Discovery and Genomic Characterisation of Novel Papillomaviruses in Australian Wild Birds
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and DNA Extraction
2.2. Next-Generation Sequencing
2.3. Bioinformatic Analyses
2.4. Comparative Genomics and Phylogenetic Analysis
3. Results
3.1. Genomes of Two Novel Avian Papillomaviruses
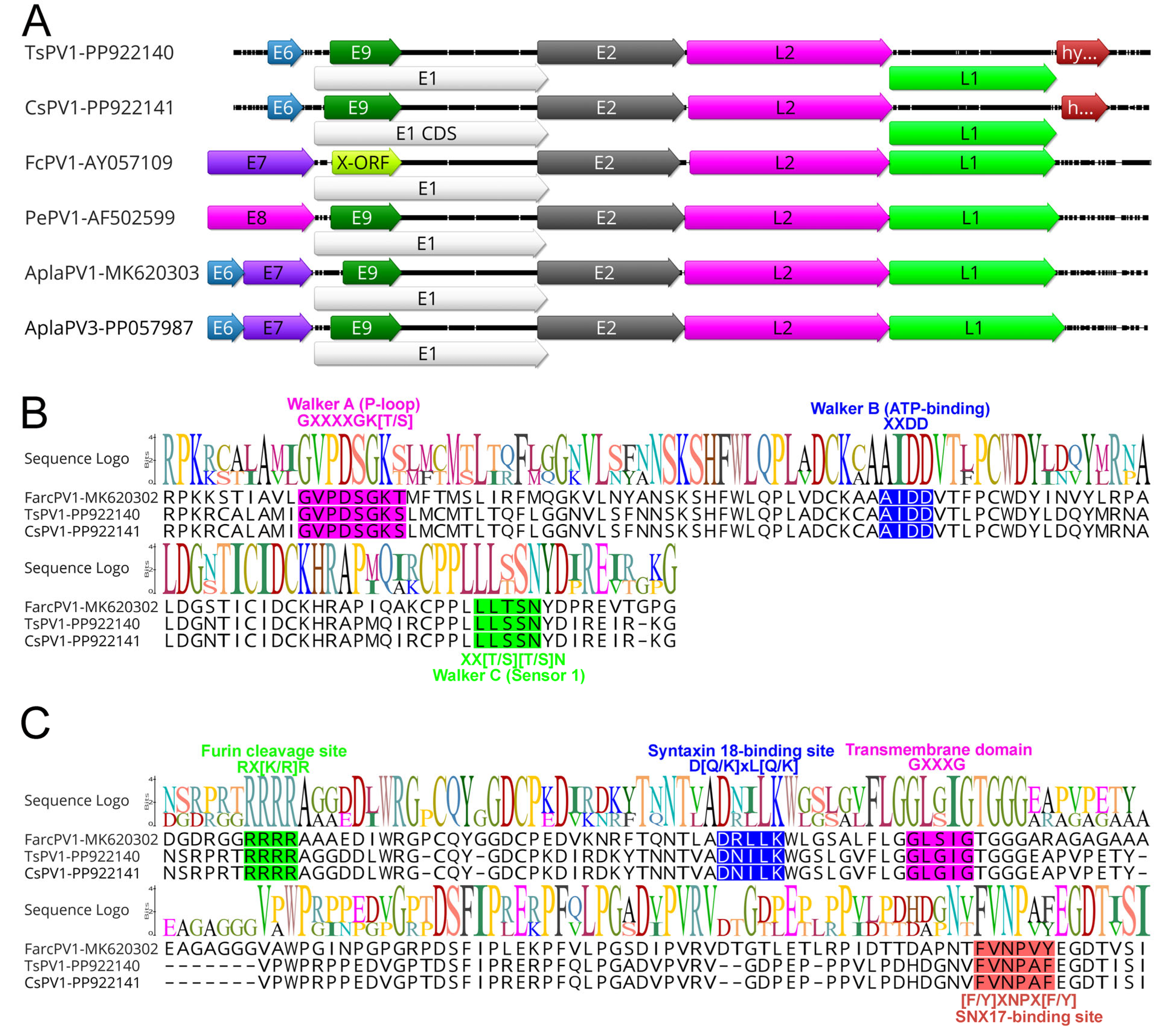
3.2. Comparative Analyses
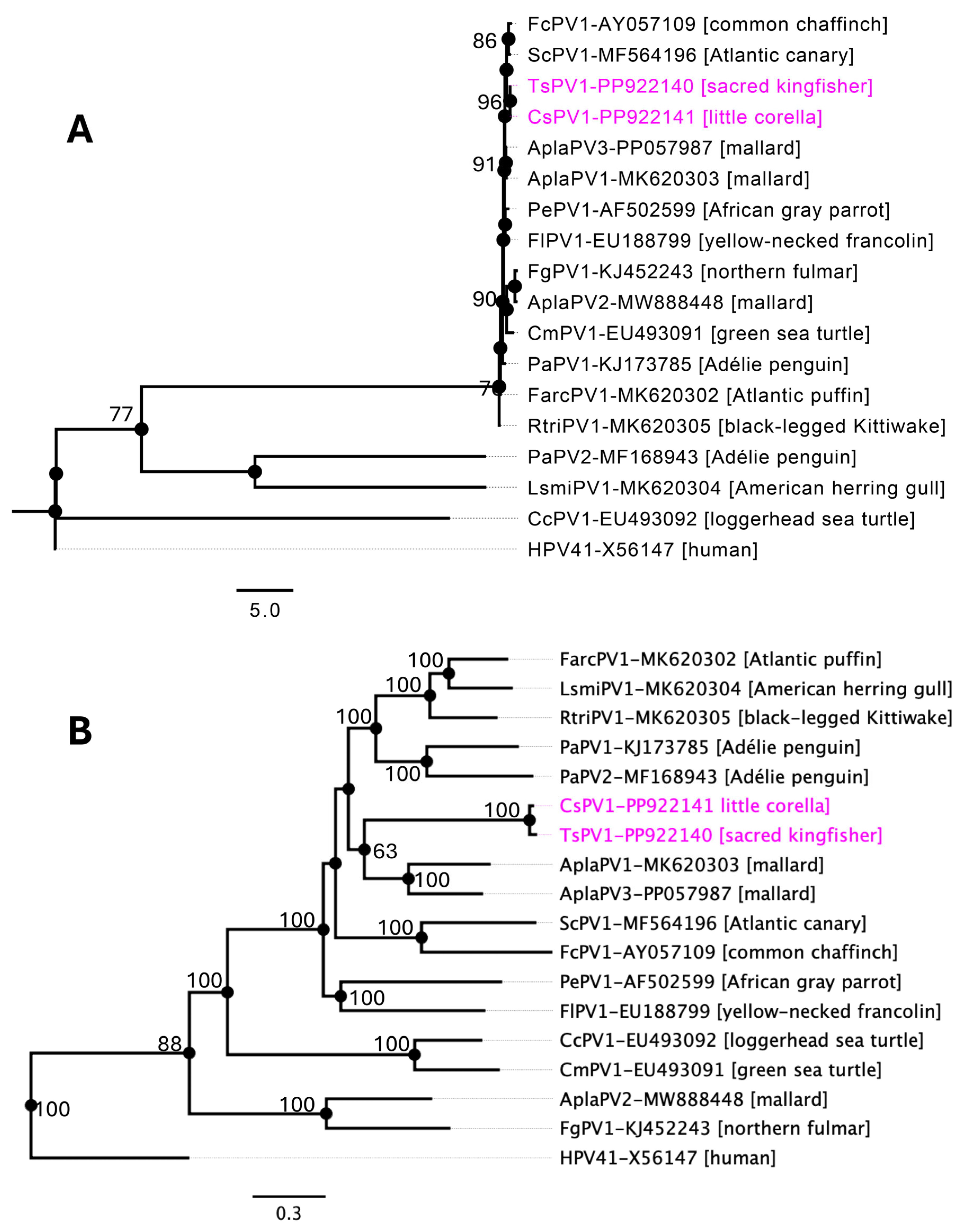
3.3. Evolutionary Relationships of APVs
3.4. Taxonomic Identification
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Varsani, A.; Kraberger, S.; Jennings, S.; Porzig, E.L.; Julian, L.; Massaro, M.; Pollard, A.; Ballard, G.; Ainley, D.G. A novel papillomavirus in Adélie penguin (Pygoscelis adeliae) faeces sampled at the Cape Crozier colony, Antarctica. J. Gen. Virol. 2014, 95, 1352–1365. [Google Scholar] [CrossRef] [PubMed]
- Nicholls, P.K.; Stanley, M.A. The immunology of animal papillomaviruses. Vet. Immunol. Immunopathol. 2000, 73, 101–127. [Google Scholar] [CrossRef]
- Campo, M.S. Papillomavirus and disease in humans and animals. Vet. Comp. Oncol. 2003, 1, 3–14. [Google Scholar] [CrossRef]
- Cubie, H.A. Diseases associated with human papillomavirus infection. Virology 2013, 445, 21–34. [Google Scholar] [CrossRef]
- Mifsud, J.C.O.; Hall, J.; Van Brussel, K.; Rose, K.; Parry, R.H.; Holmes, E.C.; Harvey, E. A novel papillomavirus in a New Zealand fur seal (Arctocephalus forsteri) with oral lesions. NPJ Viruses 2024, 2, 10. [Google Scholar] [CrossRef]
- Canuti, M.; Munro, H.J.; Robertson, G.J.; Kroyer, A.N.K.; Roul, S.; Ojkic, D.; Whitney, H.G.; Lang, A.S. New Insight into Avian Papillomavirus Ecology and Evolution from Characterization of Novel Wild Bird Papillomaviruses. Front. Microbiol. 2019, 10, 701. [Google Scholar] [CrossRef]
- Olivo, D.; Kraberger, S.; Varsani, A. New duck papillomavirus type identified in a mallard in Missouri, USA. Arch. Virol. 2024, 169, 77. [Google Scholar] [CrossRef]
- Sarker, S.; Talukder, S.; Athukorala, A.; Whiteley, P.L. The Spleen Virome of Australia’s Endemic Platypus Is Dominated by Highly Diverse Papillomaviruses. Viruses 2025, 17, 176. [Google Scholar] [CrossRef]
