google Scholar] [CrossRef]
- Siljic, M.; Cirkovic, V.; Jovanovic, L.; Antonova, A.; Lebedev, A.; Ozhmegova, E.; Kuznetsova, A.; Vinogradova, T.; Ermakov, A.; Monakhov, N.; et al. Reconstructing the Temporal Origin and the Transmission Dynamics of the HIV Subtype B Epidemic in St. Petersburg, Russia. Viruses 2022, 14, 2748. [Google Scholar] [CrossRef]
- Antonova, A.; Kazennova, E.; Lebedev, A.; Ozhmegova, E.; Kuznetsova, A.; Tumanov, A.; Bobkova, M. Recombinant Forms of HIV-1 in the Last Decade of the Epidemic in the Russian Federation. Viruses 2023, 15, 2312. [Google Scholar] [CrossRef]
- Pokrovskiĭ, V.V.; Eramova IIu Deulina, M.O.; Lipetikov, V.V.; Iashkulov, K.B.; Sliusareva, L.A.; Chemizova, N.M.; Savchenko, S.P. Vnutribol’nichnaia vspyshka VICh-infektsii v Eliste [An intrahospital outbreak of HIV infection in Elista]. Zh. Mikrobiol Epidemiol. Immunobiol. 1990, 4, 417–423. [Google Scholar]
- Bobkov, A.; Cheingsong-Popov, R.; Selimova, L.; Kazennova, E.; Karasyova, N.; Kravchenko, A.; Ladnaya, N.; Pokrovsky, V.; Weber, J. Genetic Heterogeneity of HIV Type 1 in Russia: Identification of H Variants and Relationship with Epidemiological Data. AIDS Res. Hum. Retroviruses 1996, 12, 1687–1690. [Google Scholar] [CrossRef]
- Bobkov, A.; Cheingsong-Popov, R.; Garaev, M.; Rzhaninova, A.; Kaleebu, P.; Beddows, S.; Bachmann, M.H.; Mullins, J.I.; Louwagie, J.; Janssens, W.; et al. Identification of an env G subtype and heterogeneity of HIV-1 strains in the Russian Federation and Belarus. AIDS 1994, 8, 1649–1656. [Google Scholar] [CrossRef]
- Hamers, F.F.; Batter, V.; Downs, A.M.; Alix, J.; Cazein, F.; Brunet, J.-B. The HIV epidemic associated with injecting drug use in Europe: Geographic and time trends. AIDS 1997, 11, 1365–1374. [Google Scholar] [CrossRef]
- Bobkov, A.; Cheingsong-Popov, R.; Selimova, L.; Ladnaya, N.; Kazennova, E.; Kravchenko, A.; Fedotov, E.; Saukhat, S.; Zverev, S.; Pokrovsky, V.; et al. An HIV Type 1 Epidemic among Injecting Drug Users in the Former Soviet Union Caused by a Homogeneous Subtype A Strain. AIDS Res. Hum. Retroviruses 1997, 13, 1195–1201. [Google Scholar] [CrossRef]
- Novitsky, V.A.; Montano, M.A.; Essex, M. Molecular Epidemiology of an HIV-1 Subtype A Subcluster among Injection Drug Users in the Southern Ukraine. AIDS Res. Hum. Retroviruses 1998, 14, 1079–1085. [Google Scholar] [CrossRef]
- Liitsola, K.; Tashkinova, I.; Laukkanen, T.; Korovina, G.; Smolskaja, T.; Momot, O.; Mashkilleyson, N.; Chaplinskas, S.; Brummer-Korvenkontio, H.; Vanhatalo, J.; et al. HIV-1 genetic subtype A/B recombinant strain causing an explosive epidemic in injecting drug users in Kaliningrad. AIDS 1998, 12, 1907–1919. [Google Scholar] [CrossRef]
- Bobkov, A.; Kazennova, E.; Selimova, L.; Bobkova, M.; Khanina, T.; Ladnaya, N.; Kravchenko, A.; Pokrovsky, V.; Cheingsong-Popov, R.; Weber, J. A Sudden Epidemic of HIV Type 1 among Injecting Drug Users in the Former Soviet Union: Identification of Subtype A, Subtype B, and NovelgagA/envB Recombinants. AIDS Res. Hum. Retroviruses 1998, 14, 669–676. [Google Scholar] [CrossRef]
- Zarandia, M.; Tsertsvadze, T.; Carr, J.K.; Nadai, Y.; Sanchez, J.L.; Nelson, K. HIV-1 Genetic Diversity and Genotypic Drug Susceptibility in the Republic of Georgia. AIDS Res. Hum. Retroviruses 2006, 22, 470–476, Erratum in AIDS Res. Hum. Retroviruses 2007, 23, 1066. [Google Scholar] [CrossRef]
- Kurbanov, F.; Kondo, M.; Tanaka, Y.; Zalalieva, M.; Giasova, G.; Shima, T.; Jounai, N.; Yuldasheva, N.; Ruzibakiev, R.; Mizokami, M.; et al. Human Immunodeficiency Virus in Uzbekistan: Epidemiological and Genetic Analyses. AIDS Res. Hum. Retroviruses 2003, 19, 731–738. [Google Scholar] [CrossRef]
- Baryshev, P.B.; Bogachev, V.V.; Gashnikova, N.M. HIV-1 Genetic Diversity in Russia: CRF63_02A1, a New HIV Type 1 Genetic Variant Spreading in Siberia. AIDS Res. Hum. Retroviruses 2014, 30, 592–597. [Google Scholar] [CrossRef]
- Lapovok, I.A.; Lopatukhin, A.E.; Kireev, D.E.; Kazennova, E.V.; Lebedev, A.V.; Bobkova, M.R.; Kolomeets, A.N.; Turbina, G.I.; Shipulin, G.A.; Ladnaya, N.N.; et al. Molekuliarno-Épidemiologicheskiĭ Analiz Variantov VICh-1, Tsirkulirovavshikh v Rossii v 1987–2015 gg [Molecular epidemiological analysis of HIV-1 variants circulating in Russia in 1987–2015]. Terapevticheskii Arkhiv 2017, 89, 44–49. [Google Scholar] [CrossRef]
- Kirichenko, A.; Kireev, D.; Lopatukhin, A.; Murzakova, A.; Lapovok, I.; Saleeva, D.; Ladnaya, N.; Gadirova, A.; Ibrahimova, S.; Safarova, A.; et al. Prevalence of HIV-1 drug resistance in Eastern European and Central Asian countries. PLoS ONE 2022, 17, e0257731. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Struck, D.; Lawyer, G.; Ternes, A.-M.; Schmit, J.-C.; Bercoff, D.P. COMET: Adaptive context-based modeling for ultrafast HIV-1 subtype identification. Nucleic Acids Res. 2014, 42, e144. [Google Scholar] [CrossRef]
