Abstract
Sclerotinia sclerotiorum is a globally distributed fungal pathogen responsible for significant agricultural losses across a wide range of crops. This study aimed to develop polymorphic simple sequence repeat (SSR) markers by whole-genome resequencing of three Korean isolates and a public reference genome. A total of 16,885 SSR motifs were identified, of which 368 overlapped with polymorphic insertion–deletion (InDel) sites across the four genomes. From these, 12 SSR markers were selected based on polymorphism information content and amplification quality. Validation across the 28 isolates in Korea revealed high levels of genotypic diversity, suggesting that each isolate is a unique haplotype, although S. sclerotiorum is homothallic and clonally propagated. This multi-genome approach provides robust resources for genotyping, molecular diagnostics, and epidemiological surveillance of S. sclerotiorum.
1. Introduction
Sclerotinia sclerotiorum is a cosmopolitan necrotrophic fungus that affects more than 500 plant species, including economically important crops such as canola, lettuce, oilseed rape, soybean, and sunflower [1,2,3]. Its ability to produce long-lived sclerotia enables its prolonged survival in the soil and contributes to its persistence across diverse agroecosystems, thus making it a significant threat to global agriculture [4].
Previous population studies of S. sclerotiorum isolates have used various approaches, including mycelial compatibility assays, molecular genotyping, and aggressiveness testing for selected host cultivars [5,6,7,8,9]. Microsatellite and simple sequence repeat (SSR) markers have been used for genotyping S. sclerotiorum isolates [10]. SSR markers are widely recognized for their high polymorphism, codominant inheritance, and technical ease, making them particularly useful for population genetics, gene mapping, and diagnostic applications [11,12]. In S. sclerotiorum, SSRs have been used to study clonal diversity, host adaptation, and regional differentiation [13,14,15,16,17,18,19]. Despite its broad host range and global distribution, previous population studies have frequently reported the low overall genetic diversity of S. sclerotiorum, primarily because of its clonal reproductive strategy and limited recombination [20,21]. Although most investigations suggest that S. sclerotiorum predominantly reproduces asexually, occasional genetic recombination events, often interpreted as outcrossing, have also been reported [22,23,24]. These findings underscore the need for robust molecular tools to investigate population structure and evolutionary adaptation. In addition, previous SSR-based studies relied on a small number of loci derived from expressed sequence tags (ESTs) or partial genomic libraries, limiting their resolution and genome-wide applicability. With the advent of next-generation sequencing, whole-genome data now enable the systematic and comprehensive identification of polymorphic SSR loci across different genomes.
In the present study, we performed whole-genome resequencing of three S. sclerotiorum isolates, combined with insertion–deletion (InDel) variant analysis and SSR motif discovery, to construct a new set of polymorphic SSR markers. These markers aim to support high-resolution population studies, molecular diagnostics, and the monitoring of pathogen evolution and spread in agricultural systems.
2. Materials and Methods
2.1. Fungal Isolates and G-DNA Extraction
Twenty-eight isolates of S. sclerotiorum were obtained from the Korean Agricultural Culture Collection (KACC; Rural Development Administration, Jeonju, Republic of Korea) and are listed in Table 1. Each isolate was cultured on potato dextrose agar (PDA; Difco, Detroit, MI, USA) and incubated at 25 °C for five days in the dark. Genomic DNA was extracted from mycelia harvested from the isolates using a NucleoSpin Plant II Kit (Macherey-Nagel, Düren, Germany). DNA quality and quantity were evaluated using a Qubit 1X dsDNA High Sensitivity Kit with a Qubit Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA).

Table 1.
Information on the Sclerotinia sclerotiorum isolates used in this study, including the host plant or substrate and the geographic origin.
2.2. Sequencing and Preprocessing
Whole-genome sequencing was performed on three S. sclerotiorum isolates (KACC 42223, 47260, and 410245) at Phyzen (Seongnam-si, Republic of Korea). Libraries were prepared using the TruSeq Nano DNA Kit (Illumina, San Diego, CA, USA), and paired-end sequencing (2 × 151 bp) was performed using the NovaSeq X platform. Adapter sequences and low-quality bases (Q < 20) were trimmed using Trimmomatic v0.39 [25]. Reads shorter than 50 bp were excluded from analysis. The overall pipeline for SSR marker development and analysis is illustrated in the Supplementary Data (Figure S1).
2.3. Read Alignment and Variant Calling
The high-quality reads were aligned to the S. sclerotiorum strain 1980 UF-70 reference genome (GCA_001857865.1) [26,27] using the Burrows–Wheeler Aligner (BWA-MEM) algorithm implemented in the Sentieon DNAseq pipeline v202112.01 (Sentieon, Inc., San Jose, CA, USA) [28]. The SAM files were converted to BAM and sorted, and PCR duplicates were marked. Variant calling was performed using the Sentieon Haplotyper, and the raw VCF files were filtered to retain biallelic variants with a genotype call rate of 1 and a read depth (DP) between 5× and 100×. SNP and InDel were annotated using SnpEff v5.0e [29]. False-positive SNPs were then removed from the raw variants using GATK v4.4.0.0 [30], and hard filtering was applied under the following conditions, GATK hard filtering was performed to remove low-quality variants using the following thresholds: depth (QD < 2.0), fisher strand (FS > 60.0), mapping quality (MQ < 40.0), mapping quality rank sum test (MQRankSum < −12.5), and read position rank sum test (ReadPosRankSum < −8.0), for SNP, while QD < 2.0, FS > 200.0, and ReadPosRankSum < −20.0 for InDel. Additional filtering, including allele depth (AD) and minor allele frequency (MAF), was performed using VCFtools v0.1.17 [31].
