Comprehensive Molecular and Epidemiological Characterization of Staphylococcus aureus Isolated from Bovine Mastitis in Water Buffalo of the Peshawar Division, Khyber Pakhtunkhwa, Pakistan
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area and Sample Collection
2.2. Bacterial Cultivation and Susceptibility Testing of S. aureus Strains
2.3. S. aureus Molecular Characterization
2.4. Ethics Approval
3. Results
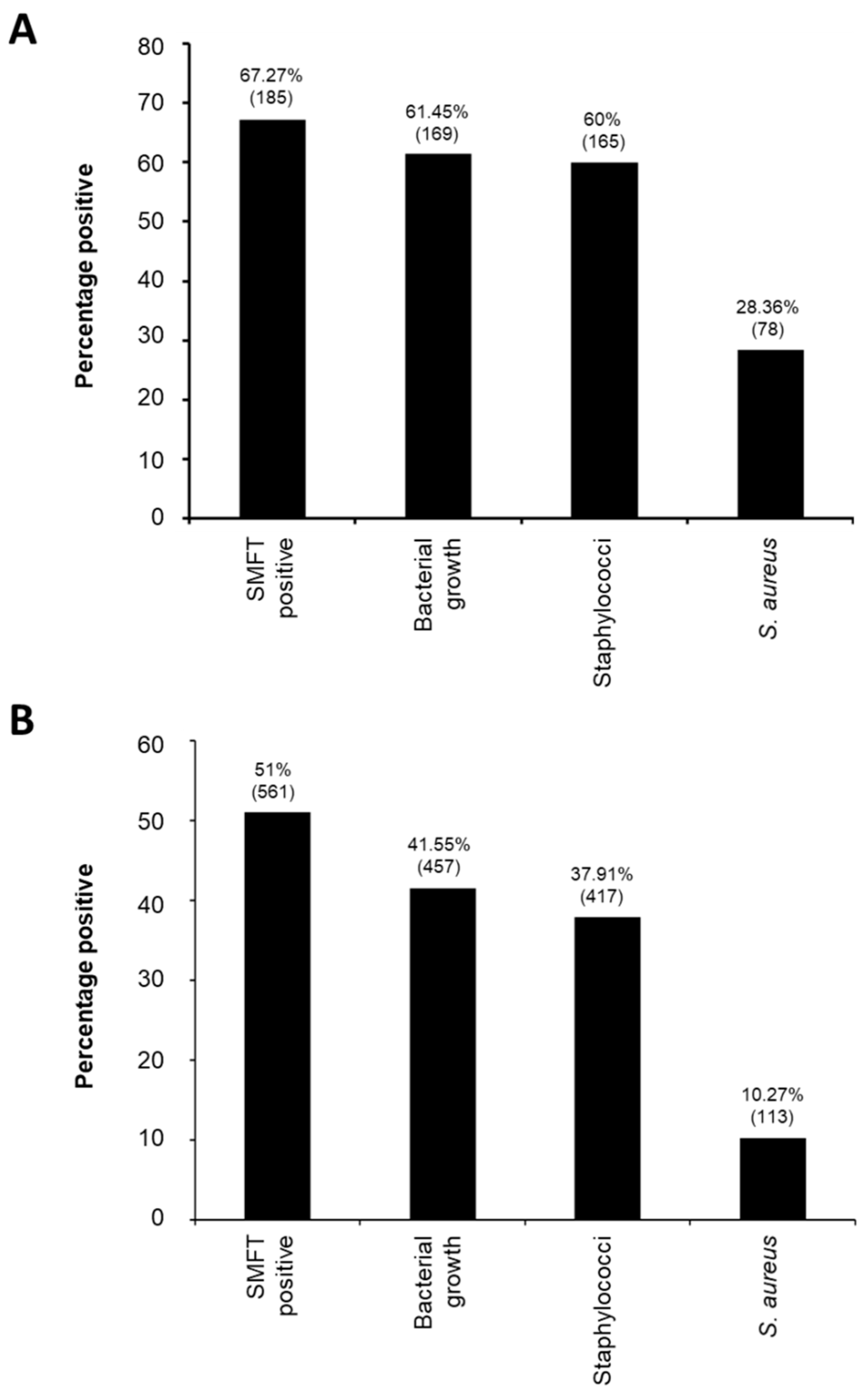
3.1. Overall Epidemiology and Molecular Characteristics of Staphylococcus aureus from Water Buffalo Milk Samples in Peshawar Division
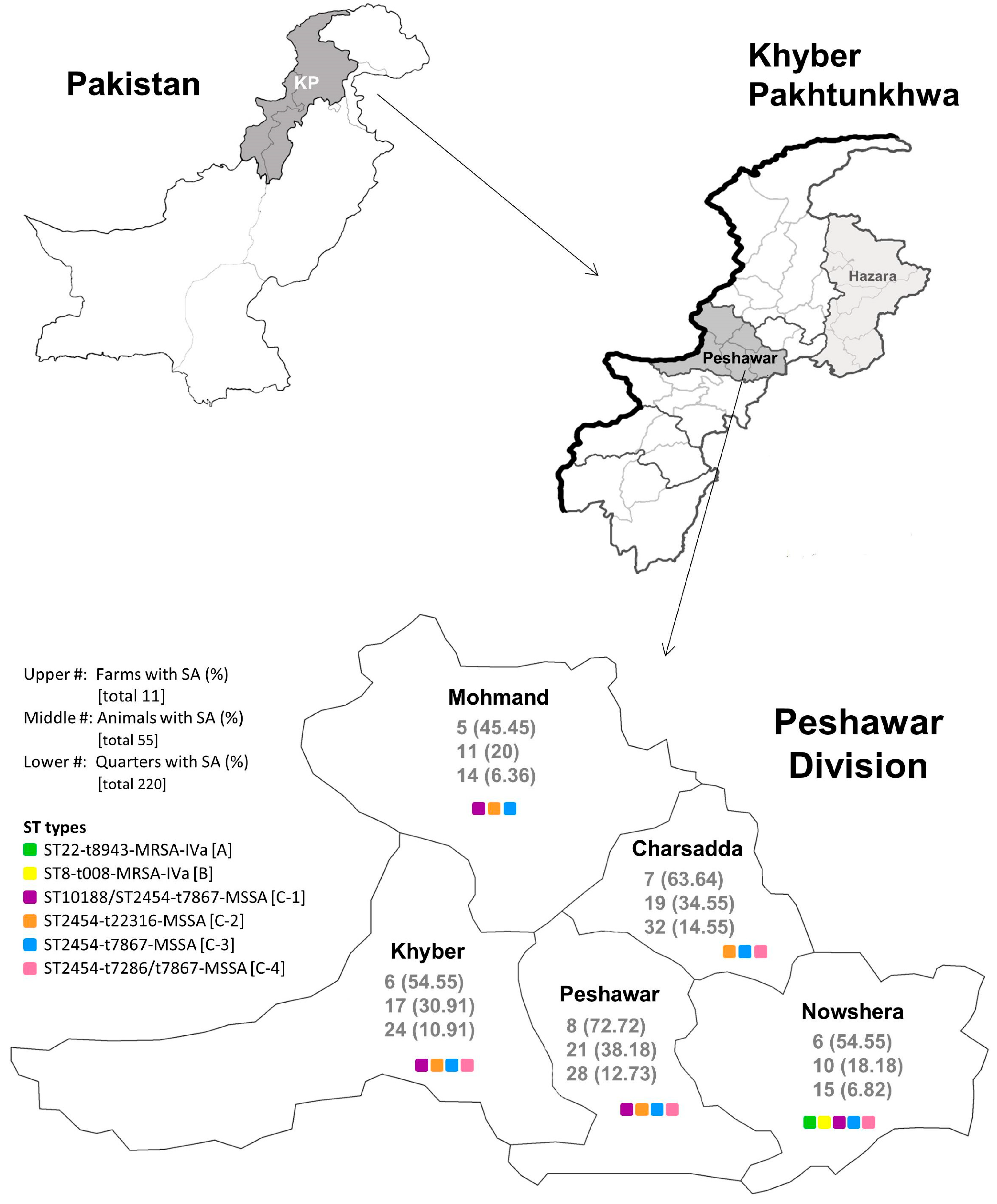
3.2. Epidemiology of S. aureus from Milk Samples in Each District of the Peshawar Division
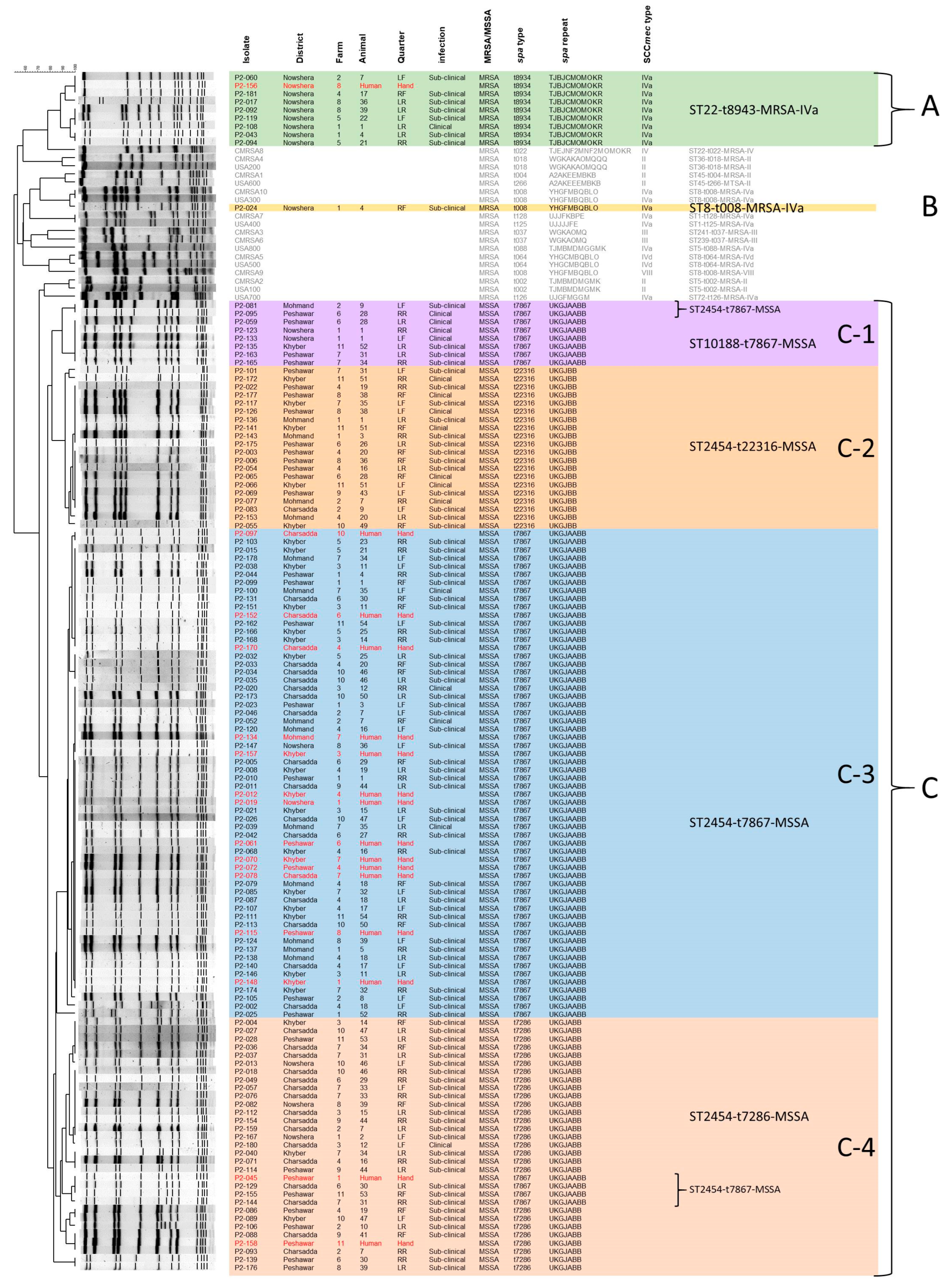
3.3. Detailed Molecular Characteristics of S. aureus Isolated from Bovine Mastitis Milk Samples
3.4. Epidemiological Investigation on the Zoonotic Transmission of S. aureus Strains from Bovine Mastitis Infected Water Buffalo to Their Caretakers
3.5. Antibiotic Resistance of S. aureus Isolated from Milk Samples in the Peshawar District
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sharma, N.; Rho, G.J.; Hong, Y.-H.; Kang, T.Y.; Lee, H.K.; Hur, T.-Y.; Jeong, D.K. Bovine Mastitis: An Asian Perspective. Asian J. Anim. Vet. Adv. 2012, 7, 454–476. [Google Scholar] [CrossRef]
- Halasa, T.; Huijps, K.; Osteras, O.; Hogeveen, H. Economic effects of bovine mastitis and mastitis management: A review. Vet. Q. 2007, 29, 18–31. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.Z.; Khan, A. Basic facts of mastitis in dairy animals: A review. Pak. Vet. J. 2006, 26, 204–208. [Google Scholar]
- Shearer, J.K.; Harris, B., Jr. Mastitis in Dairy Goats; Florida Cooperative Extension Service; University of Florida: Gainesville, FL, USA, 1992; pp. 1–7. [Google Scholar]
- Athar, M. Preparation and Evaluation of Inactivated Polyvalent Vaccines for the Control of Masitits in Dairy Buffaloes; University of Agriculture: Faisalabad, Pakistan, 2007. [Google Scholar]
- Pedersen, R.R.; Krömker, V.; Bjarnsholt, T.; Dahl-Pedersen, K.; Buhl, R.; Jørgensen, E. Biofilm Research in Bovine Mastitis. Front. Vet. Sci. 2021, 8, 656810. [Google Scholar] [CrossRef] [PubMed]
- Shahzad, M.A.; Yousaf, A.; Ahsan, A.; Irshad, H.; Riaz, A.; Khan, A.; Ullah, I.; Sattar, S.; Bostan, N.; Javed, S. Virulence and resistance profiling of Staphylococcus aureus isolated from subclinical bovine mastitis in the Pakistani Pothohar region. Sci. Rep. 2024, 14, 14569. [Google Scholar] [CrossRef] [PubMed]
- Javed, S.; McClure, J.; Syed, M.A.; Obasuyi, O.; Ali, S.; Tabassum, S.; Ejaz, M.; Zhang, K. Epidemiology and molecular characterization of Staphylococcus aureus causing bovine mastitis in water buffaloes from the Hazara division of Khyber Pakhtunkhwa, Pakistan. PLoS ONE 2022, 17, e0268152. [Google Scholar] [CrossRef] [PubMed]
- Foster, T.J. Staphylococcus. In Medical Microbiology, 4th ed.; Baron, S., Ed.; The University of Texas Medical Branch at Galveston: Galveston, TX, USA, 1996. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 34th ed.; CLSI Supplement M100; Clinical and Laboratory Standards Institute: Wayne, NY, USA, 2024. [Google Scholar]