- Pineda-Peña, A.-C.; Faria, N.R.; Imbrechts, S.; Libin, P.; Abecasis, A.B.; Deforche, K.; Gómez-López, A.; Camacho, R.J.; de Oliveira, T.; Vandamme, A.-M. Automated subtyping of HIV-1 genetic sequences for clinical and surveillance purposes: Performance evaluation of the new REGA version 3 and seven other tools. Infect. Genet. Evol. 2013, 19, 337–348. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Saad, M.D.; Shcherbinskaya, A.M.; Nadai, Y.; Kruglov, Y.V.; Antonenko, S.V.; Lyullchuk, M.G.; Kravchenko, O.N.; Earhart, K.C.; Sanchez, J.L.; Birx, D.L.; et al. Molecular Epidemiology of HIV Type 1 in Ukraine: Birthplace of an Epidemic. AIDS Res. Hum. Retroviruses 2006, 22, 709–714. [Google Scholar] [CrossRef]
- Nabatov, A.A.; Kravchenko, O.N.; Lyulchuk, M.G.; Shcherbinskaya, A.M.; Lukashov, V.V. Simultaneous Introduction of HIV Type 1 Subtype A and B Viruses into Injecting Drug Users in Southern Ukraine at the Beginning of the Epidemic in the Former Soviet Union. AIDS Res. Hum. Retroviruses 2002, 18, 891–895. [Google Scholar] [CrossRef]
- Carr, J.K.; Nadai, Y.; Eyzaguirre, L.; Saad, M.D.; Khakimov, M.M.; Yakubov, S.K.; Birx, D.L.; Graham, R.R.; Wolfe, N.D.; Earhart, K.C.; et al. Outbreak of a West African recombinant of HIV-1 in Tashkent, Uzbekistan. JAIDS J. Acquir. Immune Defic. Syndr. 2005, 39, 570–575. [Google Scholar]
- Campbell, E.M.; Boyles, A.; Shankar, A.; Kim, J.; Knyazev, S.; Cintron, R.; Switzer, W.M.; Marz, M. MicrobeTrace: Retooling molecular epidemiology for rapid public health response. PLOS Comput. Biol. 2021, 17, e1009300. [Google Scholar] [CrossRef]
- Newman, M.E.J. Assortative Mixing in Networks. Phys. Rev. Lett. 2002, 89, 208701. [Google Scholar] [CrossRef]
- Hadfield, J.; Megill, C.; Bell, S.M.; Huddleston, J.; Potter, B.; Callender, C.; Sagulenko, P.; Bedford, T.; Neher, R.A. Nextstrain: Real-time tracking of pathogen evolution. Bioinformatics 2018, 34, 4121–4123. [Google Scholar] [CrossRef]
- Rambaut, A.; Lam, T.T.; Max Carvalho, L.; Pybus, O.G. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol. 2016, 2, vew007. [Google Scholar] [CrossRef]
- ISO 3166-1; Country Codes. International Organization for Standardization: Geneva, Switzerland, 1997.
- Beloukas, A.; Psarris, A.; Giannelou, P.; Kostaki, E.; Hatzakis, A.; Paraskevis, D. Molecular epidemiology of HIV-1 infection in Europe: An overview. Infect. Genet. Evol. 2016, 46, 180–189. [Google Scholar] [CrossRef]
- Pasechnik, O.A.; Blokh, A.I. The Prevalence of HIV Recombinant Forms in Russia and Countries of the Cis: Systematic Review and Metaanalysis. Russ. J. Infect. Immun. 2018, 8, 127–138. [Google Scholar] [CrossRef]
- van de Klundert, M.A.A.; Antonova, A.; Di Teodoro, G.; Diez, R.C.; Chkhartishvili, N.; Heger, E.; Kuznetsova, A.; Lebedev, A.; Narayanan, A.; Ozhmegova, E.; et al. Molecular Epidemiology of HIV-1 in Eastern Europe and Russia. Viruses 2022, 14, 2099. [Google Scholar] [CrossRef]
- Ulyanova, Y.S.; Gashnikova, N.M.; Ivlev, V.V.; Krasnova, E.I.; Khokhlova, N.I.; Totmenin, A.V.; Melnikova, O.B.; Vorotova, M.V.; Glushko, I.R.; Ulyanov, V.V.; et al. Clinical and laboratory characterictic of acute HIV-infection in adult residents of Novosibirsk region. J. Infectology 2019, 11, 40–44. [Google Scholar] [CrossRef]
- Vasylyeva, T.I.; Liulchuk, M.; du Plessis, L.; Fearnhill, E.; Zadorozhna, V.; Babii, N.; Scherbinska, A.; Novitsky, V.; Pybus, O.G.; Faria, N.R. The Changing Epidemiological Profile of HIV-1 Subtype B Epidemic in Ukraine. AIDS Res. Hum. Retroviruses 2019, 35, 155–163. [Google Scholar] [CrossRef]
- Brenner, B.G.; Ibanescu, R.-I.; Osman, N.; Cuadra-Foy, E.; Oliveira, M.; Chaillon, A.; Stephens, D.; Hardy, I.; Routy, J.-P.; Thomas, R.; et al. The Role of Phylogenetics in Unravelling Patterns of HIV Transmission towards Epidemic Control: The Quebec Experience (2002–2020). Viruses 2021, 13, 1643. [Google Scholar] [CrossRef]
- González-Baeza, A.; Dolengevich-Segal, H.; Pérez-Valero, I.; Cabello, A.; Téllez, M.J.; Sanz, J.; Pérez-Latorre, L.; Bernardino, J.I.; Troya, J.; De La Fuente, S.; et al. Sexualized Drug Use (Chemsex) Is Associated with High-Risk Sexual Behaviors and Sexually Transmitted Infections in HIV-Positive Men Who Have Sex with Men: Data from the U-SEX GESIDA 9416 Study. AIDS Patient Care STDs 2018, 32, 112–118. [Google Scholar] [CrossRef]
- Cheung, D.H.; Samoh, N.; Jonas, K.J.; Lim, S.H.; Kongjareon, Y.; Guadamuz, T.E. Patterns of Chemsex Substance Use and Its Association with HIV Transmission Risk Among Men Who Have Sex with Men in Thailand: A Latent Class Analysis. Arch. Sex. Behav. 2024, 53, 3527–3536. [Google Scholar] [CrossRef]
- Rechel, B. HIV/AIDS in the Countries of the Former Soviet Union: Societal and Attitudinal Challenges. Central Eur. J. Public Health 2010, 18, 110–115. [Google Scholar] [CrossRef]
- Hué, S.; Brown, A.E.; Ragonnet-Cronin, M.; Lycett, S.J.; Dunn, D.T.; Fearnhill, E.; Dolling, D.I.; Pozniak, A.; Pillay, D.; Delpech, V.C.; et al. Phylogenetic analyses reveal HIV-1 infections between men misclassified as heterosexual transmissions. AIDS 2014, 28, 1967–1975. [Google Scholar] [CrossRef]