2.4. SSR Motif Discovery, Primer Design, and PCR Amplification
SSR motif discovery was first conducted by scanning the whole-genome sequences of the three Korean isolates and the reference genome using Phyzen (Seongnam-si, Republic of Korea) software. Repeat motifs ranging from 2 to 10 bp in length were identified, with minimum thresholds set at ≥5 for dinucleotides, ≥4 for trinucleotides, and ≥3 for all higher-order motifs. Polymorphic SSR candidates were defined as those exhibiting a repeat number difference of two or more between the four genomes. For each polymorphic SSR locus, primers were designed using Primer3 [32] with the following parameters: product size = 100–250 bp, primer size = 18–26 bp (optimum 22 bp), melting temperature = 55–62 °C (optimum 58 °C), and GC content ≤59%. Uniquely mapped primer pairs were selected for screening downstream polymorphisms. The PCR reaction mixture had a final volume of 25 μL, including AccuPower PCR Premix (Bioneer, Daejeon, Republic of Korea), 1 μL of G-DNA (1 ng/μL), 0.4 μM of each primer, and 0.8 μg/μL of bovine serum albumin (Biosesang, Seongnam, Republic of Korea), as well as nuclease-free water (Sigma-Aldrich; Merck, St. Louis, MO, USA). PCR amplification was performed with the following conditions: initial denaturation at 95 °C for 10 min, followed by 36 cycles of denaturation at 95 °C for 40 s, annealing at 50 °C for 40 s, elongation at 72 °C for 50 s, and a final elongation at 72 °C for 5 min. The PCR products of the SSR markers were subjected to electrophoresis and sequenced by Macrogen (Daejeon, Republic of Korea).
2.5. Detection of Polymorphic SSRs, Marker Selection, and Phylogenetic Analysis
Polymorphic SSR markers were identified by comparing SSR loci with InDel variants obtained from the whole-genome resequencing of the three S. sclerotiorum isolates against the reference genome. This comparative analysis revealed 368 SSR loci that overlapped with polymorphic InDel regions, indicating potential variations in the repeat numbers among the four genomes. To optimize the marker set for downstream applications, SSR loci were filtered using four criteria: (1) the presence of intergenomic repeat number differences of at least two, (2) even chromosomal distribution to ensure genome-wide representation, (3) the reliable performance of the designed primers under conventional PCR amplification, and (4) the presence of more than one distinct SSR motif pattern observed through the sequencing of PCR products. The candidate SSR markers were then validated across 28 S. sclerotiorum isolates by assessing amplification efficiency and sequencing quality. Finally, 12 high-quality SSR markers were retained for further genetic analysis.
2.6. Genetic and Phylogenetic Analyses
Genetic diversity among the 28 isolates was evaluated using the number of alleles (NA), expected heterozygosity (HE) [33], and polymorphism information content (PIC) [34] values. NA and HE were calculated using GenAlEx 6.51 b2 [35]. PIC values were estimated using Cervus v3.07 [36], following the classification of Botstein et al. [34], where PIC > 0.5 is considered highly informative, 0.25 < PIC ≤ 0.5 reasonably informative, and PIC < 0.25 slightly informative. Genetic relationships among the isolates were analyzed by computing the Euclidean distances based on SSR repeat number profiles. These distance values were used in order to construct an unweighted pair group method with an arithmetic mean (UPGMA) dendrogram using the genetic distance matrix generated using GenAlEx 6.51 b2. The resulting phylogenetic tree was visualized and edited using MEGA 12 [37].
3. Results
3.1. Preprocessing and Read Alignment
The whole-genome sequencing of the three S. sclerotiorum isolates (KACC42223, KACC47260, and KACC410245) generated between 18.0 and 22.4 million raw reads per sample (Table 2). After quality trimming, 84.6% to 94.1% of the reads were retained. These high-quality reads provided sufficient coverage for accurate variant calling, with average depths ranging from 48.99x to 71.14x across all three isolates. The alignment of the trimmed reads to the S. sclerotiorum reference genome (GCA_001857865.1) using BWA-MEM revealed that 74.58% to 85.67% of reads were successfully mapped, depending on the isolate. Genome-wide coverage at a minimum depth of 1× exceeded 97% for all samples, confirming uniform representation. However, coverage at higher thresholds (20× and 50×) varied among isolates, with KACC 47260 exhibiting the highest overall depth and KACC 410245 exhibiting the lowest overall depth. The genome sizes of the three S. sclerotiorum isolates were 38.18 Mb, 38.25 Mb, and 37.83 Mb, respectively.

Table 2.