- Van Doorslaer, K. Revisiting Papillomavirus Taxonomy: A Proposal for Updating the Current Classification in Line with Evolutionary Evidence. Viruses 2022, 14, 2308. [Google Scholar] [CrossRef]
- Van Doorslaer, K.; Chen, Z.; Bernard, H.U.; Chan, P.K.S.; DeSalle, R.; Dillner, J.; Forslund, O.; Haga, T.; McBride, A.A.; Villa, L.L.; et al. ICTV Virus Taxonomy Profile: Papillomaviridae. J. Gen. Virol. 2018, 99, 989–990. [Google Scholar] [CrossRef]
- Buck, C.B.; Day, P.M.; Trus, B.L. The papillomavirus major capsid protein L1. Virology 2013, 445, 169–174. [Google Scholar] [CrossRef]
- Wang, J.W.; Roden, R.B.S. L2, the minor capsid protein of papillomavirus. Virology 2013, 445, 175–186. [Google Scholar] [CrossRef]
- Rector, A.; Van Ranst, M. Animal papillomaviruses. Virology 2013, 445, 213–223. [Google Scholar] [CrossRef]
- Bravo, I.G.; Félez-Sánchez, M. Papillomaviruses: Viral evolution, cancer and evolutionary medicine. Evol. Med. Public. Health 2015, 2015, 32–51. [Google Scholar] [CrossRef]
- Gottschling, M.; Stamatakis, A.; Nindl, I.; Stockfleth, E.; Alonso, Á.; Bravo, I.G. Multiple Evolutionary Mechanisms Drive Papillomavirus Diversification. Mol. Biol. Evol. 2007, 24, 1242–1258. [Google Scholar] [CrossRef]
- Shah, S.D.; Doorbar, J.; Goldstein, R.A. Analysis of Host–Parasite Incongruence in Papillomavirus Evolution Using Importance Sampling. Mol. Biol. Evol. 2010, 27, 1301–1314. [Google Scholar] [CrossRef]
- Chambers, G.; Ellsmore, V.A.; O’brien, P.M.; Reid, S.W.J.; Love, S.; Campo, M.S.; Nasir, L. Association of bovine papillomavirus with the equine sarcoid. J. General. Virol. 2003, 84, 1055–1062. [Google Scholar] [CrossRef]
- Gil da Costa, R.M.; Peleteiro, M.C.; Pires, M.A.; DiMaio, D. An Update on Canine, Feline and Bovine Papillomaviruses. Transbound. Emerg. Dis. 2016, 64, 1371–1379. [Google Scholar] [CrossRef]
- Lina, P.H.C.; van Noord, M.J.; de Groot, F.G. Detection of Virus in Squamous Papillomas of the Wild Bird Species Fringilla coelebs. JNCI J. Natl. Cancer Inst. 1973, 50, 567–571. [Google Scholar] [CrossRef]