- Zhang, K.; Sparling, J.; Chow, B.L.; Elsayed, S.; Hussain, Z.; Church, D.L.; Gregson, D.B.; Louie, T.; Conly, J.M. New quadriplex PCR assay for detection of methicillin and mupirocin resistance and simultaneous discrimination of Staphylococcus aureus from coagulase-negative staphylococci. J. Clin. Microbiol. 2004, 42, 4947–4955. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; McClure, J.A.; Elsayed, S.; Louie, T.; Conly, J.M. Novel multiplex PCR assay for simultaneous identification of community-associated methicillin-resistant Staphylococcus aureus strains USA300 and USA400 and detection of mecA and Panton-Valentine leukocidin genes, with discrimination of Staphylococcus aureus from coagulase-negative staphylococci. J. Clin. Microbiol. 2008, 46, 1118–1122. [Google Scholar] [CrossRef] [PubMed]
- Mulvey, M.R.; Chui, L.; Ismail, J.; Louie, L.; Murphy, C.; Chang, N.; Alfa, M. Development of a Canadian standardized protocol for subtyping methicillin-resistant Staphylococcus aureus using pulsed-field gel electrophoresis. J. Clin. Microbiol. 2001, 39, 3481–3485. [Google Scholar] [CrossRef] [PubMed]
- McClure, J.A.; Zaal DeLongchamp, J.; Conly, J.M.; Zhang, K. Novel Multiplex PCR Assay for detection of chlorhexidine-quaternary ammonium, mupirocin, and methicillin resistance genes, with simultaneous discrimination of Staphylococcus aureus from coagulase-negative staphylococci. J. Clin. Microbiol. 2017, 55, 1857–1864. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; McClure, J.A.; Conly, J.M. Enhanced multiplex PCR assay for typing of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. Mol. Cell. Probes 2012, 26, 218–221. [Google Scholar] [CrossRef] [PubMed]
- Peacock, S.J.; Moore, C.E.; Justice, A.; Kantzanou, M.; Story, L.; Mackie, K.; O’Neill, G.; Day, N.P. Virulent combinations of adhesin and toxin genes in natural populations of Staphylococcus aureus. Infect. Immun. 2002, 70, 4987–4996. [Google Scholar] [CrossRef] [PubMed]
- Harmsen, D.; Claus, H.; Witte, W.; Rothganger, J.; Claus, H.; Turnwald, D.; Vogel, U. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 2003, 41, 5442–5448. [Google Scholar] [CrossRef] [PubMed]
- Enright, M.C.; Day, N.P.; Davies, C.E.; Peacock, S.J.; Spratt, B.G. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 2000, 38, 1008–1015. [Google Scholar] [CrossRef] [PubMed]
- Bartels, M.D.; Peterson, A.; Wornign, P.; Nielsen, J.B.; Larner-Svensson, H.; Johansen, H.K.; Anderson, L.P.; Jarlov, J.O.; Boye, K.; Larsen, A.R.; et al. Comparing whole-genome sequencing with Sanger sequencing for spa typing of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 2014, 52, 4305–4308. [Google Scholar] [CrossRef] [PubMed]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018, 24, 124. [Google Scholar] [CrossRef] [PubMed]
- Elsayed, M.S.; Mahmoud El-Bagoury, A.E.; Dawoud, M.A. Phenotypic and genotypic detection of virulence factors of Staphylococcus aureus isolated from clinical and subclinical mastitis in cattle and water buffaloes from different farms of Sadat City in Egypt. Vet. World 2015, 8, 1051–1058. [Google Scholar] [CrossRef] [PubMed]