Whole-genome sequencing statistics data of Sclerotinia sclerotiorum isolates.
3.2. Variant Detection and Filtering
Variant calling across the three isolates initially produced 183,126 raw variants. After stringent filtering to retain only the biallelic sites with complete genotype calls and an appropriate read depth (5×–100×), 76,603 high-confidence variants remained (Table 3). Among these, 88.8% were found to be SNPs and 11.2% were InDels. Functional annotation using SnpEff revealed that 67.3% of the variants were located in intergenic regions, and 32.7% were located within genic regions. Within the coding regions, non-synonymous (9526) and synonymous (9470) mutations were found to be most common, indicating their potential contributions to functional diversity.

Table 3.
Summary statistics of detected SNP and InDel variants of Sclerotinia sclerotiorum isolates.
3.3. SSR Motif Discovery and Marker Selection
Comparative analysis of whole-genome resequencing data from the three S. sclerotiorum isolates and a reference genome identified 16,885 SSR motifs, comprising dinucleotide (2051), trinucleotide (2267), and tetranucleotide (6556) motifs. Among them, 368 SSR loci overlapped with polymorphic InDel regions and exhibited variations in repeat numbers across the genomes (Figure 1A). Among the selected motifs, tetranucleotide repeats (85) were the most abundant, while other repeat types, including di- (51), tri- (52), hexa- (48), and higher-order motifs (89), were more evenly distributed across the loci (Figure 1A). To ensure genomic representation, SSR loci were intentionally selected across different chromosomes, resulting in a balanced chromosomal distribution of the marker set (Figure 1B), with the number of polymorphic InDel-linked SSRs ranging from 5 (e.g., CP017824.1) to 40 (e.g., CP017815.1) (Figure S2). From this pool, 36 candidate SSR markers were further selected based on two criteria: the presence of repeat number differences ≥2 among the four genomes and chromosomal distribution balance. Of these, 12 SSR markers were selected based on successful PCR amplification, high-quality sequencing, and discriminatory power across a panel of 28 isolates.
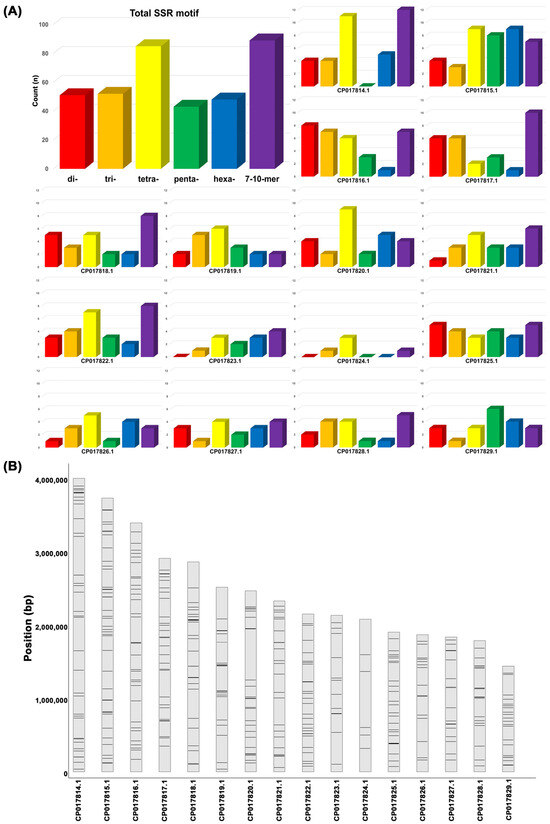
Figure 1.
The genomic distribution and abundance of SSR motifs in Sclerotinia sclerotiorum. (A) The total counts of SSR motifs grouped by repeat unit length (2–10 bp), with tetranucleotide motifs showing the highest frequency. (B) The chromosomal locations of 368 polymorphic SSR variants mapped across the 16 scaffolds (CP017814.1—CP017829.1) of the reference genome (GenBank assembly accession: GCA_001857865.1). Regions of higher density indicate potential SSR clustering hotspots.
3.4. SSR Polymorphism
Twelve SSR markers were used to evaluate genetic variation using NA, HE, and PIC (Table 4). The PIC values of the markers ranged from 0.1862 (SS29–677) to 0.9014 (SS23–1015), with four markers—SS15–1627, SS16–359, SS17–256, and SS23–1015—exhibiting high informativeness (PIC > 0.80). The most polymorphic locus, SS23–1015, had 16 alleles, indicating high allelic richness. In contrast, SS16–70 and SS28–205 exhibited the lowest NA values (3 and 2, respectively) and relatively low PIC values, suggesting limited variation among the isolates at these loci. To explore the genetic relationships among the isolates, UPGMA clustering was performed based on SSR repeat number variations (Figure 2). The resulting dendrogram revealed distinct groupings that reflect the degree of population differentiation. However, these clusters showed no correlation with geographic origins, host associations, or clonal lineages within the S. sclerotiorum population studied.

Table 4.
Properties of selected polymorphic SSR markers, including motif, PIC, and allele number.