- Di Francesco, C.E.; Profeta, F.; Romanucci, M.; Zuccarini, R.; Altea, T.; Malatesta, D.; Della Salda, L.; Marsilio, F. Evidence of avian poxvirus and papillomavirus infection in Gyps fulvus in Italy. Arch. Virol. 2019, 164, 291–295. [Google Scholar] [CrossRef]
- Gaynor, A.M.; Fish, S.; Duerr, R.S.; Cruz, F.N.D.; Pesavento, P.A. Identification of a Novel Papillomavirus in a Northern Fulmar (Fulmarus glacialis) with Viral Production in Cartilage. Vet. Pathol. 2014, 52, 553–561. [Google Scholar] [CrossRef] [PubMed]
- Tachezy, R.; Rector, A.; Havelkova, M.; Wollants, E.; Fiten, P.; Opdenakker, G.; Jenson, A.B.; Sundberg, J.P.; Van Ranst, M. Avian papillomaviruses: The parrot Psittacus erithacus papillomavirus (PePV) genome has a unique organization of the early protein region and is phylogenetically related to the chaffinch papillomavirus. BMC Microbiol. 2002, 2, 19. [Google Scholar] [CrossRef]
- Kasimov, V.; Wille, M.; Sarker, S.; Dong, Y.; Shao, R.; Hall, C.; Potvin, D.; Conroy, G.; Valenza, L.; Gillett, A.; et al. Unexpected Pathogen Diversity Detected in Australian Avifauna Highlights Potential Biosecurity Challenges. Viruses 2023, 15, 143. [Google Scholar] [CrossRef]
- Sarker, S.; Isberg, R.S.; Moran, L.J.; Araujo, D.R.; Elliott, N.; Melville, L.; Beddoe, T.; Helbig, J.K. Crocodilepox Virus Evolutionary Genomics Supports Observed Poxvirus Infection Dynamics on Saltwater Crocodile (Crocodylus porosus). Viruses 2019, 11, 1116. [Google Scholar] [CrossRef]
- Sarker, S.; Hannon, C.; Athukorala, A.; Bielefeldt-Ohmann, H. Emergence of a Novel Pathogenic Poxvirus Infection in the Endangered Green Sea Turtle (Chelonia mydas) Highlights a Key Threatening Process. Viruses 2021, 13, 219. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. A J. Comput. Mol. Cell Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Gilchrist, C.L.M.; Chooi, Y.-H. clinker & clustermap.js: Automatic generation of gene cluster comparison figures. Bioinform. 2021, 37, 2473–2475. [Google Scholar] [CrossRef]
- Hillary, W.; Lin, S.-H.; Upton, C. Base-By-Base version 2: Single nucleotide-level analysis of whole viral genome alignments. Microb. Inform. Exp. 2011, 1, 2. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Franzosa, E.A.; Sirota-Madi, A.; Avila-Pacheco, J.; Fornelos, N.; Haiser, H.J.; Reinker, S.; Vatanen, T.; Hall, A.B.; Mallick, H.; McIver, L.J.; et al. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat. Microbiol. 2019, 4, 293–305. [Google Scholar] [CrossRef] [PubMed]