- Fareed, S.K.; Memon, K.H.; Kachiwal, A.B.; Azhar, S.; Brula, M.I.; Mehmood-ul-Hasan; Ali, M.; Khan, T.A. Prevalence and economic losses of reproductive disorders and mastitis in buffaloes at Karachi, Pakistan. Indian J. Anim. Res. 2017, 51, 1130–1133. [Google Scholar] [CrossRef]
- Anwar, K.; Amin, Y.; Mujtaba, M. Prevalence and bacteriology of sub clinical mastitis in buffaloes in and around Peshawar district, Khyber Pakhtunkhwa Pakistan. Int. J. Eng. Res. Technol. 2013, 2, 364–371. [Google Scholar]
- Chishty, M.A.; Arshad, M.; Avais, M.; Hameed, S.; Ijaz, M. Cross-sectional epidemiological studies on mastitis in cattle and buffaloes of Tehsil Gojra, Pakistan. Buffalo Bull. 2007, 26, 50–55. [Google Scholar]
- Ali, T.; Kamran; Raziq, A.; Wazir, I.; Ullah, R.; Shah, P.; Ali, M.I.; Han, B.; Liu, G. Prevalence of mastitis pathogens and antimicrobial susceptibility of isolates from cattle and buffaloes in northwest of Pakistan. Front. Vet. Sci. 2021, 8, 746755. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.B.; Khan, M.A.; Ahmad, I.; Khan, F.A.; Khan, H.; Khan, S.A. Prevalence of antibiotic resistant genes in Staphylococcus aureus Isolated from bovine mastitis. Pak. J. Zool. 2022, 54, 2239–2244. [Google Scholar] [CrossRef]
- Khan, A.; Durrani, A.; Yousaf, A.; Khan, J.; Chaudhry, M.; Khan, M.; Habibunnabi, H.; Khan, A. Epidemiology of bovine sub-clinical mastitis in Pothohar region, Punjab, Pakistan in 2018. Pak. J. Zool. 2019, 51, 1667–1674. [Google Scholar] [CrossRef]
- Mustafa, Y.; Awan, F.; Zaman, T.; Chaudhry, S.R.; Zoyfro, V. Prevalence and antibacterial susceptibility in mastitis in buffalo and cow in and around the district Lahore, Pakistan. Pak. J. Pharm. 2013, 24, 29–33. [Google Scholar] [CrossRef]
- Kumar, R.; Yadav, B.R.; Singh, R.S. Antibiotic resistance and pathogenicity factors in Staphylococcus aureus isolated from mastitic Sahiwal cattle. J. Biosci. 2011, 36, 175–188. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.K.; Nam, H.M.; Jang, G.C.; Lee, H.S.; Jung, S.C.; Kim, T.S. Transmission and persistence of methicillin-resistant Staphylococcus aureus in milk, environment, and workers in dairy cattle farms. Foodborne Pathog. Dis. 2013, 10, 731–736. [Google Scholar] [CrossRef] [PubMed]
- Makovec, J.A.; Ruegg, P.L. Antimicrobial resistance of bacteria isolated from dairy cow milk samples submitted for bacterial culture: 8905 samples (1994–2001). J. Am. Vet. Med. Assoc. 2003, 222, 1582–1589. [Google Scholar] [CrossRef] [PubMed]
- Spohr, M.; Rau, J.; Friedrich, A.; Klittich, G.; Fetsch, A.; Guerra, B.; Hammerl, J.A.; Tenhagen, B.A. Methicillin-resistant Staphylococcus aureus (MRSA) in three dairy herds in southwest Germany. Zoonoses Public Health 2011, 58, 252–261. [Google Scholar] [CrossRef] [PubMed]
- Aqib, A.I.; Ijaz, M.; Anjum, A.A.; Malik, M.A.R.; Mehmood, K.; Farooqi, S.H.; Hussain, K. Antibiotic susceptibilities and prevalence of methicillin resistant Staphylococcus aureus (MRSA) isolated from bovine milk in Pakistan. Acta Trop. 2017, 176, 168–172. [Google Scholar] [CrossRef] [PubMed]