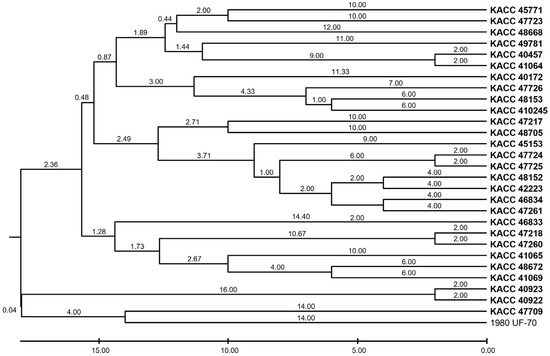
Figure 2.
UPGMA dendrogram of Sclerotinia sclerotiorum isolates based on SSR repeat variation.
4. Discussion
This study has used a genome-wide comparative approach to develop a set of polymorphic SSR markers in S. sclerotiorum by integrating repeat motif detection with InDel-based variation analysis of the whole-genome resequencing of three Korean isolates. This strategy represents a methodological advancement over previous SSR development studies that relied solely on a single reference genome (e.g., Sirjusingh and Kohn [10]), as it directly reflects inter-isolate genomic variation, enhancing marker informativeness while minimizing the inclusion of non-polymorphic loci.
The genome sizes of the three Korean isolates, viz., KACC42223, KACC47260, and KACC410245, ranged from 37.83 to 38.25 Mb, which are marginally smaller than those of previously sequenced strains such as 1980 UF-70 (38.9 Mb), ESR-01 (40.98 Mb), and WH6 (38.95 Mb) [26,27,38,39,40]. These differences suggest that the genome size of S. sclerotiorum may vary among isolates, which could be attributed to repetitive element content or structural variation.
The final set of 12 SSR markers, selected based on repeat number variation (≥2) and balanced chromosomal distribution, exhibited high amplification efficiency and polymorphisms among 28 Korean isolates. The allele count ranged from 2 to 16 and either was comparable to or surpassed those of previously reported SSR markers. In previous studies conducted by Atallah et al. [41], Dunn et al. [42], Barari et al. [43], Mahalingam et al. [22], Mert et al. [44], Tok et al. [18], and Yu et al. [19], the maximum allele number did not exceed 10. In contrast, Aldrich et al. [45], Attanayake et al. [46], Buchwaldt et al. [23], Leyronas et al. [47], Peripolli et al. [48], and Sirjusingh and Kohn [10] reported ranges from 2 to 35 alleles. Additionally, the markers in the present study exhibited PIC values ranging from 0.1862 to 0.9014, with most classified as highly informative (PIC > 0.5). For comparison, previous studies reported PIC ranges as follows: Barari et al. [43]—0.388 to 0.781 for 3 markers; Sharma et al. [49]—0.03 to 0.806 for 25 markers; Peripolli et al. [48]—0.55 to 0.77 for 10 markers; Buchwaldt et al. [23]—0.126 to 0.949 for 47 markers. Notably, the marker SS23–1015 revealed 16 alleles and a PIC value of 0.9014, underscoring its high discriminatory power. These results indicate that the SSR panel developed here is not only comparable to but, in some respects, superior to previously published markers in terms of allele diversity and PIC. Such performance makes them sufficiently informative for population genetic studies and diagnostic assay development in S. sclerotiorum.
The marker development pipeline presented in this study demonstrates a robust potential for broader applications beyond S. sclerotiorum. The generated markers were short amplicon, experimentally validated, and evenly distributed across the genome, making them suitable for high-throughput genotyping, epidemiological surveillance, and molecular diagnostics in resource-limited contexts. Nevertheless, the present marker discovery phase utilized only three Korean genomes, which may not fully capture the global allelic diversity. Future validation across more geographically diverse and host-diverse isolates is essential to confirm the broader utility of these markers and refine their application in resistance breeding and pathogen monitoring.
Despite the largely clonal reproductive nature of S. sclerotiorum [50,51,52,53], each isolate in this study exhibited a unique SSR haplotype, reflecting high genotypic diversity, which is consistent with previous studies where all 127 Canadian isolates represented unique haplotypes [23]. Although S. sclerotiorum is known to be homothallic and self-fertile, replication slippage and point mutations, particularly within SSR-rich regions, are likely the primary sources of genomic variation [23]. Our UPGMA clustering analysis revealed no clear clustering by host or geographic origin, but this may reflect the limited number and regional concentrations of the Korean isolates. Previous studies on the association between SSR marker clusters and phenotypic traits in S. sclerotiorum have yielded mixed results. Some studies found no clear correlation between pathogenicity, geographic origin, or mycelial compatibility groups [18], whereas others reported isolate-specific associations between SSR alleles and partial resistance in sunflowers [14] or fungicide-induced variation in SSR loci [54]. In contrast, several other phytopathogenic fungi have shown more consistent links between SSR genotypes and phenotypic traits. For example, Fusarium oxysporum isolates clustered in alignment with their virulence patterns [55,56], Alternaria alternata groups showed fungicide sensitivity [57], and Phytophthora capsici SSR clusters contained metalaxyl-resistant genotypes [58]. In Pyricularia oryzae, SSR variation effectively tracked pathogenicity and host specificity [59].
