Abstract
Aerococcus urinaeequi is an opportunistic pathogen that has been isolated from humans, pigs, and chickens, but with no reports in geese until now. This research aimed to isolate and identify A. urinaeequi from four geese, and establish a specific PCR detection method for A. urinaeequi. Strain E1 was identified as A. urnaeequi through a combination of Gram staining (Gram-positive coccus), colony morphology (α-hemolysis), and whole genome sequencing analysis. Comparative genomics was used to analyze the genome sequences of five reference strains of A. urinaeequi to screen for a species-specific genomic region (401 bp). Based on this region, specific primers were designed to establish the PCR detection method for A. urnaeequi, and the specificity and sensitivity of this assay were tested. The results showed that the target sequence was specifically amplified only for the genome of A. urinaeequi, and that the minimum nucleic acid detection concentration was 7.08 × 10−3 ng/μL. The mouse infection model indicated that the target fragment could be amplified from the tissue samples of dead mice in the challenge groups, verifying the applicability of PCR for clinical sample detection. Specific sequences of A. urinaeequi were detected in the lungs of three pigs using the PCR method, confirmed to be consistent through whole genome sequencing, and previously identified as A. urinaeequi or A. viridans by 16S rRNA sequencing. For the detection of fecal samples from geese, canines, and felines using the PCR method, the highest positive rate was 36.9% (31/84) of geese, followed by 21.7% (20/90) of felines, and finally 6.9% (16/230) of canines. A strain of A. urinaeequi was isolated and identified in geese for the first time, and a species-specific PCR detection method for A. urinaeequi was established with high specificity and sensitivity, which could well distinguish the bacterial species A. urinaeequi from its phylogenetically related species, A. viridans.
1. Introduction
The genus Aerococcus is a Gram-positive, microaerophilic, and hydrogen peroxide-negative bacterium [1], which was first proposed by Williams et al. [2]. There are currently 13 species in the genus of Aerococcus, including Aerococcus viridans (A. viridans) [1], A. urinaeequi [3], A. urinae [4], A. christensenii, A. sanguinicola, A. urinaehominis, A. suis, and A. vaginalis [5]. Moreover, five new species have been reported recently, including a strain of A. agrisoli sp. nov. from a paddy soil sample [6], A. loyolae sp. nov., A. mictus sp. nov., and A. tenax sp. nov., all reclassified and named separately from A. urinae [7], and A. kribbianus sp. nov., from pig manure [8]. Aerococcus is a potential opportunistic pathogen widely found in soil, air, livestock, and the medical environment. Aerococcus may cause opportunistic infection in humans [9,10,11,12]. In recent years, Aerococcus has been isolated from clinical samples of animals, such as milk produced by cows suffering from mastitis [13], the urine of pigs with a urinary system infection [14], and lobsters suffering from septicemia [15].
A. urinaeequi is a Gram-positive, spore-free, non-motile facultative anaerobic coccus in pairs, tetrads, or clusters [3]. The original taxonomic classification of this species was Pediococcus urinaeequi (genus Pediococcus). In 2005, Felis et al. reclassified it into the genus Aerococcus based on DNA–DNA hybridization data and phylogenetic analysis of 16S rRNA gene sequences, renaming it A. urinaeequi [3]. So far, A. urinaeequi has been isolated from ascites of women with chronic kidney disease [1], pig nasal swabs [16], and laying hens with foot dermatitis [17].
The sequencing of the 16S rRNA gene has been commonly used for species identification, but the method is limited as well. When the 16S rRNA sequences of bacteria are highly homologous, it is not easy to distinguish them [18]. For example, A. urinaeequi and A. viridans have been reported to exhibit a mere two-base-pair difference in their 16S rRNA gene sequences [3]. DNA–DNA hybridization (DDH) has always been considered the gold standard for identifying bacteria, but because of its cumbersome operation and the difficulty in standardizing among different laboratories, it is usually used for the identification of different strains when the 16S rRNA homology between strains is above 97% [19]. Compared with 16S rRNA sequencing, whole genome sequencing (WGS) provides more comprehensive genetic information and offers higher accuracy in bacterial species identification [20]. However, WGS requires specialized infrastructure and high-throughput sequencing platforms, which are not yet widely accessible in routine laboratories or clinical settings, coupled with their time-consuming workflow.
Current reports on the pathogenicity of Aerococcus primarily focus on A. viridans and A. urinae [9,10,11,12,13,14,15], while A. urinaeequi has rarely been investigated, and there is a risk of misidentification as A. viridans because of the 16S rRNA gene sequences of A. urinaeequi and A. viridans only differing by 1–2 bases [21]. Therefore, it is necessary to establish a simple, rapid, and accurate PCR-based method for detecting A. urinaeequi. To develop such a method, we first needed to obtain reliable genomic data of A. urinaeequi as a reference, and this opportunity arose from a natural outbreak of A. urinaeequi infection in geese.
In April 2024, four diseased and dead geese from a goose farm (Fengyang County, Chuzhou city, Anhui Province) were subjected to autopsy, and hepatosplenomegaly, turbid and thickened air sacs, and extensive intestinal bleeding were observed. Liver samples of the diseased and dead geese were collected for isolation and the identification of pathogens, as well as whole genome sequencing, average nucleotide identity (ANI) analysis, and non-redundant protein database comparison (NR). The whole genome sequences of the five reference strains of A. urinaeequi were compared and analyzed, and specific primers were designed to establish a PCR detection method, which will provide technical support for the clinical detection and identification of A. urinaeequi.