- van de Laar, T.J.; Bezemer, D.; van Laethem, K.; Vandewalle, G.; de Smet, A.; van Wijngaerden, E.; Claas, E.C.; van Sighem, A.I.; Vandamme, A.; Compernolle, V.; et al. Phylogenetic evidence for underreporting of male-to-male sex among human immunodeficiency virus–infected donors in the Netherlands and Flanders. Transfusion 2017, 57, 1235–1247. [Google Scholar] [CrossRef]
- Lazouskaya, N.V.; Eremin, V.F.; Adema, K.W.; Gasich, E.L.; Baan, E.; Lukashov, V.V. The HIV Type 1 Epidemic in Belarus: Predominance of Eastern European Subtype A Strains and Circulation of Subtype B Viruses. AIDS Res. Hum. Retroviruses 2005, 21, 830–833. [Google Scholar] [CrossRef]
- Bártolo, I.; Abecasis, A.B.; Borrego, P.; Barroso, H.; McCutchan, F.; Gomes, P.; Camacho, R.; Taveira, N.; Martin, D.P. Origin and Epidemiological History of HIV-1 CRF14_BG. PLoS ONE 2011, 6, e24130. [Google Scholar] [CrossRef]
- Baryshev, P.B.; Bogachev, V.V.; Gashnikova, N.M. Genetic characterization of an isolate of HIV type 1 AG recombinant form circulating in Siberia, Russia. Arch. Virol. 2012, 157, 2335–2341. [Google Scholar] [CrossRef]
- Fernández-García, A.; Delgado, E.; Cuevas, M.T.; Vega, Y.; Montero, V.; Sánchez, M.; Carrera, C.; López-Álvarez, M.J.; Miralles, C.; Pérez-Castro, S.; et al. Identification of an HIV-1 BG Intersubtype Recombinant Form (CRF73_BG), Partially Related to CRF14_BG, Which Is Circulating in Portugal and Spain. PLoS ONE 2016, 11, e0148549. [Google Scholar] [CrossRef]
- Vasylyeva, T.I.; Liulchuk, M.; Friedman, S.R.; Sazonova, I.; Faria, N.R.; Katzourakis, A.; Babii, N.; Scherbinska, A.; Thézé, J.; Pybus, O.G.; et al. Molecular epidemiology reveals the role of war in the spread of HIV in Ukraine. Proc. Natl. Acad. Sci. USA 2018, 115, 1051–1056. [Google Scholar] [CrossRef]
- Hemelaar, J.; Elangovan, R.; Yun, J.; Dickson-Tetteh, L.; Fleminger, I.; Kirtley, S.; Williams, B.; Gouws-Williams, E.; Ghys, P.D.; on behalf of theWHO–UNAIDS Network for HIV Isolation Characterisation. Global and regional molecular epidemiology of HIV-1, 1990–2015: A systematic review, global survey, and trend analysis. Lancet Infect. Dis. 2019, 19, 143–155, Erratum in Lancet Infect Dis. 2020, 20, e27. [Google Scholar] [CrossRef]
- Mamatkulov, A.; Kazakova, E.; Ibadullaeva, N.; Joldasova, E.; Bayjanov, A.; Musabaev, E.; Kan, N.; Mustafaeva, D.; Lebedev, A.V.; Bobkova, M.; et al. Prevalence of Antiretroviral Drug Resistance Mutations Among Pretreatment and Antiretroviral Therapy-Failure HIV Patients in Uzbekistan. AIDS Res. Hum. Retroviruses 2021, 37, 38–43. [Google Scholar] [CrossRef]
- Sivay, M.V.; Totmenin, A.V.; Zyryanova, D.P.; Osipova, I.P.; Nalimova, T.M.; Gashnikova, M.P.; Ivlev, V.V.; Meshkov, I.O.; Chokmorova, U.Z.; Narmatova, E.; et al. Characterization of HIV-1 Epidemic in Kyrgyzstan. Front. Microbiol. 2021, 12, 753675. [Google Scholar] [CrossRef]
- Sanaubarova, A.; Pujol-Hodge, E.; Dzissyuk, N.; Lemey, P.; Vermund, S.H.; Brown, A.J.L.; Ali, S. High-Level Drug-Resistant Mutations among HIV-1 Subtype A6 and CRF02_AG in Kazakhstan. Viruses 2023, 15, 1407. [Google Scholar] [CrossRef]
- Nair, M.; Gettins, L.; Fuller, M.; Kirtley, S.; Hemelaar, J. Global and regional genetic diversity of HIV-1 in 2010–21: Systematic review and analysis of prevalence. Lancet Microbe 2024, 5, 100912. [Google Scholar] [CrossRef]
- Lukashov, V.V.; Huismans, R.; Rakhmanova, A.G.; Lisitsina, Z.N.; Akhtyrskaya, N.A.; Vlasov, N.N.; Melnick, O.B.; Goudsmit, J. Circulation of Subtype A and gagA/envB Recombinant HIV Type 1 Strains among Injecting Drug Users in St. Petersburg, Russia, Correlates with Geographical Origin of Infections. AIDS Res. Hum. Retroviruses 1999, 15, 1577–1583. [Google Scholar] [CrossRef]
- Bobkov, A.; Kazennova, E.; Khanina, T.; Bobkova, M.; Selimova, L.; Kravchenko, A.; Pokrovsky, V.; Weber, J. An HIV Type 1 Subtype A Strain of Low Genetic Diversity Continues to Spread among Injecting Drug Users in Russia: Study of the New Local Outbreaks in Moscow and Irkutsk. AIDS Res. Hum. Retroviruses 2001, 17, 257–261. [Google Scholar] [CrossRef]
- Neshumaev, D.; Lebedev, A.; Malysheva, M.; Boyko, A.; Skudarnov, S.; Ozhmegova, E.; Antonova, A.; Kazennova, E.; Bobkova, M. Molecular Surveillance of HIV-1 Infection in Krasnoyarsk Region, Russia: Epidemiology, Phylodynamics and Phylogeography. Curr. HIV Res. 2019, 17, 114–125. [Google Scholar] [CrossRef]
- Lebedev, A.; Lebedeva, N.; Moskaleychik, F.; Pronin, A.; Kazennova, E.; Bobkova, M. Human Immunodeficiency Virus-1 Diversity in the Moscow Region, Russia: Phylodynamics of the Most Common Subtypes. Front. Microbiol. 2019, 10, 320. [Google Scholar] [CrossRef]
- Migration Policy Institute. Russia: A Migration System with Soviet Roots. Available online: https://www.migrationpolicy.org/article/russia-migration-system-soviet-roots (accessed on 1 December 2024).