- Bari, F.; Wazir, R.; Haroon, M.; Ali, S.; Imtiaz; Rahman, H.; Ullah, A.; Khattak, A.M.; Qasim, M. Frequency and antibiotic susceptibility profile of MRSA at Lady Reading Hospital, Peshawar. Gomal J. Med. Sci. 2015, 13, 62–65. [Google Scholar]
- Farmanullah; Abbas, W.; Parveen, F.; Zaman, H.; Shah, J.; Rafiullah. Analysis of hospital-acquired MRSA infections in pus samples: A comprehensive discussion. Insights-J. Health Rehabil. 2025, 3, 214–221. [Google Scholar] [CrossRef]
- Ullah, A.; Qasim, M.; Rahman, H.; Khan, J.; Haroon, M.; Muhammad, N.; Khan, A.; Muhammad, N. High frequency of methicillin-resistant Staphylococcus aureus in Peshawar Region of Pakistan. Springerplus 2016, 5, 600. [Google Scholar] [CrossRef] [PubMed]
- Alam, S.; Abro, S.H.; Khan, M.A.; Abro, R.; Rind, M.R.; Wagan, H.; Khan, S.A.; Maqbool, K. Emerging of methicillin resistant Staphylococcus aureus in clinical and sub-clinical bovine mastitis in Peshawar. Sci. Int. 2016, 28, 3055–3059. [Google Scholar]
- Sivakumar, R.; Pranav, P.S.; Annamanedi, M.; Chandrapriya, S.; Isloor, S.; Rajendhran, J.; Hegde, N.R. Genome sequencing and comparative genomic analysis of bovine mastitis-associated Staphylococcus aureus strains from India. BMC Genom. 2023, 24, 44. [Google Scholar] [CrossRef] [PubMed]
- Annamanedi, M.; Sheela, P.; Sundareshan, S.; Isloor, S.; Gupta, P.; Jasmeen, P.; Gargi, M.; Mallick, S.; Hegde, N.R. Molecular fingerprinting of bovine mastitis-associated Staphylococcus aureus isolates from India. Sci. Rep. 2021, 11, 15228. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.H.; El Zowalaty, M.E.; Falgenhauer, L.; Khan, M.F.R.; Alam, J.; Popy, N.N.; Ashour, H.M.; Rahman, M.B. Molecular identification and whole genome sequence analyses of methicillin-resistant and mastitis-associated Staphylococcus aureus sequence types 6 and 2454 isolated from dairy cows. J. Genom. 2024, 12, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Kahl, B.C.; Mellmann, A.; Deiwick, S.; Peters, G.; Harmsen, D. Variation of the polymorphic region X of the protein A gene during persistent airway infection of cystic fibrosis patients reflects two independent mechanisms of genetic change in Staphylococcus aureus. J. Clin. Microbiol. 2005, 43, 502–505. [Google Scholar] [CrossRef] [PubMed]
- Cvetnic, L.; Samardzija, M.; Duvnjak, S.; Habrun, B.; Cvetnic, M.; Jaki Tkalec, V.; Duricic, D.; Benic, M. Multi locus sequence typing and spa typing of Staphylococcus aureus isolated from the milk of cows with subclinical mastitis in Croatia. Microorganisms 2021, 9, 725. [Google Scholar] [CrossRef] [PubMed]
- Holt, D.C.; Harris, T.M.; Hughes, J.T.; Lilliebridge, R.; Croker, D.; Graham, S.; Hall, H.; Wilson, J.; Tong, S.Y.C.; Giffard, P.M. Longitudinal whole-genome based comparison of carriage and infection associated Staphylococcus aureus in northern Australian dialysis clinics. PLoS ONE 2021, 16, e0245790. [Google Scholar] [CrossRef] [PubMed]
- Krupa, P.; Bystron, J.; Bania, J.; Podkowik, M.; Empel, J.; Mroczkowska, A. Genotypes and oxacillin resistance of Staphylococcus aureus from chicken and chicken meat in Poland. Poult. Sci. 2014, 93, 3179–3186. [Google Scholar] [CrossRef] [PubMed]