Interestingly, the high genetic diversity in S. sclerotiorum has been documented at both micro- and macro-geographic scales in S. sclerotiorum. For instance, identical SSR haplotypes have been recovered from isolates collected more than 700 km apart [47], while distinct genotypes have been detected within a 1 m2 area [60]. Whether these patterns result from long-distance dispersal, localized selection, or host specialization remains unclear and warrants further investigation [45,61,62,63]. Although clear genotype–phenotype correlations remain elusive, the SSR markers developed in this study hold strong potential for future studies aimed at linking genetic variation to traits such as pathogenicity, fungicide resistance, and ecological adaptation.
In conclusion, the SSR markers developed in this study provide an efficient and transferable genotyping platform for S. sclerotiorum. Their high polymorphism, genome-wide coverage, and practical assay design support their applications with regard to epidemiology, population structure analysis, and pathogenicity/resistance screening.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/pathogens14070610/s1, Figure S1: A schematic workflow for the development and analysis of SSR markers in Sclerotinia sclerotiorum. The pipeline outlines the major steps including whole-genome sequencing data acquisition, variant calling, microsatellite (SSR) identification, primer design, and validation through PCR amplification and polymorphism analysis. Each stage integrates bioinformatic tools and filtering criteria to ensure the selection of robust and informative SSR loci for genetic characterization. Figure S2: Distribution of SSR motifs on each chromosome in Sclerotinia sclerotiorum.
Author Contributions
Conceptualization, Y.-J.C.; Data curation, Y.-J.C.; Formal analysis, D.J.L.; Funding acquisition, Y.-J.C.; Investigation, D.J.L.; Methodology, D.J.L. and Y.-J.C.; Project administration, Y.-J.C.; Resources, Y.-J.C.; Software, D.J.L.; Supervision, Y.-J.C.; Validation, D.J.L. and Y.-J.C.; Visualization, D.J.L.; Writing—original draft, D.J.L.; Writing—review and editing, Y.-J.C. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, and Forestry (IPET) through the Agricultural Machinery/Equipment Localization Technology Development Program, funded by the Ministry of Agriculture, Food and Rural Affairs (MAFRA) (grant number 321056–05).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The raw genome sequencing data of Illumina NovaSeq X were submitted to the NCBI SRA database in a FASTQ format with BioSample SAMN48631575, SAMN48631576, and SAMN48631577 with SRA SRR33649454, SRR33649453, and SRR33649452 under BioProject PRJNA1011199.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| AD | Allele Depth |
| BAM | Binary Alignment/Map |
| bp | Base Pair |
| BWA-MEM | Burrows–Wheeler Aligner, Maximal Exact Match Algorithm |
| DP | Read Depth |
| ESTs | Expressed Sequence Tags |
| FS | Fisher Strand |
| GATK | Genome Analysis Toolkit |
| HE | Expected Heterozygosity |
| InDel | Insertion and Deletion |
| KACC | Korean Agricultural Culture Collection |
| MAF | Minor Allele Frequency |
| Mb | Megabase |
| MQ | Mapping Quality |
| MQRankSum | Mapping Quality Rank Sum Test |
| NA | Number of Alleles |
| PIC | Polymorphism Information Content |
| PCR | Polymerase Chain Reaction |
| QD | Quality by Depth |
| ReadPosRankSum | Read Position Rank Sum Test |
| SAM | Sequence Alignment/Map |
| SNP | Single-Nucleotide Polymorphism |
| SSR | Simple Sequence Repeat |
| UPGMA | Unweighted Pair Group Method with an Arithmetic Mean |
| VCF | Variant Call Format |
References
- Boland, G.J.; Hall, R. Index of plant hosts of Sclerotinia sclerotiorum. Can. J. Plant Pathol. 1994, 16, 93–108. [Google Scholar] [CrossRef]
- Sharma, P.; Meena, P.D.; Verma, P.; Saharan, G.; Mehta, N.; Singh, D.; Kumar, A. Sclerotinia sclerotiorum (Lib.) de Bary causing sclerotinia rot in oilseed Brassicas: A review. J. Oilseed Brassica 2014, 6, 1–44. [Google Scholar]
- Purdy, L.H. Sclerotinia sclerotiorum: History, diseases and symptomatology, host range, geographic distribution, and impact. Phytopathology 1979, 69, 875–880. [Google Scholar] [CrossRef]
