Acute Respiratory and Influenza Viruses Circulating in Kazakhstan During 2018–2024
Abstract
1. Introduction
2. Materials and Methods
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| rtRT-PCR | Real-time polymerase chain reaction |
| ARVI | Acute respiratory viral infection |
| ELISA | Enzyme-linked immunosorbent assay |
| ELISA TS IgG | Enzyme-linked immunosorbent assay system for the determination of antibody class IgG |
| HI | Hemagglutinating activity inhibition |
| IDS | Influenza diagnostic serum, dry |
| hAdv | Adenovirus |
| hBov | Bocavirus |
| hCov | Coronavirus |
| hMpv | Metapneumovirus |
| hPiv | Paramyxovirus |
| hRSv | Respiratory syncytial virus |
| hRv | Rhinovirus |
References
- Turyasiima, M.; Kiconco, G.; Egesa, W.; Twesigemukama, S.; Nduwimana, M. Prevalence and Outpatient Clinical Diagnostic Approaches for Common Acute Respiratory Tract Infections in Children Under Five Years of Age: A Cross-Sectional Study. Pediatr. Health Med. Ther. 2024, 15, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Klivleyeva, N.; Lukmanova, G.; Glebova, T.; Shamenova, M.; Ongarbayeva, N.; Saktaganov, N.; Baimukhametova, A.; Baiseiit, S.; Ismagulova, D.; Kassymova, G.; et al. Spread of Pathogens Causing Respiratory Viral Diseases Before and During COVID-19 Pandemic in Kazakhstan. Indian J. Microbiol. 2023, 63, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, J.R.; Sotomayor, V.; Uez, O.C.; Oliva, O.; Bettels, D.; McCarron, M.; Bresee, J.S.; Mounts, A.W. Strategy to Enhance Influenza Surveillance Worldwide1. Emerg. Infect. Dis. 2009, 15, 1271–1278. [Google Scholar] [CrossRef] [PubMed]
- Klivleyeva, N.G.; Ongarbayeva, N.S.; Korotetskiy, I.S.; Glebova, T.I.; Saktaganov, N.T.; Shamenova, M.G.; Baimakhanova, B.B.; Shevtsov, A.B.; Amirgazin, A.; Berezin, V.E.; et al. Coding-Complete Genome Sequence of Swine Influenza Virus Isolate A/Swine/Karaganda/04/2020 (H1N1) from Kazakhstan. Microbiol. Resour. Announc. 2021, 10, e0078621. [Google Scholar] [CrossRef]
- Lukmanova, G.; Klivleyeva, N.; Glebova, T.; Ongarbayeva, N.; Shamenova, M.; Saktaganov, N.; Baimukhametova, A.; Baiseiit, S.; Ismagulova, D.; Ismailov, E.; et al. Influenza A Virus Surveillance in Domestic Pigs in Kazakhstan 2018–2021. Ciênc. Rural 2024, 54, e20230403. [Google Scholar] [CrossRef]
- Glebova, T.I.; Klivleyeva, N.G.; Baimukhametova, A.M.; Saktaganov, N.T.; Lukmanova, G.V.; Ongarbayeva, N.S.; Shamenova, M.G.; Baimakhanova, B.B. Circulation of Influenza Viruses in the Epidemic Season of 2018–2019 among People Residing in Northern and Western Kazakhstan. Infekt. Bolezn. 2021, 19, 70–75. [Google Scholar] [CrossRef]
- Klivleyeva, N.; Saktaganov, N.; Glebova, T.; Lukmanova, G.; Ongarbayeva, N.; Webby, R. Influenza A Viruses in the Swine Population: Ecology and Geographical Distribution. Viruses 2024, 16, 1728. [Google Scholar] [CrossRef]