- Schaumburg, F.; Pauly, M.; Anoh, E.; Mossoun, A.; Wiersma, L.; Schubert, G.; Flammen, A.; Alabi, A.S.; Muyembe-Tamfum, J.J.; Grobusch, M.P.; et al. Staphylococcus aureus complex from animals and humans in three remote African regions. Clin. Microbiol. Infect. 2015, 21, 345 e341–348. [Google Scholar] [CrossRef] [PubMed]
- Jamil, B.; Gawlik, D.; Syed, M.A.; Shah, A.A.; Abbasi, S.A.; Muller, E.; Reissig, A.; Ehricht, R.; Monecke, S. Hospital-acquired methicillin-resistant Staphylococcus aureus (MRSA) from Pakistan: Molecular characterisation by microarray technology. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 691–700. [Google Scholar] [CrossRef]
- Ullah, N.; Nasir, S.; Ishaq, Z.; Anwer, F.; Raza, T.; Rahman, M.; Alshammari, A.; Alharbi, M.; Bae, T.; Rahman, A.; et al. Comparative genomic analysis of a Panton-Valentine Leukocidin-positive ST22 community-acquired methicillin-resistant Staphylococcus aureus from Pakistan. Antibiotics 2022, 11, 496. [Google Scholar] [CrossRef] [PubMed]
- Haran, K.P.; Godden, S.M.; Boxrud, D.; Jawahir, S.; Bender, J.B.; Sreevatsan, S. Prevalence and characterization of Staphylococcus aureus, including methicillin-resistant Staphylococcus aureus, isolated from bulk tank milk from Minnesota dairy farms. J. Clin. Microbiol. 2012, 50, 688–695. [Google Scholar] [CrossRef]
- Madzgalla, S.; Syed, M.A.; Khan, M.A.; Rehman, S.S.; Muller, E.; Reissig, A.; Ehricht, R.; Monecke, S. Molecular characterization of Staphylococcus aureus isolates causing skin and soft tissue infections in patients from Malakand, Pakistan. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35, 1541–1547. [Google Scholar] [CrossRef] [PubMed]
- Monecke, S.; Syed, M.A.; Khan, M.A.; Ahmed, S.; Tabassum, S.; Gawlik, D.; Muller, E.; Reissig, A.; Braun, S.D.; Ehricht, R. Genotyping of methicillin-resistant Staphylococcus aureus from sepsis patients in Pakistan and detection of antibodies against staphylococcal virulence factors. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Akkou, M.; Bouchiat, C.; Antri, K.; Bes, M.; Tristan, A.; Dauwalder, O.; Martins-Simoes, P.; Rasigade, J.P.; Etienne, J.; Vandenesch, F.; et al. New host shift from human to cows within Staphylococcus aureus involved in bovine mastitis and nasal carriage of animal’s caretakers. Vet. Microbiol. 2018, 223, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Vishnupriya, S.; Antony, P.X.; Mukhopadhyay, H.K.; Pillai, R.M.; Thanislass, J.; Srinivas, M.V.; Kumar, R.S. Methicillin resistant staphylococci associated with bovine mastitis and their zoonotic importance. Vet. World 2014, 7, 422–427. [Google Scholar] [CrossRef]
- Farhan, M.; Awan, N.; Kanwal, A.; Sharif, F.; Hayyat, M.U.; Shahzad, L.; Ghafoor, G.Z. Dairy farmers’ levels of awareness of antibiotic use in livestock farming in Pakistan. Humanit. Soc. Sci. Commun. 2024, 11, 165. [Google Scholar] [CrossRef]
- Mohsin, M.; Umair, M. Trends in antimicrobial use in livestock animals in Pakistan. Int. J. Infect. Dis. 2020, 101, 17–18. [Google Scholar] [CrossRef]