2. Materials and Methods
2.1. Experimental Materials
Blood agar plates were purchased from Changde Beekman Biotechnology Co., Ltd. (Changde, China), BHI medium was purchased from Qingdao Haibo Bio-technology Co., Ltd. (Qingdao, China), the bacterial genomic DNA isolation kit was purchased from Tiangen Biochemical Technology (Beijing) Co., Ltd. (Beijing, China), and the Tissue/Cell/Blood genomic DNA extraction kit was purchased from Wuhan Saiwell Biotechnology Co., Ltd. (Wuhan, China). The strains of A. viridans AHFY, Proteus mirabilis WHZ2, Pasteurella multocida HN-1, Clostridium perfringens CQ1, Enterococcus faecalis F1, Salmonella sms, Staphylococcus aureus ptqj, Bacillus subtilis KC1, and Escherichia coli 97 were isolated and preserved by the laboratory of animal medicine, Anhui Science and Technology University. The reference strain of A. viridans ATCC 11563 was purchased from Wuhan Huizao Biotechnology Co., Ltd. (Wuhan, China). Kunming mice were purchased from Hangzhou Ziyuan Experimental Animal Technology Co., Ltd. (Hangzhou, China).
2.2. Isolation and Morphological Observation
The liver tissues of the diseased and dead geese were collected and directly streaked on a blood agar medium using a sterile inoculation loop. Single colonies with characteristics of A. urinaeequi were picked up and purified on blood agar medium after overnight culture at 35 °C. After purification, single colonies were selected for Gram staining to observe the morphological characteristics of bacteria.
2.3. 16S rRNA Amplification and Sequencing Analysis
The DNA of the isolates was extracted as templates using a rapid bacterial genomic DNA isolation kit, and universal primers of bacterial 16S rRNA (upstream 27-F: 5′-AGAGTTTGATCATGGCTCAG-3′ and downstream 1525-R: 5′-AAGGAGGTGATCCAACC-3′) were used for PCR amplification. PCR products were detected via agarose gel electrophoresis, and those with the target band were sent to General Biology (Anhui) Co., Ltd. (Chuzhou, China) for sequencing. The sequencing results of 16S rRNA were blasted in the rRNA/ITS database on NCBI, and the phylogenetic tree was constructed by the software of MEGA11 (version 11.0.13).
2.4. Whole Genome Sequencing and Analysis
2.4.1. Whole Genome Sequencing
The purified bacteria were sent to Sangon Biotech (Shanghai) Co., Ltd. (Shanghai, China) for whole genome sequencing. The Illumina NovaSeq v.3.15.4 was used to generate raw second-generation sequencing data, which was then subjected to statistical analysis and quality assessment using Fastp (version 0.23.1) [22]. Quality shearing was performed to obtain processed data with improved accuracy. Subsequently, the second-generation sequencing data was assembled into contigs via SPAdes (version 3.5.0) [23], and GAP within the contigs was supplemented using GapFiller (version 1.11) [24]. Sequence correction was performed using PrInSeS-G (version 1.0.0) to rectify base-calling errors and small insertions/deletions (indels) during the assembly process [25]. Gene elements (CDS, tRNA, and rRNA) were predicted using the NCBI-PGAP (version 1.2.1)/Prokka (version 1.10) software [26]. Finally, a circular whole genome map of strain E1 was generated with Cgview (https://proksee.ca/ (accessed on 19 June 2025)) [27].
2.4.2. Annotation of NR Database
The whole genome protein-coding gene sequences of the isolates were compared in an NR database based on Diamond software (version 2.1.8) [28], and the species information in the annotation results was statistically analyzed.
2.4.3. Analysis of ANI
The whole genome sequences of 22 reference strains of the genus Aerococcus (5 strains of A. urinaeequi, 3 strains of A. viridans, 3 strains of A. urinae, 1 strain of A. sanguinicola, 1 strain of A. urinaehominis, 2 strains of A. christensenii, 3 strains of A. loyolae, 2 strains of A. mictus, and 2 strains of A. tenax) were downloaded from the NCBI database (https://www.ncbi.nlm.nih.gov/ (accessed on 14 December 2024)), and the accession numbers of the 22 strains are shown in Table 1. The ANI of strain E1 and the above 22 strains of the genus Aerococcus were calculated using the online platform JSpeciesWS (https://jspecies.ribohost.com/jspeciesws/ (accessed on 12 March 2025)) [29]. Then, the ANI values of the 23 strains were obtained, and the ANI data were processed using TBtools-II software (version 2.310) [30] to plot the heat map of the correlation clustering labels.

Table 1.
Accession numbers of Aerococcus strains for ANI analysis.
2.5. Establishment of PCR Assay
2.5.1. Screening for Specific Sequence and Designing Primers
The complete genome sequencing maps of five reference strains of A. urinaeequi are currently available in the NCBI database, and detailed strain information is shown in Table 2. The whole genome sequences of five A. urinaeequi strains were compared and analyzed based on MAFFT 7.0 (https://mafft.cbrc.jp/alignment/server/index.html (accessed on 13 October 2024)) [31], and a species-specific sequence of the genus A. urinaeequi was identified. The screened sequence was compared and analyzed using NCBI-BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 7 December 2024)) to test the sequence specificity, and the specific primers were designed by SnapGene® software (version 7.0.2) (from Dotmatics; available at snapgene.com) to establish a PCR assay.

Table 2.
Strain information on A. urinaeequi for whole genome sequence analysis.
2.5.2. Determination of Optimal Annealing Temperature
The annealing temperature in the PCR detection method was optimized, and 9 gradients were set as 52 °C, 53 °C, 54 °C, 55 °C, 56 °C, 57 °C, 58 °C, 59 °C, and 60 °C to find the optimal annealing temperature. Also, lower temperatures from 47 °C to 51 °C were also tested for non-specific amplification.
2.5.3. PCR-Specific Detection
The genomic DNA of A. urinaeequi isolated in this research and other 10 control strains (A. viridans ATCC 11563, A. viridans AHFY, Proteus mirabilis WHZ2, Pasteurella multocida HN-1, Clostridium perfringens CQ1, Enterococcus faecalis F1, Salmonella sms, Staphylococcus aureus ptqj, Bacillus subtilis KC1, and Escherichia coli 97) were extracted as templates using a bacterial genomic DNA extraction kit to detect the specificity of the PCR assay.