- Shao, W.; Li, X.; Goraya, M.; Wang, S.; Chen, J.-L. Evolution of Influenza A Virus by Mutation and Re-Assortment. Int. J. Mol. Sci. 2017, 18, 1650. [Google Scholar] [CrossRef]
- Katz, M.A.; Schoub, B.D.; Heraud, J.M.; Breiman, R.F.; Njenga, M.K.; Widdowson, M.-A. Influenza in Africa: Uncovering the Epidemiology of a Long-Overlooked Disease. J. Infect. Dis. 2012, 206, S1–S4. [Google Scholar] [CrossRef]
- Korsun, N.; Angelova, S.; Gregory, V.; Daniels, R.; Georgieva, I.; McCauley, J. Antigenic and Genetic Characterization of Influenza Viruses Circulating in Bulgaria during the 2015/2016 Season. Infect. Genet. Evol. 2017, 49, 241–250. [Google Scholar] [CrossRef]
- Shamenova, M.G.; Glebova, T.I.; Klivleyeva, N.G.; Baiseiit, S.B.; Baimukhametova, A.M.; Saktaganov, N.T.; Ongarbayeva, N.S.; Ismagulova, D.A. Serological Studies of Influenza Infection among Population in Southern Region of Kazakhstan during the 2018–2021 Epidemic Season. J. Pak. Med. Assoc. 2023, 73, 804–807. [Google Scholar] [CrossRef]
- Polansky, L.S.; Outin-Blenman, S.; Moen, A.C. Improved Global Capacity for Influenza Surveillance. Emerg. Infect. Dis. 2016, 22, 993–1001. [Google Scholar] [CrossRef] [PubMed]
- WHO Global Influenza Surveillance Network. Manual for the Laboratory Diagnosis and Virological Surveillance of Influenza. Available online: https://www.who.int/publications/i/item/manual-for-the-laboratory-diagnosis-and-virological-surveillance-of-influenza (accessed on 13 May 2025).
- Zhang, D.; Mao, H.; Lou, X.; Pan, J.; Yan, H.; Tang, H.; Shu, Y.; Zhao, Y.; Cheng, X.; Tao, H.; et al. Clinical Evaluation of a Panel of Multiplex Quantitative Real-Time Reverse Transcription Polymerase Chain Reaction Assays for the Detection of 16 Respiratory Viruses Associated with Community-Acquired Pneumonia. Arch. Virol. 2018, 163, 2855–2860. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, E.; Stech, J.; Guan, Y.; Webster, R.G.; Perez, D.R. Universal Primer Set for the Full-Length Amplification of All Influenza A Viruses. Arch. Virol. 2001, 146, 2275–2289. [Google Scholar] [CrossRef]
- Zhou, B.; Wentworth, D.E. Influenza A Virus Molecular Virology Techniques; Humana Press: Totowa, NJ, USA, 2012; pp. 175–192. [Google Scholar]
- Hasegawa, M.; Kishino, H.; Yano, T. Dating of the Human-Ape Splitting by a Molecular Clock of Mitochondrial DNA. J. Mol. Evol. 1985, 22, 160–174. [Google Scholar] [CrossRef]
- Piantadosi, S.; Gail, M.H. A Comparison of the Power of Two Tests for Qualitative Interactions. Stat. Med. 1993, 12, 1239–1248. [Google Scholar] [CrossRef]
- Oliver-Rodríguez, J.C.; Wang, X.T. Non-parametric Three-way Mixed ANOVA with Aligned Rank Tests. Br. J. Math. Stat. Psychol. 2015, 68, 23–42. [Google Scholar] [CrossRef]
- Branche, A.; Falsey, A. Parainfluenza Virus Infection. Semin. Respir. Crit. Care Med. 2016, 37, 538–554. [Google Scholar] [CrossRef]
- GISAID. Available online: https://platform.epicov.org/epi3/cfrontend#26e134 (accessed on 13 May 2025).