- Bergvall, M.; Melendy, T.; Archambault, J. The E1 proteins. Virology 2013, 445, 35–56. [Google Scholar] [CrossRef]
- Kosugi, S.; Hasebe, M.; Tomita, M.; Yanagawa, H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc. Natl. Acad. Sci. USA 2009, 106, 10171–10176. [Google Scholar] [CrossRef]
- McBride, A.A. The Papillomavirus E2 proteins. Virology 2013, 445, 57–79. [Google Scholar] [CrossRef]
- Fawaz, M.; Vijayakumar, P.; Mishra, A.; Gandhale, P.N.; Dutta, R.; Kamble, N.M.; Sudhakar, S.B.; Roychoudhary, P.; Kumar, H.; Kulkarni, D.D.; et al. Duck gut viral metagenome analysis captures snapshot of viral diversity. Gut Pathog. 2016, 8, 30. [Google Scholar] [CrossRef]
- Truchado, D.A.; Williams, R.A.J.; Benítez, L. Natural history of avian papillomaviruses. Virus Res. 2018, 252, 58–67. [Google Scholar] [CrossRef]
- Williams, R.A.J.; Tolf, C.; Waldenström, J. Molecular identification of papillomavirus in ducks. Sci. Rep. 2018, 8, 9096. [Google Scholar] [CrossRef]
- McBride, A.A. Human papillomaviruses: Diversity, infection and host interactions. Nat. Rev. Microbiol. 2022, 20, 95–108. [Google Scholar] [CrossRef]

| Papillomavirus Name | Genus | Species | GenBank Accession Numbers | Length (nt) | Host | Abbreviation | Pairwise Genomic Identity (%) _TsPV1 | Pairwise Genomic Identity (%) _CsPV1 |
|---|---|---|---|---|---|---|---|---|
| Todiramphus sanctus papillomavirus 1 | unclassified | unclassified | PP922140.1 | 7883 | Todiramphus sanctus (Sacred kingfisher) | TsPV1 | 99.69 | |
| Cacatua sanguinea papillomavirus 1 | unclassified | unclassified | PP922141.1 | 7825 | Cacatua sanguinea (Little corella) | CsPV1 | 99.69 | |
| Psittacus erithacus papillomavirus 1 | Thetapapillomavirus | Thetapapillomavirus 1 | AF502599.1 | 7304 | Psittacus erithacus (African grey parrot) | PePV1 | 52.98 | 52.17 |
| Fringilla coelebs papillomavirus 1 | Etapapillomavirus | Etapapillomavirus 1 | AY057109.1 | 7729 | Fringilla coelebs (Common chaffinch) | FcPV1 | 50.30 | 49.76 |