2.5.4. PCR Sensitivity Test
The bacterial genomic DNA extraction kit was used for extracting the genomic DNA of A. urinaeequi, and the nucleic acid concentration was detected to be 70.8 ng/μL via spectrophotometer. The DNA template of A. urinaeequi was diluted by a 10-fold gradient (10−1~10−7) to test the sensitivity of this PCR method.
2.5.5. Artificial Infection of Mice and PCR Detection for A. urinaeequi
Twenty-four Kunming mice were divided into 4 groups randomly, and there were six mice in each group, which were housed in separate cages and provided with sufficient water and food. Groups 1, 2, and 3 were set as the challenge groups, while group 4 was the control group. A. urinaeequi was cultured in BHI medium for 12 h at 35 °C, and the bacterial suspension was adjusted to three different concentrations, 1.05 × 107, 1.05 × 108, and 1.05 × 109 CFU/mL. The mice in groups 1, 2, and 3 had intraperitoneal injection with a 0.5 mL bacterial suspension of 1.05 × 107, 1.05 × 108, and 1.05 × 109 CFU/mL, respectively. In contrast, the control group received 0.5 mL of BHI medium. The mice were continuously observed for 7 days after the injection, and statistical analysis on the survival rate of the mice in challenge groups was carried out. The organs and tissues of the heart, liver, spleen, lung, kidney, and small intestine were collected from the dead and healthy mice, and the DNA was extracted using the tissue genomic DNA extraction kit for the PCR detection of A. urinaeequi.
2.5.6. Detection of A. urinaeequi from Clinical Samples by PCR
Lung tissue samples of three pigs were collected for the identification of the pathogen, which was previously identified as A. urinaeequi or A. viridans via 16S rRNA sequencing, or could not be diagnosed. Therefore, the PCR method established in this research for identification was used, and as well as whole genome sequencing to confirm the results.
A total of 230, 92, and 84 fecal samples were collected from canines, felines, and geese, respectively. Feces were diluted 1000-fold with sterile physiological saline, and then added to LB broth for shaking culture at 37 °C for 12 h. Bacterial DNA was extracted from the culture using a bacterial genomic DNA isolation kit for the subsequent PCR detection of A. urinaeequi.
3. Results
3.1. Colony and Morphology Observation
The isolated bacterium was designated as strain E1, the colony characteristics of which were round, small, gray-white, smooth-surfaced, translucent, and with α-hemolysis on the blood agar medium, as shown in Figure 1A. The Gram staining result of strain E1 was Gram-positive cocci in pairs, tetrads, or clusters, which is shown in Figure 1B.
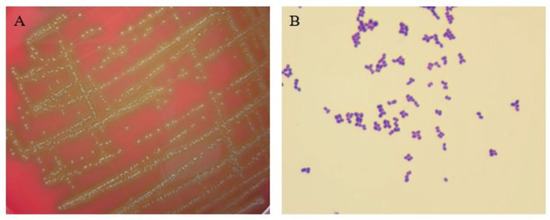
Figure 1.
Colony features and morphology observation of the strain. (A) Colony characteristics on blood agar medium. (B) Morphology observation via Gram staining (1000×).
3.2. 16S rRNA Sequencing and Analysis Results
The phylogenetic tree was constructed using the software MEGA11 (version 11.0.13) [32], based on the 16S rRNA of strain E1 and 10 strains of Aerococcus, including A. viridans, A. urinaeequi, A. suis, A. sanguinicola, A. christensenii, A. urinae, A. urinaehominis and A. vaginalis, and it is shown in Figure 2. The results indicated that strain E1 belonged to the same cluster as A. viridans and A. urinaeequi, and that the similarity of the 16S rRNA of strain E1 to that of strains NBRC 12219 and ATCC 11563 of A. viridans and strain IFO12173 of A. urinaeequi was 99.65%, 99.24%, and 99.86%, respectively. In view of the sequence similarity of 16S rRNA being above 99% between strain E1 and both A. urinaeequi and A. viridans, strain E1 could only be identified at the genus level as Aerococcus.
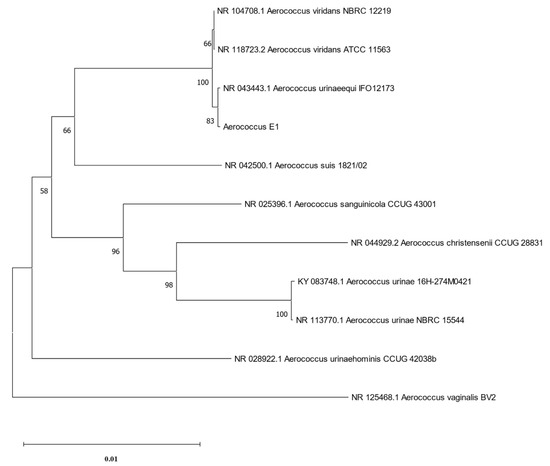
Figure 2.
Phylogenetic tree of 16S rRNA. The phylogenetic tree was constructed based on 16S rRNA sequences of strain E1 and 10 strains of Aerococcus.
3.3. WGS Analysis Results
The whole genome sequence of strain E1 was uploaded to DDBJ/ENA/GenBank, and the registration number was JBLEBE000000000. The whole genome size of E1 was 2 015 846 bp, and the GC content was 39.1%. A genome circle plot was drawn based on the Cgview (https://proksee.ca/ (accessed on 19 June 2025)) [27] online website according to genomic data, which is shown in Figure 3.