- Kuatbaeva, A.M.; Esmagambetova, A.S.; Akhmetova, Z.D.; Smagul, M.A.; Utegenova, E.S.; Stolyarov, K.A.; Kasabekova, L.K.; Nusupbaeva, G.E.; Smagulova, M.K.; Amrin, M.K. Epidemiological Characteristics of Acute Respiratory Viral Infections and Influenza in Kazakhstan during Epidemic Seasons from 2018 to 2022. Infekc. Bolezn. 2023, 21, 80–94. [Google Scholar] [CrossRef]
- Wang, J.; Wang, R.; Xie, Z. Rethinking Paediatric Respiratory Infections: The Role of Mixed Pathogen Infections. Rev. Med. Virol. 2025, 35, e70021. [Google Scholar] [CrossRef]
- Burtseva, E.I.; Panova, A.D.; Kolobukhina, L.V.; Ignatjeva, A.V.; Kirillova, E.S.; Breslav, N.V.; Trushakova, S.V.; Mukasheva, E.A.; Feodoritova, E.L.; Krasnoslobodtsev, K.G.; et al. Epidemic Season 2021–2022: Frequency of Co-Infection by Respiratory Viral Pathogens. Epidemiol. Infect. Dis. 2023, 28, 67–77. [Google Scholar] [CrossRef]
- Elhakim, M.; Hafiz Rasooly, M.; Fahim, M.; Sheikh Ali, S.; Haddad, N.; Cherkaoui, I.; Hjaija, D.; Nadeem, S.; Assiri, A.; Aljifri, A.; et al. Epidemiology of Severe Cases of Influenza and Other Acute Respiratory Infections in the Eastern Mediterranean Region, July 2016 to June 2018. J. Infect. Public Health 2020, 13, 423–429. [Google Scholar] [CrossRef] [PubMed]
- Matias, G.; Haguinet, F.; Lustig, R.L.; Edelman, L.; Chowell, G.; Taylor, R.J. Model Estimates of the Burden of Outpatient Visits Attributable to Influenza in the United States. BMC Infect. Dis. 2016, 16, 641. [Google Scholar] [CrossRef] [PubMed]
- Kazakhstan Population Reaches 19.17 Million in 2021. Available online: https://astanatimes.com/2021/12/kazakhstan-population-reaches-19-17-million-in-2021/ (accessed on 12 March 2025).
- Wratil, P.R.; Schmacke, N.A.; Karakoc, B.; Dulovic, A.; Junker, D.; Becker, M.; Rothbauer, U.; Osterman, A.; Spaeth, P.M.; Ruhle, A.; et al. Evidence for Increased SARS-CoV-2 Susceptibility and COVID-19 Severity Related to Pre-Existing Immunity to Seasonal Coronaviruses. Cell Rep. 2021, 37, 110169. [Google Scholar] [CrossRef]
- Contes, K.M.; Liu, B.M. Epidemiology, Clinical Significance, and Diagnosis of Respiratory Viruses and Their Co-Infections in the Post-COVID Era. Pathogens 2025, 14, 262. [Google Scholar] [CrossRef]
- Amanat, F.; Clark, J.; Carreño, J.M.; Strohmeier, S.; Yellin, T.; Meade, P.S.; Bhavsar, D.; Muramatsu, H.; Sun, W.; Coughlan, L.; et al. Immunity to Seasonal Coronavirus Spike Proteins Does Not Protect from SARS-CoV-2 Challenge in a Mouse Model but Has No Detrimental Effect on Protection Mediated by COVID-19 MRNA Vaccination. J. Virol. 2023, 97, e0166422. [Google Scholar] [CrossRef]
- Krumbein, H.; Kümmel, L.S.; Fragkou, P.C.; Thölken, C.; Hünerbein, B.L.; Reiter, R.; Papathanasiou, K.A.; Renz, H.; Skevaki, C. Respiratory Viral Co-infections in Patients with COVID-19 and Associated Outcomes: A Systematic Review and Meta-analysis. Rev. Med. Virol. 2023, 33, e2365. [Google Scholar] [CrossRef]