- Bolton, M.D.; Thomma, B.P.H.J.; Nelson, B.D. Sclerotinia sclerotiorum (Lib.) de Bary: Biology and molecular traits of a cosmopolitan pathogen. Mol. Plant Pathol. 2006, 7, 1–16. [Google Scholar] [CrossRef]
- Baturo-Ciesniewska, A.; Groves, C.L.; Albrecht, K.A.; Grau, C.R.; Willis, D.K.; Smith, D.L. Molecular Identification of Sclerotinia trifoliorum and Sclerotinia sclerotiorum Isolates from the United States and Poland. Plant Dis. 2016, 101, 192–199. [Google Scholar] [CrossRef]
- Nanjunadappa, M.; Prameela Devi, T.; Narayanasamy, P.; Navali, G.V.; Patil, S.S. Morphological and molecular diversity of Sclerotinia sclerotiorum (Lib.) de Bary isolates of India. Bioscan 2014, 9, 1763–1767. [Google Scholar]
- Rathi, A.S.; Jattan, M.; Punia, R.; Singh, S.; Kumar, P.; Avtar, R. Morphological and molecular diversity of Sclerotinia sclerotiorum infecting Indian mustard. Indian Phytopathol. 2018, 71, 407–413. [Google Scholar] [CrossRef]
- Mandal, A.K.; Dubey, S.C. Genetic diversity analysis of Sclerotinia sclerotiorum causing stem rot in chickpea using RAPD, ITS-RFLP, ITS sequencing and mycelial compatibility grouping. World J. Microbiol. Biotechnol. 2012, 28, 1849–1855. [Google Scholar] [CrossRef]
- Petrofeza, S.; Nasser, L.C.B. Case study: Sclerotinia sclerotiorum: Genetic diversity and disease control. In The Molecular Basis of Plant Genetic Diversity; Çalışkan, M., Ed.; IntechOpen: Rijeka, Croatia, 2012. [Google Scholar]
- Sirjusingh, C.; Kohn, L.M. Characterization of microsatellites in the fungal plant pathogen, Sclerotinia sclerotiorum. Mol. Ecol. Notes 2001, 1, 267–269. [Google Scholar] [CrossRef]
- Neha Mittal, N.M.; Dubey, A.K. Microsatellite markers—A new practice of DNA based markers in molecular genetics. Pharmacogn. Rev. 2009, 3, 235–246. [Google Scholar]
- Alves, S.I.A.; Dantas, C.W.D.; Macedo, D.B.; Ramos, R.T.J. What are microsatellites and how to choose the best tool: A user-friendly review of SSR and 74 SSR mining tools. Front. Genet. 2024, 15, 1474611. [Google Scholar] [CrossRef] [PubMed]
- Arahana, V.S.; Graef, G.L.; Specht, J.E.; Steadman, J.R.; Eskridge, K.M. Identification of QTLs for Resistance to Sclerotinia sclerotiorum in soybean. Crop Sci. 2001, 41, 180–188. [Google Scholar] [CrossRef]
- Darvishzadeh, R. Association of SSR markers with partial resistance to Sclerotinia sclerotiorum isolates in sunflower (‘Helianthus annuus’ L.). Aust. J. Crop Sci. 2012, 6, 276–282. [Google Scholar]
- Sexton, A.C.; Howlett, B.J. Microsatellite markers reveal genetic differentiation among populations of Sclerotinia sclerotiorum from Australian canola fields. Curr. Genet. 2004, 46, 357–365. [Google Scholar] [CrossRef]
- Gyawali, S.; Harrington, M.; Durkin, J.; Horner, K.; Parkin, I.A.P.; Hegedus, D.D.; Bekkaoui, D.; Buchwaldt, L. Microsatellite markers used for genome-wide association mapping of partial resistance to Sclerotinia sclerotiorum in a world collection of Brassica napus. Mol. Breed. 2016, 36, 72. [Google Scholar] [CrossRef]
- Gomes, E.V.; Do Nascimento, L.B.; De Freitas, M.A.; Nasser, L.C.B.; Petrofeza, S. Microsatellite markers reveal genetic variation within Sclerotinia sclerotiorum populations in irrigated dry bean crops in Brazil. J. Phytopathol. 2011, 159, 94–99. [Google Scholar] [CrossRef]
- Tok, F.M.; Sibel, D.; Arslan, M. Analysis of genetic diversity of Sclerotinia sclerotiorum from eggplant by mycelial compatibility, random amplification of polymorphic DNA (RAPD) and simple sequence repeat (SSR) analyses. Biotechnol. Biotechnol. Equip. 2016, 30, 921–928. [Google Scholar] [CrossRef]
- Yu, Y.; Cai, J.; Ma, L.; Huang, Z.; Wang, Y.; Fang, A.; Yang, Y.; Qing, L.; Bi, C. Population structure and aggressiveness of Sclerotinia sclerotiorum from rapeseed (Brassica napus) in Chongqing city. Plant Dis. 2019, 104, 1201–1206. [Google Scholar] [CrossRef]
- Attanayake, R.N.; Carter, P.A.; Jiang, D.; del Río-Mendoza, L.; Chen, W. Sclerotinia sclerotiorum populations infecting canola from China and the United States are genetically and phenotypically distinct. Phytopathology 2013, 103, 750–761. [Google Scholar] [CrossRef]
- Winton, L.M.; Krohn, A.L.; Leiner, R.H. Genetic diversity of Sclerotinia species from Alaskan vegetable crops. Can. J. Plant Pathol. 2006, 28, 426–434. [Google Scholar] [CrossRef]