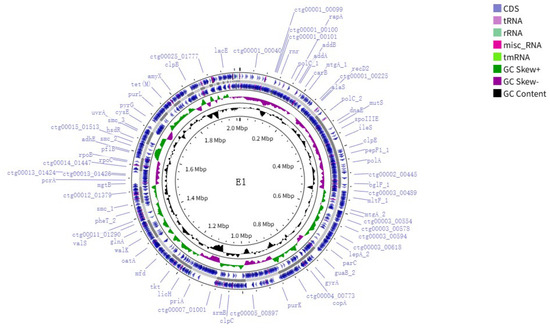
Figure 3.
Whole genome circular map of strain E1. The representation from inside to outside is as follows: the first circle is the scale, the second circle is the GC content, the third circle is the GC skew, the fifth circle is the framework of the whole genome sequence, and the fourth and sixth circles are the positions of CDS sequences, tRNAs, rRNAs, misc_RNAs, and tmRNA in the genome.
3.3.1. NR Annotation Results
The results of the NR database comparison are shown in Figure 4, and a total of 1788 genes are annotated. Among them, 1320 genes were annotated in A. urinaeequi, and 287 genes in A. viridans. The annotation genes of A. urinaeequi accounted for the highest proportion in the whole genome of strain E1, which further supports its identification as A. urinaeequi.
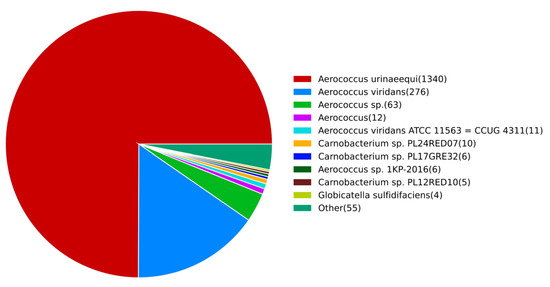
Figure 4.
NR annotation results. The number of species-related genes annotated in NR database was listed on the right side, arranged in descending order from high to low.
3.3.2. ANI Analysis Results
The whole genome sequence between strain E1 and 22 other strains of Aerococcus was analyzed based on the ANI, and the similarity between strain E1 and 5 strains of A. urinaeequi was found to be higher than 95%, such as in the strains T43, K79-1, 020-HN-1, CCUG28094, and USDA-ARS-USMARC-56713, which are shown in Figure 5; the darker the color of the grid, the higher the value.
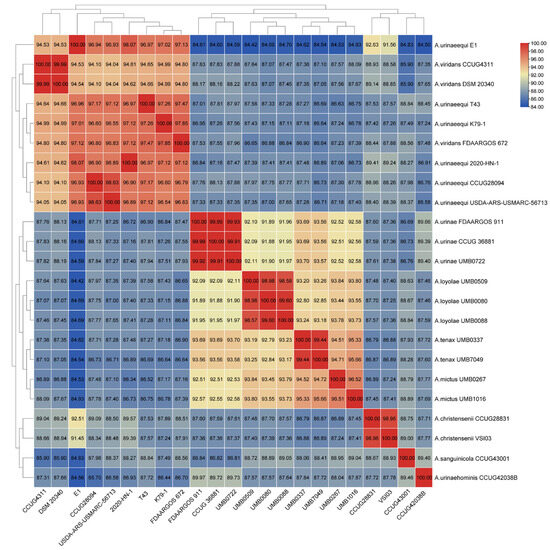
Figure 5.
The correlation cluster heat map. The correlation cluster heat map was drawn by the ANI value between strain E1 and 22 strains of Aerococcus, which included specific ANI values in each grid; the darker the color of the grid, the higher the value.
3.4. Establishment of the PCR Method
3.4.1. Specific Sequence Screening Results and PCR Establishment
A specific sequence of 401 bp was screened out as the target sequence based on the online software MAFFT 7.0, which was located between the nucleotide 1,852,008 and 1,852,408 of A. urinaeequi T43 (GenBank accession number is CP063065, locus tag is IMX20_08495), and belonged to a segment of the gene abc-f, which encoded the ABC transporter of type F. A comparison of the specific sequences of 401 bp in 5 strains of A. urinaeequi and 2 strains of A. viridans is shown in Figure 6. Based on the sequences, a pair of specific primers, upstream primer (AU-F: 5′-AAAGCTTACGAAAAGCAGCA-3′) and downstream primer (AU-R: 5′-CCAATTTGTAAGTGAGCGC-3′), were designed using SnapGene software and synthesized via General Biology (Anhui) Co., Ltd. (Chuzhou, China).
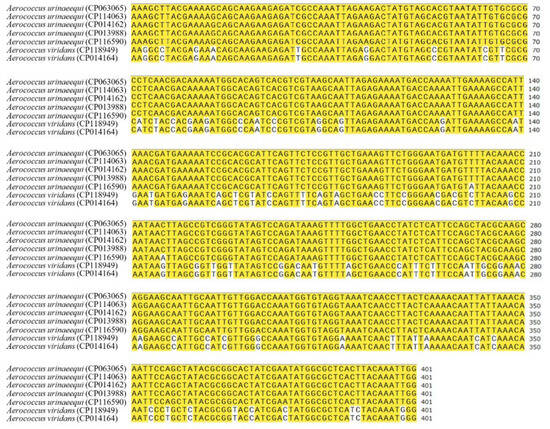
Figure 6.
Comparison of the specific sequences. The left side is the strain name and their accession number, and the right side is the specific sequences. The yellow color indicates the same nucleotide in different strains, while the bases without color markings represent differences.
The genomic DNA of A. urinaeequi E1 was extracted as a template using a bacterial genomic DNA extraction kit. The PCR reaction system had a total of 50 µL, including 2 μL of DNA template, 2 μL of upstream primer and downstream primer separately, 25 μL of PCR mixture, and 19 μL of sterile water. The annealing temperature was recommended as 54 °C, and the PCR reaction conditions were as follows: pre-denaturation at 95 °C for 5 min, 30 cycles of 95 °C for 30 s, 54 °C for 30 s, and 72 °C for 1 min, and a final extension at 72 °C for 10 min.