- Marshall, N.C.; Kariyawasam, R.M.; Zelyas, N.; Kanji, J.N.; Diggle, M.A. Broad Respiratory Testing to Identify SARS-CoV-2 Viral Co-Circulation and Inform Diagnostic Stewardship in the COVID-19 Pandemic. Virol. J. 2021, 18, 93. [Google Scholar] [CrossRef]
- Blyth, C.C.; Webb, S.A.R.; Kok, J.; Dwyer, D.E.; van Hal, S.J.; Foo, H.; Ginn, A.N.; Kesson, A.M.; Seppelt, I.; Iredell, J.R. The Impact of Bacterial and Viral Co-Infection in Severe Influenza. Influenza Other Respir. Viruses 2013, 7, 168–176. [Google Scholar] [CrossRef]
- Basile, K.; Kok, J.; Dwyer, D.E. Point-of-Care Diagnostics for Respiratory Viral Infections. Expert. Rev. Mol. Diagn. 2018, 18, 75–83. [Google Scholar] [CrossRef]
- Dao, T.L.; Hoang, V.T.; Colson, P.; Million, M.; Gautret, P. Co-Infection of SARS-CoV-2 and Influenza Viruses: A Systematic Review and Meta-Analysis. J. Clin. Virol. Plus 2021, 1, 100036. [Google Scholar] [CrossRef] [PubMed]
- Inma, P.; Suntronwong, N.; Sinsulpsiri, S.; Srimaneewiroon, S.; Poovorawan, Y. Viral Etiology Associated with Acute Respiratory Tract Infection Patients in Bangkok, Thailand. Cureus 2024, 16, e66897. [Google Scholar] [CrossRef]
- Lei, C.; Lou, C.T.; Io, K.; SiTou, K.I.; Ip, C.P.; U, H.; Pan, B.; Ung, C.O.L. Viral Etiology among Children Hospitalized for Acute Respiratory Tract Infections and Its Association with Meteorological Factors and Air Pollutants: A Time-Series Study (2014–2017) in Macao. BMC Infect. Dis. 2022, 22, 588. [Google Scholar] [CrossRef]
- Al-Romaihi, H.E.; Smatti, M.K.; Al-Khatib, H.A.; Coyle, P.V.; Ganesan, N.; Nadeem, S.; Farag, E.A.; Al Thani, A.A.; Al Khal, A.; Al Ansari, K.M.; et al. Molecular Epidemiology of Influenza, RSV, and Other Respiratory Infections among Children in Qatar: A Six Years Report (2012–2017). Int. J. Infect. Dis. 2020, 95, 133–141. [Google Scholar] [CrossRef]
- Liu, T.; Li, Z.; Zhang, S.; Song, S.; Julong, W.; Lin, Y.; Guo, N.; Xing, C.; Xu, A.; Bi, Z.; et al. Viral Etiology of Acute Respiratory Tract Infections in Hospitalized Children and Adults in Shandong Province, China. Virol. J. 2015, 12, 168. [Google Scholar] [CrossRef]
- Garcia-Garcia, M.L.; Rey, C.C.; del Rosal Rabes, T. Asma y Virus En El Niño. Arch. Bronconeumol. 2016, 52, 269–273. [Google Scholar] [CrossRef]
- Pisareva, M.M.; Eder, V.A.; Buzitskaya, Z.V.; Musaeva, T.D.; Afanaseva, V.S.; Go, A.A.; Obraztsova, E.A.; Sukhovetskaya, V.F.; Komissarov, A.B. Etiological Structure of Influenza and Other Arvi in St. Petersburg during Epidemic Seasons 2012–2016. Probl. Virol. 2018, 63, 233–239. [Google Scholar] [CrossRef]
- Oh, D.-Y.; Buda, S.; Biere, B.; Reiche, J.; Schlosser, F.; Duwe, S.; Wedde, M.; von Kleist, M.; Mielke, M.; Wolff, T.; et al. Trends in Respiratory Virus Circulation Following COVID-19-Targeted Nonpharmaceutical Interventions in Germany, January–September 2020: Analysis of National Surveillance Data. Lancet Reg. Health-Eur. 2021, 6, 100112. [Google Scholar] [CrossRef]
- Ljubin-Sternak, S.; Meštrović, T.; Lukšić, I.; Mijač, M.; Vraneš, J. Seasonal Coronaviruses and Other Neglected Respiratory Viruses: A Global Perspective and a Local Snapshot. Front. Public Health 2021, 9, 691163. [Google Scholar] [CrossRef]