| Francolinus leucoscepus papillomavirus 1 | Dyoepsilonpapillomavirus | Dyoepsilonpapillomavirus 1 | EU188799.1 | 7498 | Pternistis leucoscepus (Yellow-necked francolin) | FlPV1 | 50.78 | 50.46 |
| Chelonia mydas papillomavirus 1 | Dyozetapapillomavirus | Dyozetapapillomavirus 1 | EU493091.1 | 6953 | Chelonia mydas (Green sea turtle) | CmPV1 | 46.02 | 45.34 |
| Caretta caretta papillomavirus 1 | Dyozetapapillomavirus | Dyozetapapillomavirus 1 | EU493092.1 | 7020 | Caretta caretta (Loggerhead sea turtle) | CcPV1 | 46.22 | 45.43 |
| Pygoscelis adeliae papillomavirus 1 | Treisepsilonpapillomavirus | Treisepsilonpapillomavirus 1 | KJ173785.1 | 7637 | Pygoscelis adeliae (Adélie penguin) | PaPV1 | 51.71 | 51.47 |
| Fulmarus glacialis papillomavirus 1 | Treiszetapapillomavirus | Treiszetapapillomavirus 1 | KJ452243.1 | 8132 | Fulmarus glacialis (Northern fulmar) | FgPV1 | 45.32 | 44.19 |
| Pygoscelis adeliae papillomavirus 2 | Treisepsilonpapillomavirus | Treisepsilonpapillomavirus 1 | MF168943.1 | 7654 | Pygoscelis adeliae (Adélie penguin) | PaPV2 | 51.41 | 51.74 |
| Serinus canaria papillomavirus 1 | Etapapillomavirus | unclassified Etapapillomavirus | MF564196.1 | 8071 | Serinus canaria (Atlantic canary) | ScPV1 | 50.35 | 50.13 |
| Fratercula arctica papillomavirus 1 | unclassified | unclassified | MK620302.1 | 7703 | Fratercula arctica (Atlantic puffin) | FarcPV1 | 54.50 | 54.17 |
| Duck papillomavirus 3 | unclassified | unclassified | MK620303.1 | 7887 | Anas platyrhynchos (Mallard) | AplaPV1 | 54.30 | 53.89 |
| Larus smithsonianus papillomavirus 1 | unclassified | unclassified | MK620304.1 | 7699 | Larus smithsonianus (American herring gull) | LsmiPV1 | 54.30 | 54.04 |
| Rissa tridactyla papillomavirus 2 | unclassified | unclassified | MK620305.1 | 7763 | Rissa tridactyla (Black-legged Kittiwake) | RtriPV1 | 54.57 | 54.20 |
| Anas platyrhynchos papillomavirus 2 | unclassified | unclassified | MW888448.1 | 8350 | Anas platyrhynchos (Mallard or wild duck) | AplaPV2 | 45.28 | 45.79 |
| Anas platyrhynchos papillomavirus 3 | unclassified | unclassified | PP057987.1 | 7839 | Anas platyrhynchos (Mallard or wild duck) | AplaPV3 | 56.66 | 56.39 |