- Amirkhanian, Y.A.; Kuznetsova, A.V.; Kelly, J.A.; DiFranceisco, W.J.; Musatov, V.B.; Avsukevich, N.A.; Chaika, N.A.; McAuliffe, T.L. Male Labor Migrants in Russia: HIV Risk Behavior Levels, Contextual Factors, and Prevention Needs. J. Immigr. Minor. Health 2010, 13, 919–928. [Google Scholar] [CrossRef]
- Zakharov, V.Y.; Ivanova, A.N.; Suzdaleva, T.R. Labor migration in modern Russia: Features, problems and ways to solve them. Rev. Amaz. Investig. 2023, 12, 198–207. [Google Scholar] [CrossRef]
- Yılmaz, G.; Midilli, K.; Türkoğlu, S.; Bayraktaroğlu, Z.; Kuşkucu, A.M.; Özkan, E.; Atasever, L.; Çalangu, S.; Altaş, K. Genetic subtypes of human immunodeficiency virus type 1 (HIV-1) in Istanbul, Turkey. Int. J. Infect. Dis. 2006, 10, 286–290. [Google Scholar] [CrossRef]
- Salemi, M.; Goodenow, M.M.; Montieri, S.; de Oliveira, T.; Santoro, M.M.; Beshkov, D.; Alexiev, I.; Elenkov, I.; Yakimova, T.; Varleva, T.; et al. The HIV Type 1 Epidemic in Bulgaria Involves Multiple Subtypes and Is Sustained by Continuous Viral Inflow from West and East European Countries. AIDS Res. Hum. Retroviruses 2008, 24, 771–779. [Google Scholar] [CrossRef]
- Lai, A.; Bozzi, G.; Franzetti, M.; Binda, F.; Simonetti, F.R.; De Luca, A.; Micheli, V.; Meraviglia, P.; Bagnarelli, P.; Di Biagio, A.; et al. HIV-1 A1 Subtype Epidemic in Italy Originated from Africa and Eastern Europe and Shows a High Frequency of Transmission Chains Involving Intravenous Drug Users. PLoS ONE 2016, 11, e0146097. [Google Scholar] [CrossRef][Green Version]
- Moskaleychik, F.F.; Laga, V.Y.; Delgado, E.; Vega, Y.; Fernandez-Garcia, A.; Kornilaeva, G.V.; Pronin, A.Y.; Zhernov, Y.V.; Thomson, M.M.; Bobkova, M.R.; et al. Rapid spread of the HIV-1 circular recombinant CRF02-AG in Russia and neighboring countries. Voprosy Virusologii 2015, 60, 14–19. [Google Scholar]
- Mir, D.; Jung, M.; Delatorre, E.; Vidal, N.; Peeters, M.; Bello, G. Phylodynamics of the major HIV-1 CRF02_AG African lineages and its global dissemination. Infect. Genet. Evol. 2016, 46, 190–199. [Google Scholar] [CrossRef]
- Lebedev, A.; Kuznetsova, A.; Kim, K.; Ozhmegova, E.; Antonova, A.; Kazennova, E.; Tumanov, A.; Mamatkulov, A.; Kazakova, E.; Ibadullaeva, N.; et al. Identifying HIV-1 Transmission Clusters in Uzbekistan through Analysis of Molecular Surveillance Data. Viruses 2022, 14, 1675. [Google Scholar] [CrossRef]
- Niculescu, I.; Paraschiv, S.; Paraskevis, D.; Abagiu, A.; Batan, I.; Banica, L.; Otelea, D. Recent HIV-1 Outbreak Among Intravenous Drug Users in Romania: Evidence for Cocirculation of CRF14_BG and Subtype F1 Strains. AIDS Res. Hum. Retroviruses 2015, 31, 488–495. [Google Scholar] [CrossRef]
- Paraskevis, D.; Nikolopoulos, G.; Fotiou, A.; Tsiara, C.; Paraskeva, D.; Sypsa, V.; Lazanas, M.; Gargalianos, P.; Psichogiou, M.; Skoutelis, A.; et al. Economic Recession and Emergence of an HIV-1 Outbreak among Drug Injectors in Athens Metropolitan Area: A Longitudinal Study. PLoS ONE 2013, 8, e78941. [Google Scholar] [CrossRef]

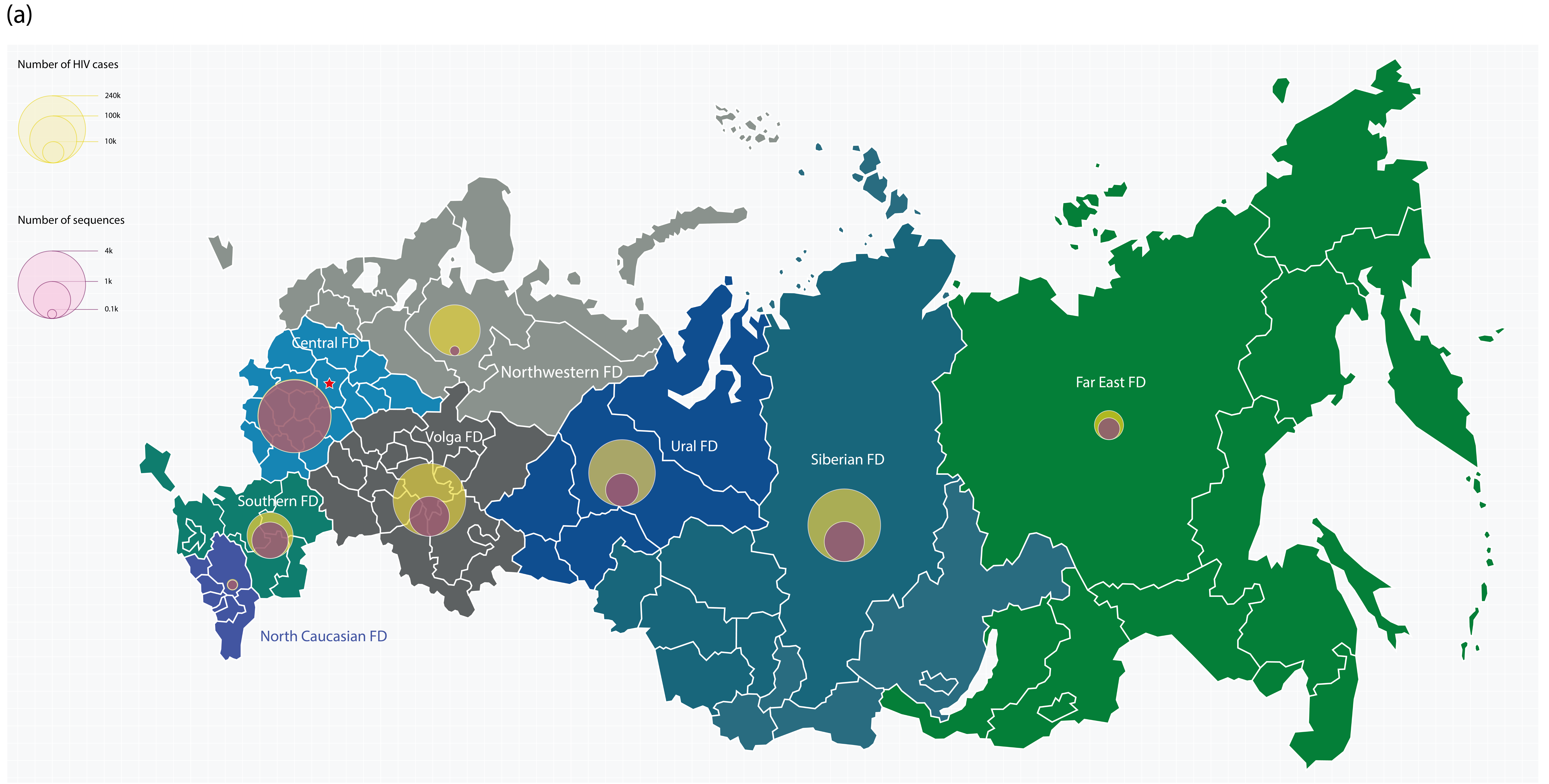

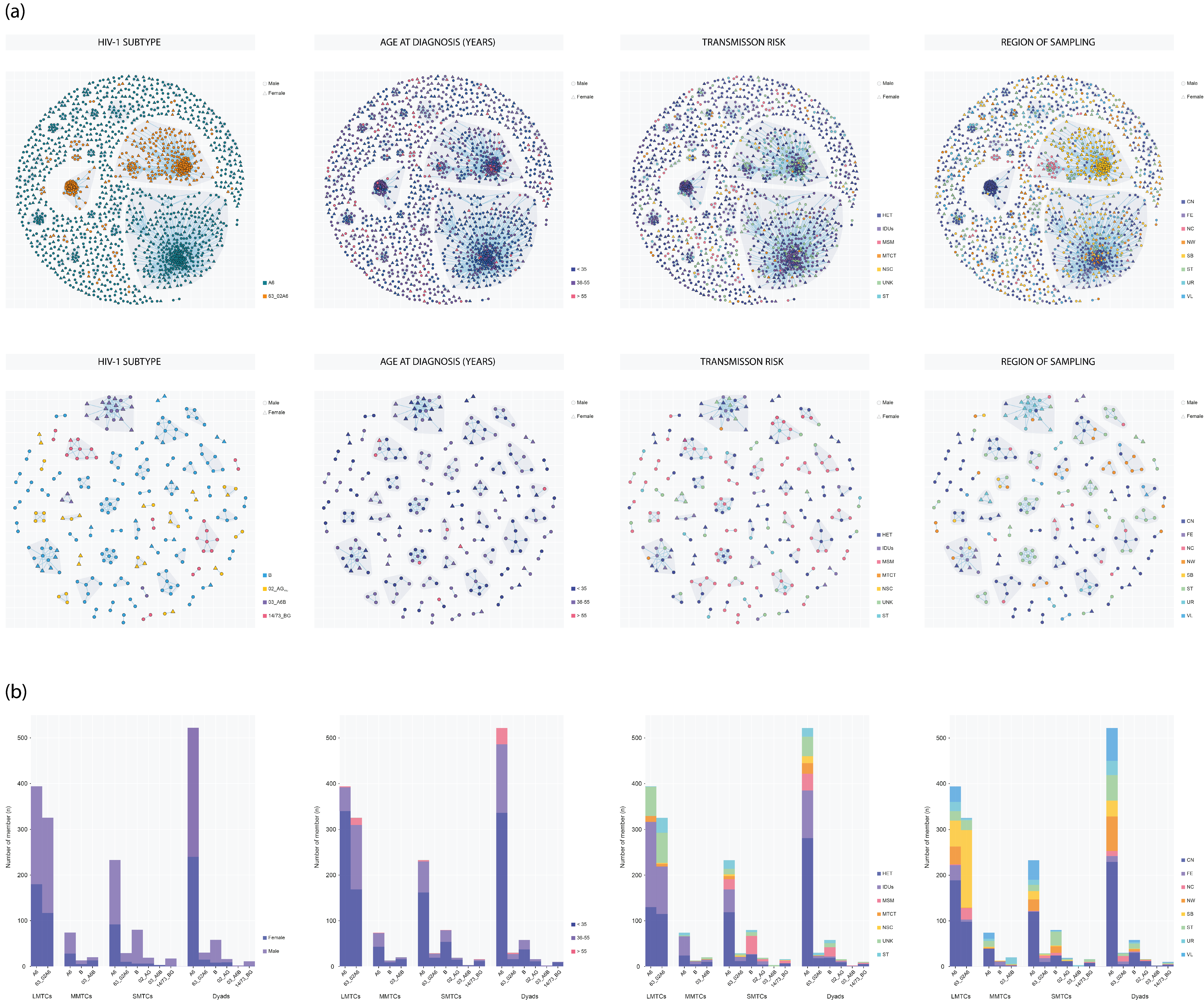
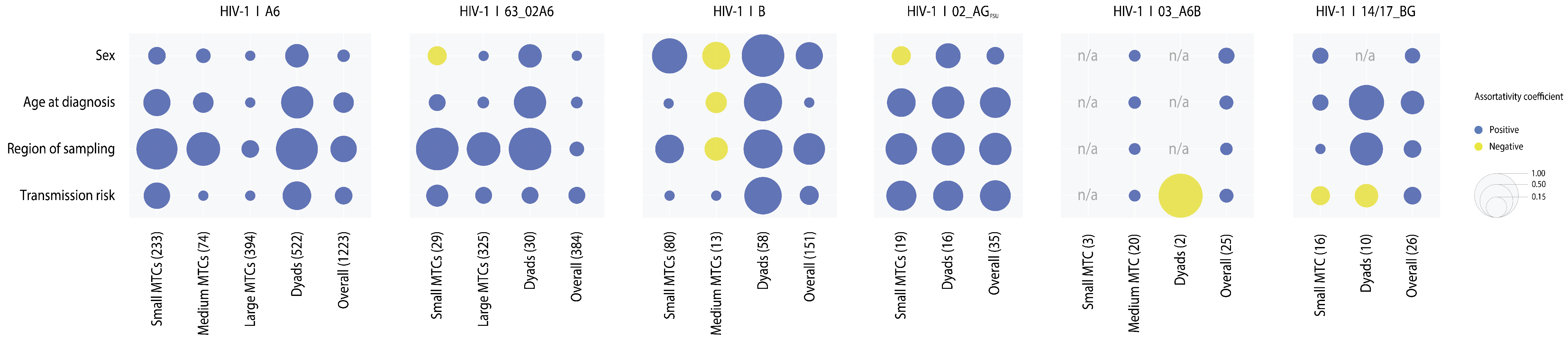
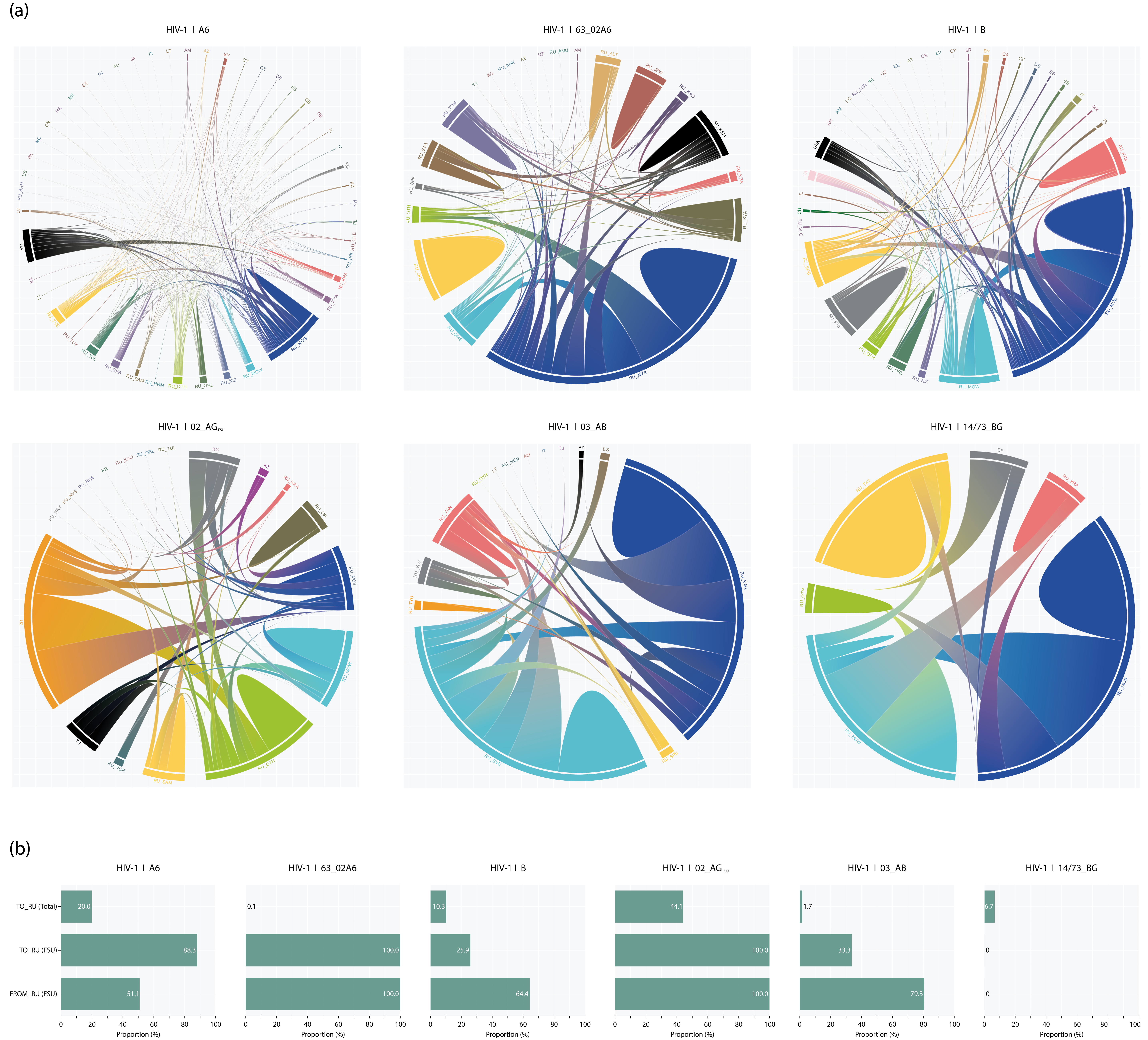
| HIV-1 Subtype | ||||||||
|---|---|---|---|---|---|---|---|---|