- Mahalingam, T.; Chen, W.; Rajapakse, C.S.; Somachandra, K.P.; Attanayake, R.N. Genetic diversity and recombination in the plant pathogen Sclerotinia sclerotiorum detected in Sri Lanka. Pathogens 2020, 9, 306. [Google Scholar] [CrossRef]
- Buchwaldt, L.; Garg, H.; Puri, K.D.; Durkin, J.; Adam, J.; Harrington, M.; Liabeuf, D.; Davies, A.; Hegedus, D.D.; Sharpe, A.G.; et al. Sources of genomic diversity in the self-fertile plant pathogen, Sclerotinia sclerotiorum, and consequences for resistance breeding. PLoS ONE 2022, 17, e0262891. [Google Scholar] [CrossRef] [PubMed]
- Attanayake, R.N.; Xu, L.; Chen, W. Sclerotinia sclerotiorum populations: Clonal or recombining? Trop. Plant Pathol. 2019, 44, 23–31. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Indian Phytopathol. 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Amselem, J.; Cuomo, C.A.; van Kan, J.A.L.; Viaud, M.; Benito, E.P.; Couloux, A.; Coutinho, P.M.; de Vries, R.P.; Dyer, P.S.; Fillinger, S.; et al. Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLOS Genetics 2011, 7, e1002230. [Google Scholar] [CrossRef]
- Derbyshire, M.; Denton-Giles, M.; Hegedus, D.; Seifbarghy, S.; Rollins, J.; van Kan, J.; Seidl, M.F.; Faino, L.; Mbengue, M.; Navaud, O.; et al. The complete genome sequence of the phytopathogenic fungus Sclerotinia sclerotiorum reveals insights into the genome architecture of broad host range pathogens. Genome Biol. Evol. 2017, 9, 593–618. [Google Scholar] [CrossRef]
- Freed, D.; Aldana, R.; Weber, J.A.; Edwards, J.S. The Sentieon Genomics Tools—A fast and accurate solution to variant calling from next-generation sequence data. bioRxiv 2017, 115717. [Google Scholar] [CrossRef]
- Cingolani, P.; Adrian, P.; Lily, W.L.; Melissa, C.; Tung, N.; Luan, W.; Land, S.J.; Xiangyi, L.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [PubMed]
- Nei, M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 1978, 89, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314. [Google Scholar]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Kalinowski, S.; Taper, M.; Marshall, T. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 2007, 16, 1099–1106. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Suleski, M.; Sanderford, M.; Sharma, S.; Tamura, K. MEGA12: Molecular evolutionary genetic analysis version 12 for adaptive and green computing. Mol. Biol. Evol. 2024, 41, msae263. [Google Scholar] [CrossRef]
- Gupta, N.C.; Yadav, S.; Arora, S.; Mishra, D.C.; Budhlakoti, N.; Gaikwad, K.; Rao, M.; Prasad, L.; Rai, P.K.; Sharma, P. Draft genome sequencing and secretome profiling of Sclerotinia sclerotiorum revealed effector repertoire diversity and allied broad-host range necrotrophy. Sci. Rep. 2022, 12, 21855. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Cheng, X.; Liu, L.; Liu, S. Genome sequence resource for the plant pathogen Sclerotinia sclerotiorum WH6 Isolated in China. Plant Dis. 2021, 105, 3720–3722. [Google Scholar] [CrossRef]
- Peng, F.; Li, X.; Wei, Z.; Han, G. The complete genome sequence of Sclerotinia sclerotiorum (S1), one of the pathogens causing sclerotinosis in mulberry fruit. PhytoFrontiers 2023, 4, 416–418. [Google Scholar] [CrossRef]
- Atallah, Z.; Larget, B.; Chen, X.; Johnson, D. High genetic diversity, phenotypic uniformity, and evidence of outcrossing in Sclerotinia sclerotiorum in the Columbia Basin of Washington state. Phytopathology 2004, 94, 737–742. [Google Scholar] [CrossRef]
- Dunn, A.R.; Kikkert, J.R.; Pethybridge, S.J. Genotypic characteristics in populations of Sclerotinia sclerotiorum from New York State, USA. Ann. Appl. Biol. 2017, 170, 219–228. [Google Scholar] [CrossRef]
- Barari, H.; Dalili, S.A.; Hassani, H.M. Genetic diversity among different isolates of Sclerotinia sclerotiorum in north of Iran. J. Bacteriol. Mycol. Open Access 2017, 5, 387–389. [Google Scholar] [CrossRef][Green Version]
- Mert-Türk, F.; Ipek, M.; Mermer, D.; Nicholson, P. Microsatellite and morphological markers reveal genetic variation within a population of Sclerotinia sclerotiorum from oilseed rape in the Çanakkale Province of Turkey. J. Phytopathol. 2007, 155, 182–187. [Google Scholar] [CrossRef]
- Aldrich-Wolfe, L.; Travers, S.; Nelson, B.D., Jr. Genetic variation of Sclerotinia sclerotiorum from multiple crops in the North Central United States. PLoS ONE 2015, 10, e0139188. [Google Scholar] [CrossRef] [PubMed]