| Gene Synteny | Genome Coordinates | nt Size | AA Size | Best Blast Hits (GenBank Accession Number) | Product | Similarity (%) | Note |
|---|---|---|---|---|---|---|---|
| Todiramphus sanctus papillomavirus 1 (TsPV1, GenBank accession no. PP922140.1) | |||||||
| ORF01 | 318–629 | 312 | 103 | Duck papillomavirus 3 (QBR99466.1) | E6 | 44.44 | |
| ORF02 | 693–2801 | 2109 | 702 | Duck papillomavirus 3 (QBR99468.1) | E1 | 64.84 | |
| ORF03 | 823–1440 | 618 | 205 | Serinus canaria papillomavirus 1 (YP_009551917.1) | E9 | 29.85 | |
| ORF04 | 2722–3918 | 1197 | 398 | Gull papillomavirus 1 (QBR99477.1) | E2 | 38.65 | |
| ORF07 | 3939–5570 | 1632 | 543 | Duck papillomavirus (ANN29878.1) | L2 | 45.36 | |
| ORF08 | 5558–7108 | 1551 | 516 | Duck papillomavirus 3 (QBR99472.1) | L1 | 65.05 | |
| ORF06 | 7110–7556 | 447 | 148 | hypothetical gene | |||
| Cacatua sanguinea papillomavirus 1 (CsPV1, GenBank accession no. PP922141.1) | |||||||
| ORF01 | 318–629 | 312 | 103 | Duck papillomavirus 3 (QBR99466.1) | E6 | 44.44 | |
| ORF02 | 717–2825 | 2109 | 702 | Duck papillomavirus 3 (QBR99468.1) | E1 | 64.84 | |
| ORF03 | 808–1464 | 657 | 218 | Serinus canaria papillomavirus 1 (YP_009551917.1) | E9 | 29.85 | |
| ORF04 | 2746–3942 | 1197 | 398 | Gull papillomavirus 1 (QBR99477.1) | E2 | 38.90 | |
| ORF05 | 3950–5554 | 1605 | 534 | Duck papillomavirus (ANN29878.1) | L2 | 44.90 | |
| ORF06 | 5542–7092 | 1551 | 516 | Duck papillomavirus 3 (QBR99472.1) | L1 | 65.05 | |
| ORF07 | 7136–7516 | 381 | 126 | hypothetical gene | |||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | TsPV1-PP922140 | |||||||||||||||||
| 2 | CsPV1-PP922141 | 100 | ||||||||||||||||
| 3 | AplaPV1-MK620303 | 63.14 | 63.14 | |||||||||||||||
| 4 | AplaPV2-MW888448 | 49.89 | 49.89 | 50.17 | ||||||||||||||
| 5 | AplaPV3-PP057987 | 64.24 | 64.24 | 72.73 | 51.68 | |||||||||||||
| 6 | FlPV1-EU188799 | 55 | 55 | 61.75 | 49.10 | 61.84 | ||||||||||||
| 7 | PePV1-AF502599 | 59.65 | 59.65 | 60.71 | 47.70 | 61.65 | 59.66 | |||||||||||
| 8 | RtriPV1-MK620305 | 60.13 | 60.13 | 60.42 | 48.55 | 59.75 | 57.16 | 58.16 | ||||||||||