| Total (n = 9500, 100.0%) | A6 (n = 7659; 80.6% (95% CI, 79.8–81.4) | 63_02A6 (n = 747; 7.9%; (95% CI, 7.3–8.4) | B (n = 531; 5.6% (95% CI, 5.1–6.1) | 02_AGFSU (n = 111; 1.2% (95% CI, 0.1–1.4) | 03_A6B (n = 68; 0.7% (95% CI, 0.6–0.9) | 14/73_BG (n = 59; 0.6% (95% CI, 0.5–0.8) | Others 1 (n = 325; 3.4% (95% CI, 3.1–3.8) | |
| Age at diagnosis (in years) | ||||||||
| <20 | 1034 (10.9) | 887 (11.6) | 55 (7.4) | 33 (6.2) | 6 (5.4) | 10 (14.7) | 2 (3.4) | 41 (12.6) |
| 20–29 | 3406 (35.9) | 2817 (36.8) | 165 (22.1) | 215 (40.5) | 40 (36.0) | 29 (42.6) | 29 (49.2) | 111 (34.1) |
| 30–39 | 3084 (32.5) | 2422 (31.6) | 299 (40.0) | 185 (34.8) | 38 (34.2) | 22 (32.4) | 14 (23.7) | 104 (32.0) |
| 40–49 | 1336 (14.1) | 1025 (13.4) | 158 (21.2) | 68 (12.8) | 21 (18.9) | 6 (8.8) | 8 (13.6) | 50 (15.4) |
| ≥50 | 640 (6.7) | 508 (6.6) | 70 (9.4) | 30 (5.6) | 6 (5.4) | 1 (1.5) | 6 (10.2) | 19 (5.8) |
| Median (IQR) | 30.0 (24.0–38.0) | 30.0 (23.0–37.0) | 35.0 (28.0–41.0) | 30.0 (25.0–38.0) | 32.0 (25.0–39.0) | 26.0 (22.0–34.0) | 28.0 (24.0–38.0) | 30.0 (24.0–37.0) |
| Sex | ||||||||
| Female | 3864 (40.7) | 3310 (43.2) | 294 (39.4) | 63 (11.9) | 41 (36.9) | 42 (61.8) | 6 (10.2) | 108 (33.2) |
| Male | 5636 (59.3) | 4349 (56.8) | 453 (60.6) | 468 (88.1) | 70 (63.1) | 26 (38.2) | 53 (89.8) | 217 (66.8) |
| Transmission risk | ||||||||
| HET | 4140 (43.6) | 3482 (45.5) | 281 (37.6) | 169 (31.8) | 49 (44.1) | 27 (39.7) | 21 (35.6) | 111 (34.1) |
| IDUs | 2541 (26.7) | 2195 (28.7) | 213 (28.5) | 38 (7.2) | 19 (17.1) | 12 (17.6) | 2 (3.4) | 62 (19.1) |
| MSM | 535 (5.6) | 226 (3.0) | 5 (0.7) | 202 (38.0) | 19 (17.1) | 1 (1.5) | 22 (37.3) | 60 (18.5) |
| MTCT | 315 (3.3) | 278 (3.6) | 18 (2.4) | 4 (0.8) | 3 (2.7) | 3 (4.4) | 0 (0) | 9 (2.8) |
| NSC | 62 (0.7) | 40 (0.5) | 6 (0.8) | 0 (0) | 0 (0) | 0 (0) | 1 (1.7) | 15 (4.6) |
| UNK | 1275 (13.4) | 985 (12.9) | 140 (18.7) | 60 (11.3) | 14 (12.6) | 22 (32.4) | 6 (10.2) | 48 (14.8) |
| ST | 632 (6.7) | 453 (5.9) | 84 (11.2) | 58 (10.9) | 7 (6.3) | 3 (4.4) | 7 (11.9) | 20 (6.2) |
| Diagnosis date | ||||||||
| before 2000 | 536 (5.6) | 452 (5.9) | 0 (0) | 51 (9.6) | 0 (0) | 3 (4.4) | 0 (0) | 30 (9.2) |
| 2001–2005 | 1188 (12.5) | 1105 (14.4) | 10 (1.3) | 37 (7.0) | 4 (3.6) | 9 (13.2) | 1 (1.7) | 22 (6.8) |
| 2006–2010 | 1902 (20.0) | 1644 (21.5) | 64 (8.6) | 95 (17.9) | 21 (19.0) | 31 (45.6) | 4 (6.8) | 43 (13.2) |
| 2011–2015 | 2214 (23.3) | 1767 (23.0) | 186 (24.9) | 144 (27.1) | 21 (18.9) | 14 (20.6) | 18 (30.5) | 64 (19.7) |
| 2016–2020 | 2270 (24.0) | 1805 (23.6) | 180 (24.1) | 142 (26.7) | 33 (29.7) | 10 (14.7) | 24 (40.7) | 76 (23.4) |
| after 2020 | 1390 (14.6) | 886 (11.6) | 307 (41.1) | 62 (11.7) | 32 (28.8) | 1 (1.5) | 12 (20.3) | 90 (27.7) |
| Characteristics | Estimate | St. Error | t Value | p Value | Odds Ratio (95% CI) |
|---|---|---|---|---|---|
| Sex (female vs. male) | 0.414 | 0.058 | 7.103 | <0.001 | 1.51 (1.35–1.70) |
| Age at diagnosis (in years) | 0.014 | 0.002 | 6.693 | <0.001 | 3.60 (2.47–5.24) |
| Region of sampling (Central FD vs. Other) 1 | 0.221 | 0.054 | 4.076 | <0.001 | 1.25 (1.12–1.39) |
| Transmission category | |||||
| HET vs. IDUs | −0.134 | 0.079 | −1.700 | 0.089 | 0.87 (0.74–1.02) |
| HET vs. MSM | 0.996 | 0.057 | 17.483 | <0.001 | 7.33 (5.86–9.17) |