- Umair, M.; Abdullah, R.M.; Aslam, B.; Nawaz, M.H.; Ali, Q.; Fatima, F.; Ali, J.; Zahoor, M.A.; Mohsin, M. First case report on quantification of antimicrobial use in corporate dairy farms in Pakistan. Front. Vet. Sci. 2020, 7, 575848. [Google Scholar] [CrossRef] [PubMed]
- Haq, I.U.; Kamal, M.; Swelum, A.A.; Khan, S.; Rios-Escalante, P.R.L.; Usman, T. Alarming multidrug resistance in Staphylococcus aureus isolated from raw milk of cows with subclinical mastitis: Antibiotic resistance patterns and occurrence of selected resistance genes. PLoS ONE 2024, 19, e0301200. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Cheung, A.; Bayer, A.S.; Chen, L.; Abdelhady, W.; Kreiswirth, B.N.; Yeaman, M.R.; Xiong, Y.Q. The global regulon sarA regulates β-Lactam antibiotic resistance in methicillin-resistant Staphylococcus aureus in vitro and in endovascular infections. J. Infect. Dis. 2016, 214, 1421–1429. [Google Scholar] [CrossRef] [PubMed]
- Dunman, P.M.; Murphy, E.; Haney, S.; Palacios, D.; Tucker-Kellogg, G.; Wu, S.; Brown, E.L.; Zagursky, R.J.; Shlaes, D.; Projan, S.J. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J. Bacteriol. 2001, 183, 7341–7353. [Google Scholar] [CrossRef] [PubMed]
- Luong, T.T.; Dunman, P.M.; Murphy, E.; Projan, S.J.; Lee, C.Y. Transcription profiling of the mgrA regulon in Staphylococcus aureus. J. Bacteriol. 2006, 188, 1899–1910. [Google Scholar] [CrossRef] [PubMed]
| Farm No (%) | Animal No (%) | Quarter No (%) | |
|---|---|---|---|
| S. aureus positive | 32 (58.18) | 78 (28.36) | 113 (10.27) |
| Sub-Clinical | 32 (100.00) | 71 (91.03) | 96 (84.96) |
| Clinical | 7 (21.88) | 7 (8.97) | 17(15.04) |
| MSSA | 29 (90.63) | 73 (93.59) | 104 (92.04) |
| Sub-Clinical | 29 (100.00) | 66 (90.41) | 88 (84.62) |
| Clinical | 7 (24.14) | 7 (9.59) | 16 (15.38) |
| MRSA | 5 (15.63) | 8 (10.26) | 9 (7.96) |
| Sub-Clinical | 5 (100.00) | 7 (87.50) | 8 (88.89) |
| Clinical | 1 (20.00) | 1 (12.50) | 1 (11.11) |
| District | |||||
|---|---|---|---|---|---|
| Charsadda No. (%) | Khyber No. (%) | Mohmand No. (%) | Nowshera No. (%) | Peshawar No. (%) | |
| Farm | |||||
| S. aureus positive | 7 (63.64) | 6 (54.55) | 5 (45.45) | 6 (54.55) | 8 (72.72) |
| MSSA | 7 (100) | 6 (100) | 5 (100) | 3 (50) | 8 (100) |
| Sub-clinical | 7 (100) | 6 (100) | 5 (100) | 3 (100) | 8 (100) |
| Clinical | 1 (14.29) | 1 (16.67) | 2 (40) | 1 (33.33) | 2 (25.00) |
| MRSA | 0 | 0 | 0 | 5 (83.33) | 0 |
| Sub-clinical | 0 | 0 | 0 | 5 (100) | 0 |
| Clinical | 0 | 0 | 0 | 1 (20) | 0 |
| Animal | |||||
| S. aureus positive | 19 (34.55) | 17 (30.91) | 11 (20) | 10 (18.18) | 21 (38.18) |
| MSSA | 19 (100) | 17 (100) | 11 (100) | 5 (50) | 21 (100) |
| Sub-clinical | 18 (94.74) | 16 (94.12) | 9 (81.82) | 4 (80) | 19 (90.48) |
| Clinical | 1 (5.26) | 1 (5.88) | 2 (18.18) | 1 (20) | 2 (9.52) |
| MRSA | 0 | 0 | 0 | 8 (80) | 0 |
| Sub-clinical | 0 | 0 | 0 | 7 (87.5) | 0 |
| Clinical | 0 | 0 | 0 | 1 (12.5) | 0 |
| Quarter | |||||
| S. aureus positive | 32 (14.55) | 24 (10.91) | 14 (6.36) | 15 (6.82) | 28 (12.73) |
| MSSA | 32 (100) | 24 (100) | 14 (100) | 6 (40) | 28 (100) |
| Sub-clinical | 30 (93.75) | 21 (87.50) | 10 (71.43) | 4 (66.67) | 23 (82.14) |
| Clinical | 2 (6.25) | 3 (12.50) | 4 (28.57) | 2 (33.33) | 5 (17.86) |
| MRSA | 0 | 0 | 0 | 9 (60) | 0 |