| 9 | LsmiPV1-MK620304 | 57.81 | 57.81 | 60.32 | 49.23 | 59.97 | 57.49 | 57.12 | 68.38 | |||||||||
| 10 | PaPV2-MF168943 | 56.19 | 56.19 | 58.70 | 49.44 | 58.23 | 55.47 | 56.91 | 59.06 | 60.51 | ||||||||
| 11 | FarcPV1-MK620302 | 58.94 | 58.94 | 60.71 | 49.40 | 60.53 | 59.59 | 58.49 | 70.04 | 69.45 | 59.87 | |||||||
| 12 | PaPV1-KJ173785 | 56.74 | 56.74 | 60.89 | 51.35 | 59.02 | 59.04 | 57.20 | 59.53 | 59.41 | 65.15 | 62.60 | ||||||
| 13 | FcPV1-AY057109 | 59.72 | 59.72 | 59.95 | 50.35 | 59.20 | 55.33 | 56.57 | 55.44 | 54.84 | 54.48 | 57.07 | 54.38 | |||||
| 14 | ScPV1-MF564196 | 60.08 | 60.08 | 59.37 | 49.41 | 60.01 | 54.62 | 56.84 | 56.57 | 55.08 | 54.34 | 56.65 | 54.64 | 64.86 | ||||
| 15 | CmPV1-EU493091 | 49.83 | 49.83 | 51.77 | 47.95 | 50.71 | 48.60 | 50.96 | 48.44 | 47.88 | 50.24 | 50.07 | 50.07 | 47.81 | 49.62 | |||
| 16 | CcPV1-EU493092 | 51.72 | 51.72 | 52.92 | 48.69 | 52.95 | 50.27 | 49.83 | 50.85 | 48.59 | 50.83 | 51.68 | 50.31 | 49.21 | 51.58 | 70.91 | ||
| 17 | FgPV1-KJ452243 | 49.86 | 49.86 | 50.21 | 63.18 | 50.70 | 49.31 | 50.25 | 49.68 | 48.77 | 50.63 | 48.87 | 49.25 | 48.63 | 50.49 | 46.79 | 47.67 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sarker, S.; Kasimov, V.; Rahaman, M.M.; Nath, B.K.; Jelocnik, M. Discovery and Genomic Characterisation of Novel Papillomaviruses in Australian Wild Birds. Pathogens 2025, 14, 514. https://doi.org/10.3390/pathogens14060514
Sarker S, Kasimov V, Rahaman MM, Nath BK, Jelocnik M. Discovery and Genomic Characterisation of Novel Papillomaviruses in Australian Wild Birds. Pathogens. 2025; 14(6):514. https://doi.org/10.3390/pathogens14060514
Chicago/Turabian StyleSarker, Subir, Vasilli Kasimov, Md. Mizanur Rahaman, Babu Kanti Nath, and Martina Jelocnik. 2025. "Discovery and Genomic Characterisation of Novel Papillomaviruses in Australian Wild Birds" Pathogens 14, no. 6: 514. https://doi.org/10.3390/pathogens14060514
APA StyleSarker, S., Kasimov, V., Rahaman, M. M., Nath, B. K., & Jelocnik, M. (2025). Discovery and Genomic Characterisation of Novel Papillomaviruses in Australian Wild Birds. Pathogens, 14(6), 514. https://doi.org/10.3390/pathogens14060514