| HET vs. Other 2 | 0.119 | 0.024 | 5.032 | <0.001 | 1.34 (1.24–1.64) |
| Attribute | Category | Total, n | Non-Clustering, n (%) | Clustering, n (%) | Adjusted Odds Ratio (95% CI) | p-Value |
|---|---|---|---|---|---|---|
| Age at diagnosis (in years) | ||||||
| ≥50 | 621 | 504 (81.2) | 117 (18.8) | Ref | ||
| 30–49 | 4266 | 3400 (79.7) | 866 (20.3) | 1.17 (0.93–1.46) | 0.180 | |
| <30 | 4288 | 3427 (79.9) | 861 (20.1) | 1.31 (1.03–1.66) | 0.028 | |
| Sex | ||||||
| Female | 3756 | 3023 (80.5) | 733 (19.5) | Ref | ||
| Male | 5419 | 4308 (79.5) | 1111 (20.5) | 1.09 (0.97–1.22) | 0.143 | |
| Transmission risk | ||||||
| HET | 4029 | 3247 (80.6) | 782 (19.4) | Ref | ||
| IDUs | 2479 | 1954 (78.8) | 525 (21.2) | 1.28 (1.01–1.62) | 0.043 | |
| MSM | 475 | 338 (71.2) | 137 (28.8) | 1.11 (0.86–1.44) | 0.410 | |
| MTCT | 306 | 254 (83.0) | 52 (17.0) | 1.35 (0.96–1.90) | 0.083 | |
| NSC | 47 | 21 (44.7) | 26 (55.3) | 5.26 (2.83–9.76) | <0.001 | |
| UNK | 1228 | 1003 (81.7) | 225 (18.3) | 0.83 (0.70–1.00) | 0.054 | |
| ST | 611 | 514 (84.1) | 97 (15.9) | 0.62 (0.48–0.82) | 0.001 | |
| ART status | ||||||
| Treated | 5084 | 4414 (86.8) | 670 (13.2) | Ref | ||
| Naive | 4091 | 2917 (71.3) | 1174 (28.7) | 2.70 (2.40–3.04) | <0.001 | |
| HIV-1 subtype | ||||||
| A6 | 7659 | 6436 (84.0) | 1223 (16.0) | Ref | ||
| B | 531 | 380 (71.6) | 151 (28.4) | 2.16 (1.76–2.66) | <0.001 | |
| 63_02A6 | 747 | 363 (48.6) | 384 (51.4) | 5.70 (4.82–6.73) | <0.001 | |
| 02_AGFSU | 111 | 76 (68.5) | 35 (31.5) | 2.37 (1.56–3.60) | <0.001 | |
| 03_A6B | 68 | 43 (63.2) | 25 (36.8) | 2.81 (1.68–4.70) | <0.001 | |
| 14/73_BG | 59 | 33 (55.9) | 26 (44.1) | 3.54 (2.06–6.06) | <0.001 | |
| Region of sampling 1 | ||||||
| CN | 3840 | 3057 (79.6) | 783 (20.4) | Ref | ||
| FE | 411 | 331 (80.5) | 80 (19.5) | 0.84 (0.64–1.09) | 0.193 | |
| NC | 267 | 182 (68.2) | 85 (31.8) | 1.81 (1.35–2.43) | <0.001 | |
| NW | 834 | 787 (94.4) | 47 (5.6) | 0.28 (0.20–0.39) | <0.001 | |
| SB | 1111 | 794 (71.5) | 317 (28.5) | 1.31 (1.10–1.56) | 0.002 | |
| ST (FD) | 870 | 576 (66.2) | 294 (33.8) | 1.55 (1.31–1.84) | <0.001 | |
| UR | 764 | 667 (87.3) | 97 (12.7) | 0.49 (0.39–0.63) | <0.001 | |
| VL | 1078 | 937 (86.9) | 141 (13.1) | 0.62 (0.50–0.75) | <0.001 | |
| Diagnosis date | ||||||
| 1988–2004 | 1476 | 1192 (80.8) | 284 (19.2) | Ref | ||
| 2005–2015 | 4205 | 3478 (82.7) | 727 (17.3) | 0.60 (0.51–0.72) | <0.001 | |
| 2016–2023 | 3494 | 2661 (76.2) | 833 (23.8) | 0.76 (0.61–0.93) | 0.009 |
| Subtype | Phylogenetic Cluster (Subcluster, #) | Size, n 1 | Sampling Year | Likely Phylogenetic Origin | Geographic Location (Proportion, %) 2 | TMRCA (95% CI) |
|---|---|---|---|---|---|---|
| A6 | Overall | 12,396 | 1997–2024 | Ukraine | Mixed | 1993.2 (1992.0–1994.9) |
| Cluster 1 | 540 | 2001–2023 | Russia, Moscow oblast | Mixed [Russian (76.1)] | 1996.8 (1995.9–1998.3) | |
| Cluster 2 | 203 | 2007–2022 | Belarus | Belarus (99.5) [Russian (0.5)] | 1999.1 (1997.8–2000.6) | |
| Cluster 3 | 140 | 2011–2023 | Poland | Poland (86.4%) [Russian (3.6)] | 2002.8 (2001.6–2004.1) | |
| Cluster 4 | 101 | 2010–2021 | Ukraine | Poland (95.0) [Russian (1.0)] | 2005.6 (2003.8–2007.4) | |
| Cluster 5 | 93 | 2005–2023 | Ukraine | Krasnoyarsk krai (78.5) [Russian (94.6)] | 1999.1 (1997.6–2000.8) | |