3.4.2. Optimization of Annealing Temperature
The results of PCR amplification at different annealing temperatures are shown in Figure 7. The target bands were successfully amplified at a temperature of 52 °C~60 °C, and there was no non-specific amplification. Among them, the amplified target band was the brightest at 52 °C, so 52 °C was selected as the optimal annealing temperature. Lower temperatures from 47 °C to 51 °C were also tested for non-specific amplification, and there was no non-specific amplification in any conditions.
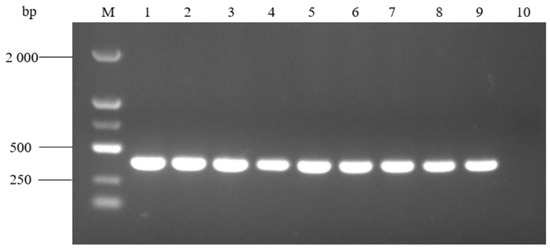
Figure 7.
PCR amplification at different annealing temperatures. M: molecular weight marker (2000 bp ladder); lanes 1–9: annealing temperature at 52 °C, 53 °C, 54 °C, 55 °C, 56 °C, 57 °C, 58 °C, 59 °C, and 60 °C, respectively; and lane 10: negative control.
3.4.3. Specificity Detection Results
The specificity detection showed that the target band with 401 bp was amplified from the template of A. urinaeequi (Figure 8), while the other 9 bacteria species and the negative control were negative and did not amplify the target band. This indicated that the PCR assay established in this research has a high specificity for A. urinaeequi.
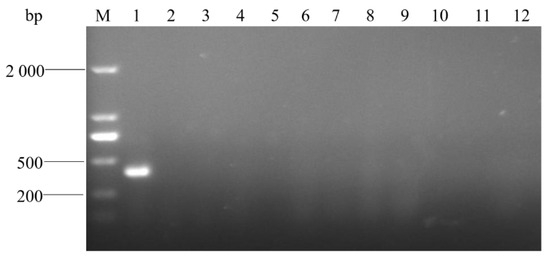
Figure 8.
Electrophoresis detection for PCR products. M: molecular weight marker (2000 bp ladder); lanes 1–11: strains of A. urinaeequi E1, A. viridans ATCC 11563, A. viridans AHFY, Proteus mirabilis WHZ2, Pasteurella multocida HN-1, Clostridium perfringens CQ1, Enterococcus faecalis F1, Salmonella sms, Staphylococcus aureus ptqj, Bacillus subtilis KC1, and Escherichia coli 97, respectively; and lane 12: negative control.
3.4.4. Sensitivity Test Results
PCR amplification of A. urinaeequi E1 with different dilutions of DNA templates (10−1~10−7) showed that the specific bands were clear and bright at a concentration of 10−1~10−4 dilution, and that there was also a faint target band at 10−5 dilution, so the minimum nucleic acid concentration of this PCR method was 7.08 × 10−3 ng/μL (Figure 9).
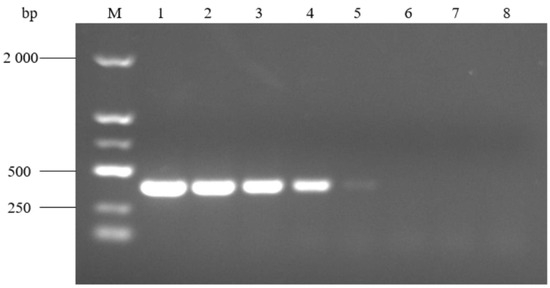
Figure 9.
Sensitivity of the PCR detection assay. M: molecular weight marker (2000 bp ladder); lanes 1–7: different dilutions of 10−1, 10−2, 10−3, 10−4, 10−5, 10−6, and 10−7, respectively; and lane 8: negative control.
3.4.5. PCR Detection for A. urinaeequi from Artificially Infected Mice
After artificial infection, mice in group 1 were lethargic and huddled together for 24 h, but all returned to normal thereafter. One mouse died within 24 h in group 2, and two mice died within 24 h and one additional mouse in the next 12 h in group 3. The rest of the mice in the challenge and control groups did not die. In the challenge groups 1, 2, and 3, the survival rates of the mice within 36 h were 100%, 83.33%, and 50%, respectively, while the survival rate of the control group mice was 100%. In autopsy of the dead mice and control group mice, the heart, liver, spleen, lung, kidney, and small intestine were all collected. Then, the DNA from the samples was extracted using the tissue DNA extraction kit and detected via the PCR assay established in this study. As shown in Figure 10, the target bands could be amplified in all samples of the dead mice, but were not detected in the samples of the control group mice. The results indicated that the PCR method could be directly used to detect A. urinaeequi from tissue samples of mice, and that there was no need for further isolation or culture.
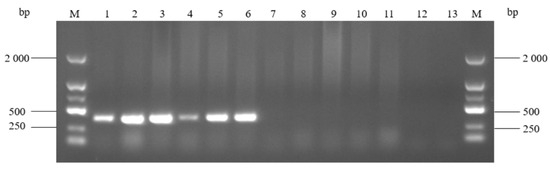
Figure 10.
PCR detection of samples from artificially infected mice. M: molecular weight marker (2000 bp ladder); lanes 1–6: heart, liver, spleen, lung, kidney, and small intestine of the dead mice; lanes 7–12: heart, liver, spleen, lung, kidney, and small intestine of the control group mice; and lane 13: negative control.