| Sub-clinical | 0 | 0 | 0 | 8 (88.89) | 0 |
| Clinical | 0 | 0 | 0 | 1 (11.11) | 0 |
| District | Farm | Human Isolate (PFGE Group) | Mastitis Isolates (FPGE Group) |
|---|---|---|---|
| Charsadda | 4 | ST2454-t7867-MSSA (C-3) * | ST2454-t7867-MSSA (C-3) * ST2454-t7286-MSSA (C-4) |
| 6 | ST2454-t7867-MSSA (C-3) * | ST2454-t7867-MSSA (C-3) * ST2454-t7286-MSSA (C-4) | |
| 7 | ST2454-t7867-MSSA (C-3) | ST2454-t7867-MSSA (C-4) ST2454-t7286-MSSA (C-4) | |
| 10 | ST2454-t7867-MSSA (C-3) * | ST2454-t7867-MSSA (C-3) * ST2454-t7286-MSSA (C-4) | |
| Khyber | 3 | ST2454-t7867-MSSA (C-3) * | ST2454-t7867-MSSA (C-3) * ST2454-t7286-MSSA (C-4) |
| 4 | ST2454-t7867-MSSA (C-3) * | ST2454-t7867-MSSA (C-3) * | |
| 7 | ST2454-t7867-MSSA (C-3) * | ST2454-t7867-MSSA (C-3) * ST2454-t7286-MSSA (C-4) ST2454-t22316-MSSA (C-2) | |
| 11 | ST2454-t7867-MSSA (C-3) * | ST2454-t7867-MSSA (C-3) * ST2454-t22316-MSSA (C-2) ST2454-t7867-MSSA (C-1) | |
| Mohmand | 7 | ST2454-t7867-MSSA (C-3) * | ST2454-t7867-MSSA (C-3) * |
| Nowshera | 1 | ST2454-t7867-MSSA (C-3) | ST2454-t7867-MSSA (C-1) ST22-t8934-MRSA (A) ST2454-t7286-MSSA (C-4) ST8-t008-MRSA (B) |
| 8 | ST22-t8934-MRSA (A) * | ST22-t8934-MRSA (A) * ST2454-t7867-MSSA (C-3) ST2454-t7286-MSSA (C-4) | |
| Peshawar | 1 | ST2454-t7867-MSSA (C-4) | ST2454-t7867-MSSA (C-3) |
| 4 | ST2454-t7867-MSSA (C-3) | ST2454-t22316-MSSA (C-2) ST2454-t7286-MSSA (C-4) | |
| 6 | ST2454-t7867-MSSA (C-3) | ST2454-t22316-MSSA (C-2) ST10188-t7867-MSSA (C-1) ST2454-t7867-MSSA (C-1) ST2454-t7286-MSSA (C-4) | |
| 8 | ST2454-t7867-MSSA (C-3) | ST2454-t22316-MSSA (C-2) ST2454-t7286-MSSA (C-4) | |
| 11 | ST2454-t7286-MSSA (C-4) * | ST2454-t7286-MSSA (C-4) * ST2454-t7867-MSSA (C-3) ST2454-t7867-MSSA (C-4) |
| District | |||||||
|---|---|---|---|---|---|---|---|
| Overall % | Charsadda % | Khyber % | Mohmand % | Nowshera % (MRSA) | Peshawar % | ||
| Ampicillin | S | 78.76 | 84.38 | 91.67 | 64.29 | 46.67 (11.11) | 85.71 |
| I | 0 | 0 | 0 | 0 | 0 | 0 | |
| R | 21.24 | 15.63 | 8.33 | 35.71 | 53.33 (88.89) | 14.28 | |
| Cefoxitin | S | 92.92 | 100 | 100 | 100 | 46.67 (11.11) | 100 |
| I | 0 | 0 | 0 | 0 | 0 | 0 | |
| R | 7.08 | 0 | 0 | 0 | 53.33 (88.89) | 0 | |
| Clindamycin | S | 80.53 | 84.38 | 87.5 | 92.86 | 40 (11.11) | 85.71 |
| I | 9.73 | 6.25 | 12.5 | 7.14 | 6.67 (0) | 14.29 | |
| R | 9.73 | 9.38 | 0 | 0 | 53.33 (88.89) | 0 | |
| Gentamycin | S | 78.76 | 84.38 | 87.5 | 78.57 | 33.33 (11.11) | 89.29 |
| I | 10.62 | 9.38 | 8.33 | 14.29 | 20.00 (11.11) | 7.14 | |
| R | 10.62 | 6.25 | 4.17 | 7.14 | 46.67 (77.78) | 3.57 | |
| Amoxicillin | S | 76.11 | 81.25 | 79.17 | 78.57 | 40 (11.11) | 85.71 |
| I | 0 | 0 | 0 | 0 | 0 | 0 | |
| R | 23.89 | 18.75 | 20.83 | 21.43 | 60 (88.89) | 14.28 | |
| Doxycycline | S | 80.53 | 90.63 | 83.33 | 78.57 | 40 (22.22) | 89.29 |
| I | 14.16 | 9.38 | 16.67 | 14.29 | 26.67 (33.33) | 10.71 | |
| R | 5.31 | 0 | 0 | 7.14 | 33.33 (44.44) | 0 | |
| Lincomycin | S | 80.53 | 84.38 | 91.67 | 64.29 | 46.67 (11.11) | 92.86 |
| I | 10.62 | 12.5 | 8.33 | 21.43 | 6.67 (11.11) | 7.14 | |
| R | 8.85 | 3.13 | 0 | 14.29 | 46.67 (77.78) | 0 | |
| Ceftazidime | S | 0 | 0 | 0 | 0 | 0 | 0 |