| Cluster 6 | 84 | 2005–2014 | Ukraine | Latvia (88.1) [Russian (3.6)] | 2000.1 (1998.8–2001.6) | |
| Cluster 7 | 61 | 2018–2023 | Ukraine | Orel oblast (98.4) [Russian (100.0)] | 2004.4 (2002.7–2006.1) | |
| Cluster 8 | 55 | 2002–2022 | Ukraine | Mixed [Russian (81.8)] | 2000.0 (1998.4–2001.5) | |
| B | Overall | 3034 | 1978–2024 | United States | Mixed | 1972.5 (1967.5–1976.1) |
| Cluster 1 | 192 | 1995–2023 | United States | Mixed [Russian (72.4)] | 1978.1 (1977.3–1978.8) | |
| #1.1 | 33 | 1995–2023 | Russia, Moscow (city) | Mixed [Russian (85.3)] | 1982.7 (1981.7–1984.1) | |
| #1.2 | 16 | 1995–2023 | Russia, St. Petersburg (city) | Mixed [Russian (93.7)] | 1988.3 (1986.8–1990.2) | |
| #1.3 | 15 | 2006–2017 | Russia, Moscow oblast | Krasnodar krai (73.3) [Russian (100.0)] | 1987.4 (1985.6–1989.5) | |
| Cluster 2 | 273 | 1987–2023 | Poland | Mixed [Russian (20.5)] | 1982.9 (1980.8–1985.2) | |
| #2.1 | 106 | 1996–2023 | Poland | Mixed [Russian (51.9)] | 1988.7 (1987.4–1990.3) | |
| Cluster 3 | 87 | 1995–2023 | Russia, Moscow oblast | Mixed [Russian (87.3)] | 1981.8 (1982.0–1982.8) | |
| #3.1 | 29 | 1995–2022 | Russia, Moscow (city) | Mixed [Russian (93.1)] | 1987.2 (1985.8–1988.8) | |
| #3.2 | 32 | 1999–2023 | Russia, Moscow oblast | Mixed [Russian (96.9)] | 1984.3 (1983.3–1985.4) | |
| Cluster 4 | 80 | 1995–2023 | Russia, Moscow oblast | Mixed [Russian (66.2)] | 1980.9 (1979.6–1982.7) | |
| Cluster 5 | 70 | 1995–2023 | Russia, Moscow oblast | Mixed [Russian (68.6)] | 1983.0 (1982.0–1984.5) | |
| #5.1 | 24 | 2004–2020 | Russia, Moscow oblast | Mixed [Russian (54.2)] | 1987.0 (1985.6–1988.8) | |
| #5.2 | 14 | 2008–2023 | Russia, Moscow oblast | Mixed [Russian (85.7)] | 1988.7 (1986.5–1991.4) | |
| Cluster 6 | 46 | 2003–2021 | Italy | Mixed [Russian (43.5)] | 1986.8 (1985.2–1988.8) | |
| #6.1 | 21 | 2009–2020 | Italy/Russia, Moscow oblast | Mixed [Russian (90.5)] | 1992.3 (1990.4–1994.3) | |
| Cluster 7 | 41 | 1995–2023 | Russia, Moscow oblast | Mixed [Russian (85.4)] | 1985.7 (1981.0–1986.7) | |
| Cluster 8 | 17 | 2007–2020 | United States/Russia, St. Petersburg (city) | Mixed [Russian (76.5)] | 1990.5 (1988.6–1993.1) | |
| Cluster 9 | 14 | 2002–2022 | United States/Russia, St. Petersburg (city) | Mixed [Russian (50.0)] | 1985.8 (1984.7–1987.7) | |
| Cluster 10 | 12 | 1999–2021 | United Kingdom | Mixed [Russian (63.6)] | 1987.3 (1985.5–1989.0) | |
| 63_02A6 | Overall | 1372 | 2008–2024 | Russia, Novosibirsk oblast | Mixed | 2004.3 (2002.8–2005.7) |
| Cluster 1 | 129 | 2016–2023 | Russia, Tomsk oblast | Tomsk oblast (44.2) [Russian (99.2)] | 2010.6 (2009.9–2011.6) | |
| #1.1 | 62 | 2016–2023 | Russia, Stavropol krai/Krasnodar krai | Karachay-Cherkess rep. (46.8) [Russian (100.0)] | 2012.2 (2011.7–2012.8) | |
| Cluster 2 | 108 | 2016–2023 | Russia, Novosibirsk oblast | Orel region (98.1) [Russian (100.0)] | 2012.1 (2011.2–2013.1) | |
| Cluster 3 | 92 | 2011–2023 | Russia, Kemerovo oblast | Kemerovo oblast (37.0) [Russian (100.0)] | 2009.3 (2008.7–2009.9) | |
| Cluster 4 | 54 | 2015–2023 | Russia, Novosibirsk oblast | Krasnoyarsk krai (48.1) [Russian (100.0)] | 2010.9 (2010.3–2012.1) | |
| Cluster 5 | 37 | 2015–2024 | Russia, Novosibirsk oblast | Jewish Aut. oblast (97.3) [Russian (100.0)] | 2010.7 (2010.1–2011.6) | |