3.4.6. PCR Detection of A. urinaeequi from Clinical Samples
Three strains isolated from lung tissue samples of three dead pigs were further identified via the PCR method established in this study, and specific sequences with 401 bp were all detected (Figure 11). WGS of the three strains identified them as A. urinaeequi, consistent with the PCR results, and the relevant sequences were uploaded to the NCBI database with the accession numbers JBPDXK000000000, JBPDXL000000000, and JBPDXM000000000.
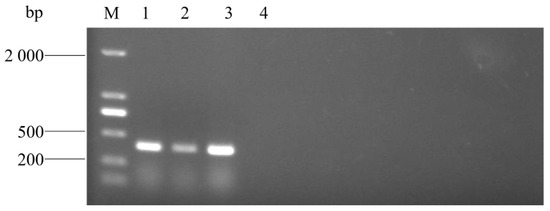
Figure 11.
PCR detection of pig samples. M: molecular weight marker (2000 bp ladder); lanes 1–4: AU1, AU2, and AU3; and lanes 4: negative control.
Using the PCR method established in this research, A.urinaeequi was detected from fecal samples of geese, canines, and felines in different areas, as shown in Table 3. Notably, the highest positive rate was in geese in Anhui province, reaching 36.9% (31/84), followed by felines in Beijing, and finally canines in Beijing and Nantong city of Jiangsu province. For different regions of Anhui, the highest positive rate was 55.6% (5/9) in Dingyuan county, while the lowest positive rate was 30.6% (19/62) in Fengyang county. For canine fecal samples, the positive rate was 8.2% (12/146) in Beijing, whereas in Nantong city, the positivity rate was 4.8% (4/84). For feline fecal samples, the positive rate was 21.7% (20/92) in Beijing. These results indicate that the positive rate of A.urinaeequi among different animals in different regions varies to some extent.

Table 3.
Positive rates of A.urinaeequi in fecal samples.
4. Discussion
In clinical practice, the rapid and accurate detection and diagnosis of bacteria is crucial for disease prevention, control, and treatment. The genus Aerococcus is often misidentified as Streptococcus, Enterococcus, or Staphylococcus due to its numerous physiological and biochemical similarities with them [33]. This not only reduces the detection frequency of Aerococcus, but also underestimates its potential pathogenicity.
The traditional detection method of A. urinaeequi mainly relies on isolation, culture, and biochemical detection. These methods are not only time-consuming, but also prone to interference from surrounding environmental factors, and cannot meet the clinical detection requirements [34]. The identification of A. urinaeequi is usually based on biochemical reactions, and commercial biochemical identification systems are widely employed, such as API 20 Strep, Vitek 2 Compact, and BBL-Crystal-GP. However, these systems may lead to misidentification at the species level [35] or insufficient adequate data for the genus Aerococcus in their databases. For example, A. urinaeequi AV208 was misidentified as A. viridans by the API 20 Strep and Vitek 2 Compact systems due to insufficient database coverage [21]. When the API 20 Strep system was used to identify 18 strains of A. sanguinicola, there was a misjudgment, as they were incorrectly classified as A. viridans [36].
Moreover, it is difficult for 16S rRNA sequencing to effectively distinguish between the strains A. urinaeequi and A. viridans. For example, the 16S rRNA sequences of strain AV208 of A. urinaeequi and strain CCUG4311T of A. viridans differed by only one base [21]. Moreover, A. urinaeequi IFO 12173 and A. viridans ATCC 11563 differed by two bases [3]. In this research, the phylogenetic tree constructed based on the 16S rRNA sequences of 10 strains of the genus Aerococcus show that the isolated strain E1 belonged to the same cluster as A. urinaeequi and A. viridans, so could only be identified as Aerococcus spp.
WGS of bacteria has a stronger species classification capability and genomic coverage than 16S rRNA sequencing; it has been applied in most microbiological laboratories and given rise to ANI technology, which is used to achieve an identification function similar to that of DDH [20]. WGS is accurate but time-consuming, costly, and impractical for rapid clinical application. Consequently, developing an alternative diagnostic approach with improved accessibility and efficiency becomes clinically crucial.
The whole genome sequence of E1 consisted of 2 015 846 bp, with a GC content of 39.1%, which was consistent with the GC content of the strains AV208 (39.1%) and 2020-HN-1 (39.04%) of A. urinaeequi. The ANI values between E1 and the five strains of A. urinaeequi were all higher than 95%, and the highest ANI value was found with the strain 2020-HN-1 (98.07%) of A. urinaeequi, while the ANI values of E1 were 94.53% compared with the strains DSM 20340 and CCUG4311 of A. viridans [37]. The whole genome sequence of E1 was annotated with a total of 1788 genes in the NR database, among which the highest number of genes (1340) were related to A. urinaeequi. In summary, the isolate E1 was identified as A. urinaeequi. In addition, the ANI values between the strain FDAARGOS_672 of A. viridans and strains DSM 20340 and CCUG4311 of A. viridans were both 94.80%. In comparison, the ANI values between the strain FDAARGOS_672 and strains E1 and T43 of A. urinaeequi were 97.13% and 97.47%, respectively, which is shown in Figure 5, indicating that the strain FDAARGOS_672 may be reclassified as A. urinaeequi rather than A. viridans.