- Attanayake, R.N.; Tennekoon, V.; Johnson, D.A.; Porter, L.D.; del Río-Mendoza, L.; Jiang, D.; Chen, W. Inferring outcrossing in the homothallic fungus Sclerotinia sclerotiorum using linkage disequilibrium decay. Heredity 2014, 113, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Leyronas, C.; Bardin, M.; Berthier, K.; Duffaud, M.; Troulet, C.; Torres, M.; Villeneuve, F.; Nicot, P.C. Assessing the phenotypic and genotypic diversity of Sclerotinia sclerotiorum in France. Eur. J. Plant Pathol. 2018, 152, 933–944. [Google Scholar] [CrossRef]
- Peripolli, M.; Martinelli, J.A.; Delatorre, C.A. Avaliação da agressividade e da diversidade genética de Sclerotinia sclerotiorum em tabaco no sul do Brasil. Summa Phytopathol. 2018, 44, 170–177. [Google Scholar] [CrossRef]
- Sharma, P.; Samkumar, A.; Rao, M.; Singh, V.V.; Prasad, L.; Mishra, D.C.; Bhattacharya, R.; Gupta, N.C. Genetic diversity studies based on morphological variability, pathogenicity and molecular phylogeny of the Sclerotinia sclerotiorum population from indian mustard (Brassica juncea). Front. Microbiol. 2018, 9, 1169. [Google Scholar] [CrossRef]
- Kohli, Y.; Kohn, L.M. Random association among alleles in clonal populations of Sclerotinia sclerotiorum. Fungal Genet. Biol. 1998, 23, 139–149. [Google Scholar] [CrossRef]
- Kohli, Y.; Morrall, R.; Anderson, J.; Kohn, L. Local and trans-Canadian clonal distribution of Sclerotinia sclerotiorum on canola. Phytopathology 1992, 82, 875–880. [Google Scholar] [CrossRef]
- Kohli, Y.; Brunner, L.J.; Yoell, H.; Milgroom, M.G.; Anderson, J.B.; Morrall, R.A.A.; Kohn, L.M. Clonal dispersal and spatial mixing in populations of the plant pathogenic fungus, Sclerotinia sclerotiorum. Mol. Ecol. 1995, 4, 69–77. [Google Scholar] [CrossRef]
- Kohn, L.M. The clonal dynamic in wild and agricultural plant–pathogen populations. Can. J. Bot. 1995, 73, 1231–1240. [Google Scholar] [CrossRef]
- Amaradasa, B.S.; Everhart, S.E. Effects of sublethal fungicides on mutation rates and genomic variation in fungal plant pathogen, Sclerotinia sclerotiorum. PLoS ONE 2016, 11, e0168079. [Google Scholar] [CrossRef]
- Datta, J.; Lal, N. Application of molecular markers for genetic discrimination of Fusarium wilt pathogen races affecting chickpea and pigeonpea in major regions of India. Cell. Mol. Biol. (Noisy-le-grand) 2012, 58, 55–65. [Google Scholar] [CrossRef]
- Datta, J.; Lal, N. Genetic variability assessment of Fusarium wilt pathogen races affecting chickpea using molecular markers. J. Microbiol. Biotechnol. Food Sci. 2013, 2, 2392–2397. [Google Scholar]
- He, M.-H.; Wang, Y.-P.; Wu, E.J.; Shen, L.-L.; Yang, L.-N.; Wang, T.; Shang, L.-P.; Zhu, W.; Zhan, J. Constraining evolution of Alternaria alternata resistance to a demethylation inhibitor (DMI) fungicide difenoconazole. Front. Microbiol. 2019, 10, 1609. [Google Scholar] [CrossRef]
- Chen, X.-R.; Zhang, Y.; Huang, S.-X.; Liu, T.-T.; Qiao, G.-H. Investigation of the genetic diversity of Phytophthora capsici in China using a universal fluorescent labelling method. J. Phytopathol. 2019, 167, 111–122. [Google Scholar] [CrossRef]
- Utami, D.W.; Afandi, A.; Yuriyah, S.; Terryana, R.T.; Ambarawati, A.D.; Apriana, A.; Sisharmini, A. The pathogenicity and genetic diversity of the Indonesian blast pathogen from wide host ranges of rice sub-species. J. Plant Pathol. 2025, 107, 661–673. [Google Scholar] [CrossRef]
- Attanayake, R.N.; Porter, L.; Johnson, D.A.; Chen, W. Genetic and phenotypic diversity and random association of DNA markers of isolates of the fungal plant pathogen Sclerotinia sclerotiorum from soil on a fine geographic scale. Soil Biol. Biochem. 2012, 55, 28–36. [Google Scholar] [CrossRef]
- Faraghati, M.; Abrinbana, M.; Ghosta, Y. Genetic structure of Sclerotinia sclerotiorum populations from sunflower and cabbage in West Azarbaijan province of Iran. Sci. Rep. 2022, 12, 9263. [Google Scholar] [CrossRef]
- Clarkson, J.P.; Coventry, E.; Kitchen, J.; Carter, H.E.; Whipps, J.M. Population structure of Sclerotinia sclerotiorum in crop and wild hosts in the UK. Plant Pathol. 2013, 62, 309–324. [Google Scholar] [CrossRef]
- Hemmati, R.; Javan-Nikkhah, M.; Linde, C.C. Population genetic structure of Sclerotinia sclerotiorum on canola in Iran. Eur. J. Plant Pathol. 2009, 125, 617–628. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).