| I | 25.66 | 34.38 | 37.5 | 28.57 | 6.67 (0) | 14.29 | |
| R | 74.34 | 65.63 | 62.5 | 71.43 | 93.33 (100) | 85.71 | |
| Rifampin | S | 77.88 | 81.25 | 83.33 | 64.29 | 40 (22.22) | 96.43 |
| I | 16.81 | 18.75 | 12.5 | 28.57 | 33.33 (33.33) | 3.57 | |
| R | 5.31 | 0 | 4.17 | 7.14 | 26.67 (44.44) | 0 | |
| Trimethoprim-sulfamet-hoxazole | S | 79.65 | 87.5 | 83.33 | 64.29 | 53.33 (22.2) | 89.29 |
| I | 11.50 | 9.38 | 8.33 | 14.29 | 20 (33.33) | 10.71 | |
| R | 8.85 | 3.13 | 8.33 | 21.43 | 26.67 (44.44) | 0 | |
| Linezolid | S | 92.04 | 93.75 | 95.83 | 92.86 | 80 (66.67) | 21.43 |
| I | 6.19 | 6.25 | 4.17 | 7.14 | 13.33 (22.22) | 3.57 | |
| R | 1.77 | 0 | 0 | 0 | 6.67 (11.11) | 3.57 | |
| Azithromycin | S | 84.07 | 84.38 | 95.83 | 92.86 | 46.67 (11.11) | 89.29 |
| I | 7.08 | 9.38 | 4.17 | 7.14 | 6.67 (11.11) | 7.14 | |
| R | 8.85 | 6.25 | 0 | 0 | 46.67 (77.78) | 3.57 | |
| Ceftriaxone | S | 76.99 | 84.38 | 75 | 78.57 | 33.33 (11.11) | 92.86 |
| I | 15.04 | 15.63 | 20.83 | 21.43 | 13.33 (0) | 7.14 | |
| R | 7.96 | 0 | 4.17 | 0 | 53.33 (88.89) | 0 | |
| Tetracycline | S | 81.42 | 93.75 | 87.5 | 78.57 | 26.67 (11.11) | 92.86 |
| I | 9.73 | 3.13 | 8.33 | 14.29 | 26.67 (22.22) | 7.14 | |
| R | 8.85 | 3.13 | 4.17 | 7.14 | 46.67 (66.67) | 0 | |
| Norfloxacin | S | 87.61 | 93.75 | 91.67 | 92.86 | 46.67 (11.11) | 96.43 |
| I | 7.08 | 6.25 | 8.33 | 7.14 | 13.33 (22.22) | 3.57 | |
| R | 5.31 | 0 | 0 | 0 | 40 (66.67) | 0 | |
| Erythromycin | S | 69.91 | 78.13 | 79.17 | 64.29 | 33.33 (0) | 75 |
| I | 14.16 | 6.25 | 16.67 | 21.43 | 20 (22.22) | 14.29 | |
| R | 15.93 | 15.63 | 4.17 | 14.29 | 46.67 (77.78) | 10.71 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Javed, S.; McClure, J.-A.; Ullah, I.; Ali, S.; Ejaz, M.; Tabassum, S.; Syed, M.A.; Zhang, K. Comprehensive Molecular and Epidemiological Characterization of Staphylococcus aureus Isolated from Bovine Mastitis in Water Buffalo of the Peshawar Division, Khyber Pakhtunkhwa, Pakistan. Pathogens 2025, 14, 735. https://doi.org/10.3390/pathogens14080735
Javed S, McClure J-A, Ullah I, Ali S, Ejaz M, Tabassum S, Syed MA, Zhang K. Comprehensive Molecular and Epidemiological Characterization of Staphylococcus aureus Isolated from Bovine Mastitis in Water Buffalo of the Peshawar Division, Khyber Pakhtunkhwa, Pakistan. Pathogens. 2025; 14(8):735. https://doi.org/10.3390/pathogens14080735
Chicago/Turabian StyleJaved, Salma, Jo-Ann McClure, Irfan Ullah, Shahzad Ali, Mohammad Ejaz, Sadia Tabassum, Muhammad Ali Syed, and Kunyan Zhang. 2025. "Comprehensive Molecular and Epidemiological Characterization of Staphylococcus aureus Isolated from Bovine Mastitis in Water Buffalo of the Peshawar Division, Khyber Pakhtunkhwa, Pakistan" Pathogens 14, no. 8: 735. https://doi.org/10.3390/pathogens14080735
APA StyleJaved, S., McClure, J.-A., Ullah, I., Ali, S., Ejaz, M., Tabassum, S., Syed, M. A., & Zhang, K. (2025). Comprehensive Molecular and Epidemiological Characterization of Staphylococcus aureus Isolated from Bovine Mastitis in Water Buffalo of the Peshawar Division, Khyber Pakhtunkhwa, Pakistan. Pathogens, 14(8), 735. https://doi.org/10.3390/pathogens14080735