| Cluster 6 | 31 | 2015–2023 | Russia, Novosibirsk oblast | Mixed [Russian (100.0)] | 2008.8 (2008.4–2009.3) | |
| 02_AGFSU | Overall | 309 | 2002–2023 | Uzbekistan | Mixed | 1999.6 (1997.2–2001.8) |
| Cluster 1 | 25 | 2014–2023 | Russia, Moscow (city) | Mixed [Russian (96.0)] | 2005.1 (2004.0–2007.3) | |
| Cluster 2 | 24 | 2009–2021 | Uzbekistan | Kyrgyzstan (75.0) [Russian (25.0)] | 2006.7 (2005.7–2008.0) | |
| Cluster 3 | 9 | 2015–2023 | Uzbekistan | Lipetsk oblast (77.8) [Russian (100.0)] | 2006.0 (2004.6–2007.7) | |
| Cluster 4 | 7 | 2018–2022 | Kazakhstan/Kyrgyzstan | Moscow (city) (57.1) [Russian (85.7)] | 2010.4 (2008.3–2012.9) | |
| Cluster 5 | 6 | 2017–2023 | Kyrgyzstan | Kyrgyzstan (83.3) [Russian (16.7)] | 2012.6 (2011.0–2014.3) | |
| 03_A6B | Overall | 300 | 1997–2024 | Russia, Kaliningrad oblast | Mixed | 1990.8 (1987.6–1994.0) |
| Cluster 1 | 290 | 1997–2024 | Russia, Kaliningrad oblast | Mixed [Russian (88.6)] | 1993.4 (1991.1–1995.9) | |
| Cluster 2 | 10 | 1998–2017 | Russia, Kaliningrad oblast | Mixed [Russian (100.0)] | 1996.3 (1993.9–1997.0) | |
| 14/73_BG | Overall | 143 | 1998–2023 | Spain | Mixed | 1991.6 (1987.3–1993.3) |
| Cluster 1 | 56 | 2009–2023 | Spain | Mixed [Russian (100.0)] | 1999.9 (1998.1–2002.3) | |
| #1.1 | 10 | 2010–2022 | Russia, Moscow (city) | Moscow (city) (40.0%) [Russian (100.0)] | 2008.0 (2006.8–2009.3) | |
| #1.2 | 14 | 2009–2022 | Russia, Moscow (city)/Moscow oblast | Tatarstan (71.4%) [Russian (100.0)] | 2003.3 (2001.4–2005.2) | |
| Cluster 2 | 3 | 2008–2023 | Spain | Bashkortostan (66.7%) [Russian (100.0)] | 2005.8 (2004.0–2008.0) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lebedev, A.; Kireev, D.; Kirichenko, A.; Mezhenskaya, E.; Antonova, A.; Bobkov, V.; Lapovok, I.; Shlykova, A.; Lopatukhin, A.; Shemshura, A.; et al. The Molecular Epidemiology of HIV-1 in Russia, 1987–2023: Subtypes, Transmission Networks and Phylogenetic Story. Pathogens 2025, 14, 738. https://doi.org/10.3390/pathogens14080738
Lebedev A, Kireev D, Kirichenko A, Mezhenskaya E, Antonova A, Bobkov V, Lapovok I, Shlykova A, Lopatukhin A, Shemshura A, et al. The Molecular Epidemiology of HIV-1 in Russia, 1987–2023: Subtypes, Transmission Networks and Phylogenetic Story. Pathogens. 2025; 14(8):738. https://doi.org/10.3390/pathogens14080738
Chicago/Turabian StyleLebedev, Aleksey, Dmitry Kireev, Alina Kirichenko, Ekaterina Mezhenskaya, Anastasiia Antonova, Vyacheslav Bobkov, Ilya Lapovok, Anastasia Shlykova, Alexey Lopatukhin, Andrey Shemshura, and et al. 2025. "The Molecular Epidemiology of HIV-1 in Russia, 1987–2023: Subtypes, Transmission Networks and Phylogenetic Story" Pathogens 14, no. 8: 738. https://doi.org/10.3390/pathogens14080738
APA StyleLebedev, A., Kireev, D., Kirichenko, A., Mezhenskaya, E., Antonova, A., Bobkov, V., Lapovok, I., Shlykova, A., Lopatukhin, A., Shemshura, A., Kulagin, V., Kovelenov, A., Cherdantseva, A., Filoniuk, N., Turbina, G., Ermakov, A., Monakhov, N., Piterskiy, M., Semenov, A., ... Bobkova, M. (2025). The Molecular Epidemiology of HIV-1 in Russia, 1987–2023: Subtypes, Transmission Networks and Phylogenetic Story. Pathogens, 14(8), 738. https://doi.org/10.3390/pathogens14080738