Based on comparative genomics, a specific region of 401 bp of A. urinaeequi was screened out, and the specific PCR detection method of A. urinaeequi was established. The specific detection of the PCR assay showed that only strain E1 of A. urinaeequi was successfully amplified to produce the corresponding target band, and that none of the other species, including the strain AHFY of A. viridans, were amplified to the target band; therefore, the PCR method could meet the clinical detection requirements for A. urinaeequi. Rubel et al. developed a conventional PCR technique for the detection of Xanthomonas campestris pv. raphanin that could detect the pathogen directly from artificially infected samples with a minimum nucleic acid detection limit of 7.0 × 10−4 ng/μL [38]. Alvandi et al. designed a pair of specific PCR primers and established a PCR detection method for Xanthomonas translucens pv. undulosa via comparative genomics analysis, with a minimum detection limit of 4.5 × 10−3 ng/μL [39]. In this research, the PCR detection assay established for A. urinaeequi could be directly detected from the tissues of artificially infected dead mice and the lung tissues of pigs, as well as fecal samples from geese, canines, and felines, and the minimum detection concentration of nucleic acid was 7.08 × 10−3 ng/μL. Compared with the above assays, the sensitivity was lower than that of Rubel, and higher than that of Alvandi. Compared with the high cost, long duration, and complex data analysis of WGS, the PCR method for A. urinaeequi established in this research has high specificity and sensitivity, low cost, and rapid detection, which can be completed within 3 h from the DNA extraction of tissue samples to the observation of results using a gel imaging system, and the detection efficiency has been greatly improved.
5. Conclusions
In this study, a strain of A. urinaeequi was isolated and identified from four dead geese through methods such as isolation and culture, microscopic examination, 16S rRNA sequencing, and whole genome sequencing and analysis. Based on comparative genomics analysis, a specific sequence of A. urinaeequi was screened out. A pair of specific primers were designed, and a PCR detection method was established. The specificity and sensitivity detection results of this PCR method showed that it had strong specificity and high sensitivity, and could complete the detection within 3 h and effectively distinguish between the bacterial species A. urinaeequi and A. viridans. Moreover, the PCR method was effective in clinical samples from different animals such as pigs, geese, canines, and felines.
Author Contributions
H.W.: investigation, writing—original draft, and writing—review and editing. H.L.: writing—review and editing. Z.L.: writing—review and editing and resources. W.L.: writing—review and editing. W.G.: conceptualization, writing—review and editing, supervision, and funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Key Natural Science Research Project of Anhui Provincial Higher Education Institution (grant number KJ2020A0087), the Stable Talent Project of Anhui Science and Technology University (grant number DKWD201801), and the Veterinary Science Peak Discipline Project of Anhui Science and Technology University (grant number XK-XJGF002).
Institutional Review Board Statement
Protocols for collecting samples and animal experiments in this study were reviewed and approved by the Institutional Animal Care and Use Committee of Anhui Science and Technology University, China (Approval No. 2022011, approved 2 March 2022). To minimize the suffering of the animals in the animal experiment, the less painful infection route—intraperitoneal inoculation—was chosen, the minimum number of mice selected in each group was 6 through statistical calculations, and the method of causing death was the rapid and painless one—cervical dislocation.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in this study are included in the article; further inquiries can be directed to the corresponding author.
Conflicts of Interest
The authors declare that this research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
References
- Zhou, W.; Niu, D.; Zhang, Z.; Liu, Y.; Ning, M.; Cao, X.; Zhang, C.; Shen, H. Complete genome sequence of Aerococcus urinaeequi strain AV208. Genome Announc. 2016, 4, e01218-16. [Google Scholar] [CrossRef]
- Williams, R.E.; Hirch, A.; Cowan, S.T. Aerococcus, a new bacterial genus. J. Gen. Microbiol. 1953, 8, 475–480. [Google Scholar] [CrossRef] [PubMed]
- Felis, G.E.; Torriani, S.; Dellaglio, F. Reclassification of Pediococcus urinaeequi (ex Mees 1934) Garvie 1988 as Aerococcus urinaeequi comb. nov. Int. J. Syst. Evol. Microbiol. 2005, 55, 1325–1327. [Google Scholar] [CrossRef] [PubMed]
- Siam, T.; Stephen, T.; Munsiff, S. Case report of Aerococcus urinae tricuspid valve endocarditis, New York, USA. Emerg. Infect. Dis. 2025, 31, 1055–1056. [Google Scholar] [CrossRef] [PubMed]
- Tohno, M.; Kitahara, M.; Matsuyama, S.; Kimura, K.; Ohkuma, M.; Tajima, K. Aerococcus vaginalis sp. nov., isolated from the vaginal mucosa of a beef cow, and emended descriptions of Aerococcus suis, Aerococcus viridans, Aerococcus urinaeequi, Aerococcus urinaehominis, Aerococcus urinae, Aerococcus christensenii and Aerococcus sanguinicola. Int. J. Syst. Evol. Microbiol. 2014, 64, 1229–1236. [Google Scholar] [CrossRef]
- Sun, L.; Qiu, W.; Gao, X.; Cheng, Z.; Huang, K.; Chen, W. Aerococcus agrisoli sp. nov., isolated from paddy soil. Int. J. Syst. Evol. Microbiol. 2023, 73, 6069. [Google Scholar] [CrossRef]
- Choi, B.I.; Ene, A.; Du, J.; Johnson, G.; Putonti, C.; Schouw, C.H.; Dargis, R.; Senneby, E.; Christensen, J.J.; Wolfe, A.J. Taxonomic considerations on Aerococcus urinae with proposal of subdivision into Aerococcus urinae, Aerococcus tenax sp. nov., Aerococcus mictus sp. nov., and Aerococcus loyolae sp. nov. Int. J. Syst. Evol. Microbiol. 2023, 73, 9. [Google Scholar] [CrossRef]
- Bai, L.; Paek, J.; Shin, Y.; Kim, H.; Kim, S.H.; Shin, J.H.; Chang, Y.H. Aerococcus kribbianus sp. nov., a facultatively anaerobic bacterium isolated from pig faeces. Int. J. Syst. Evol. Microbiol. 2024, 74, 2. [Google Scholar] [CrossRef]
- Zabel, M.; Longfellow, L. Aerococcus urinae urinary tract infection in a hospitalised patient: An increasingly common infection. BMJ Case Rep. 2023, 16, e257496. [Google Scholar] [CrossRef]
- Saeed Al-Asad, K.; Mazhar, N.; Srivastava, S.; Quadri, S.; Mitra, S. Aerococcus urinae endocarditis: A case report and literature review. Cureus 2022, 14, e29853. [Google Scholar] [CrossRef]
- Chhibber, A.V.; Muttaiyah, S.; Hill, A.A.; Roberts, S.A. Aerococcus urinae aortitis: A case report. Open Forum Infect. Dis. 2019, 6, 453. [Google Scholar] [CrossRef] [PubMed]
- Varshini, K.; Ganesan, V.; Charles, J. Aerococcus viridans bacteremia: A rare case report from India. Indian J. Crit. Care Med. 2022, 26, 127–128. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Gao, J.; Ali, T.; Yu, D.; Zhang, S.; Khan, S.U.; Fanning, S.; Han, B. Characteristics of Aerococcus viridans isolated from bovine subclinical mastitis and its effect on milk SCC, yield, and composition. Trop. Anim. Health Prod. 2017, 49, 843–849. [Google Scholar] [CrossRef] [PubMed]
- Moreno, L.Z.; Matajira, C.E.; Gomes, V.T.; Silva, A.P.; Mesquita, R.E.; Christ, A.P.; Sato, M.I.; Moreno, A.M. Molecular and antibiotic susceptibility characterization of Aerococcus viridans isolated from porcine urinary infection. Vet. Microbiol. 2016, 184, 7–10. [Google Scholar] [CrossRef]
- Stewart, J.E.; Cornick, J.W.; Zwicker, B.M.; Arie, B. Studies on the virulence of Aerococcus viridans var. homari, the causative agent of gaffkemia, a fatal disease of homarid lobsters. Dis. Aquat. Org. 2004, 60, 149–155. [Google Scholar] [CrossRef]
- Yang, Q.; Zhu, Y.; Schwarz, S.; Wang, L.; Liu, W.; Yang, W.; Luan, T.; Liu, S.; Zhang, W. A novel plasmid from Aerococcus urinaeequi of porcine origin co-harboring the tetracycline resistance genes tet(58) and tet(61). Vet. Microbiol. 2021, 257, 109065. [Google Scholar] [CrossRef]
- Heidemann Olsen, R.; Christensen, H.; Kabell, S.; Bisgaard, M. Characterization of prevalent bacterial pathogens associated with pododermatitis in table egg layers. Avian Pathol. 2018, 47, 281–285. [Google Scholar] [CrossRef]
- Devanga Ragupathi, N.K.; Muthuirulandi Sethuvel, D.P.; Inbanathan, F.Y.; Veeraraghavan, B. Accurate differentiation of Escherichia coli and Shigella serogroups: Challenges and strategies. New Microbes New Infect. 2017, 21, 58–62. [Google Scholar] [CrossRef]
- Tu, Q.; Lin, L. Gene content dissimilarity for subclassification of highly similar microbial strains. BMC Genom. 2016, 17, 647. [Google Scholar] [CrossRef]
- Ranjan, R.; Rani, A.; Metwally, A.; McGee, H.S.; Perkins, D.L. Analysis of the microbiome: Advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem. Biophys. Res. Commun. 2016, 469, 967–977. [Google Scholar] [CrossRef]
- Zhou, W.; Gao, S.; Zheng, J.; Zhang, Y.; Zhou, H.; Zhang, Z.; Cao, X.; Shen, H. Identification of an Aerococcus urinaeequi isolate by whole genome sequencing and average nucleotide identity analysis. J. Glob. Antimicrob. Resist. 2022, 29, 353–359. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Boetzer, M.; Pirovano, W. Toward almost closed genomes with GapFiller. Genome Biol. 2012, 13, 56. [Google Scholar] [CrossRef]
- Massouras, A.; Hens, K.; Gubelmann, C.; Uplekar, S.; Decouttere, F.; Rougemont, J.; Cole, S.T.; Deplancke, B. Primer-initiated sequence synthesis to detect and assemble structural variants. Nat. Methods 2010, 7, 485–486. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.Y.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 2023, 51, 484–492. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R.; Oliver Glöckner, F.; Peplies, J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, M. Aerococcus: An increasingly acknowledged human pathogen. Clin. Microbiol. Infect. 2016, 22, 22–27. [Google Scholar] [CrossRef]
- Wang, W.; Chauhan, V.; Luo, Y.; Sharma, S.; Li, C.; Chen, H. Comparing NGS-Based identification of bloodstream infections to traditional culture methods for enhanced ICU care: A comprehensive study. Front. Cell. Infect. Microbiol. 2024, 14, 1454549. [Google Scholar] [CrossRef]
- Cattoir, V.; Kobal, A.; Legrand, P. Aerococcus urinae and Aerococcus sanguinicola, two frequently misidentified uropathogens. Scand. J. Infect. Dis. 2010, 42, 775–780. [Google Scholar] [CrossRef]
- Senneby, E.; Nilson, B.; Petersson, A.C.; Rasmussen, M. Matrix-assisted laser desorption ionization-time of flight mass spectrometry is a sensitive and specific method for identification of aerococci. J. Clin. Microbiol. 2013, 51, 1303–1304. [Google Scholar] [CrossRef]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef]
- Rubel, M.H.; Hossain, M.R.; Nath, U.K.; Natarajan, S.; Lee, J.H.; Jung, H.J.; Kim, T.H.; Park, J.I.; Nou, I.S. Development of a PCR test for detection of Xanthomonas campestris pv. raphani. Australas. Plant Pathol. 2019, 48, 179–182. [Google Scholar] [CrossRef]
- Alvandi, H.; Taghavi, S.M.; Khojasteh, M.; Rahimi, T.; Dutrieux, C.; Taghouti, G.; Jacques, M.A.; Portier, P.; Osdaghi, E. Pathovar-specific PCR method for detection and identification of Xanthomonas translucens pv. undulosa. Plant Dis. 2023, 107, 2279–2287. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).