- L’vov, D.K.; Burtseva, E.I.; Kolobukhina, L.V.; Fedyakina, I.T.; Bovin, N.V.; Ignatjeva, A.V.; Krasnoslobodtsev, K.G.; Feodoritova, E.L.; Trushakova, S.V.; Breslav, N.V.; et al. Peculiarities of the Influenza and ARVI Viruses Circulation during Epidemic Season 2019–2020 in Some Regions of Russia. Probl. Virol. 2021, 65, 335–349. [Google Scholar] [CrossRef]
- Bashir, U.; Nisar, N.; Mahmood, N.; Alam, M.M.; Sadia, H.; Zaidi, S.S.Z. Molecular Detection and Characterization of Respiratory Syncytial Virus B Genotypes Circulating in Pakistani Children. Infect. Genet. Evol. 2017, 47, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Naz, R.; Gul, A.; Javed, U.; Urooj, A.; Amin, S.; Fatima, Z. Etiology of Acute Viral Respiratory Infections Common in Pakistan: A Review. Rev. Med. Virol. 2018, 29, e2024. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Xu, D.; Zhang, Y.; Wang, T.; Zhang, L.; Gu, W.; Shen, M. Epidemiological Characteristics of Four Common Respiratory Viral Infections in Children. Virol. J. 2021, 18, 10. [Google Scholar] [CrossRef]
- Khalid, S.; Ghani, E. Study on Etiology of Viral Lower Respiratory Tract Infections in Children Under 10 Years of Age. J. Virol. Antivir. Res. 2016, 4–5. [Google Scholar] [CrossRef]
- Ji, L.; Chen, L.; Xu, D.; Wu, X. Molecular Typing and Epidemiologic Profiles of Human Metapneumovirus Infection among Children with Severe Acute Respiratory Infection in Huzhou, China. Mol. Biol. Rep. 2021, 48, 7697–7702. [Google Scholar] [CrossRef] [PubMed]
- Divarathna, M.V.M.; Rafeek, R.A.M.; Noordeen, F. A Review on Epidemiology and Impact of Human Metapneumovirus Infections in Children Using TIAB Search Strategy on PubMed and PubMed Central Articles. Rev. Med. Virol. 2020, 30, e2090. [Google Scholar] [CrossRef]
- Çiçek, C.; Arslan, A.; Karakuş, H.S.; Yalaz, M.; Saz, E.U.; Pullukçu, H.; Çok, G. Prevalence and Seasonal Distribution of Respiratory Viruses in Patients with Acute Respiratory Tract Infections, 2002–2014. Mikrobiyol. Bul. 2015, 49, 188–200. [Google Scholar] [CrossRef]
- Brestovac, B.; Lawrence, C.; Speers, D.J.; Sammels, L.M.; Mulrennan, S. Respiratory Viral Infections in Western Australians with Cystic Fibrosis. Respir. Med. 2020, 161, 105854. [Google Scholar] [CrossRef]
- Klivleyeva, N.G.; Ongarbayeva, N.S.; Baimukhametova, A.M.; Saktaganov, N.T.; Lukmanova, G.V.; Glebova, T.I.; Sayatov, M.K.; Berezin, V.E.; Nusupbaeva, G.E.; Aikimbayev, A.M. Detection of Influenza Virus and Pathogens of Acute Respiratory Viral Infections in Population of Kazakhstan during 2018–2019 Epidemic Season. Russ. J. Infect. Immun. 2021, 11, 137–147. [Google Scholar] [CrossRef]
- Klivleyeva, N.G.; Lukmanova, G.V.; Saktaganov, N.T.; Sayatov, M.K.; Glebova, T.I.; Ongarbayeva, N.S.; Baimukhametova, A.M.; Shamenova, M.G.; Berezin, V.E.; Nusupbaeva, G.E.; et al. Acute Respiratory Viral Infections in Kazakhstan in 2017–2019. Bulletin 2020, 3, 29–35. [Google Scholar] [CrossRef]
- Abdel-Whab, M.; Kolosova, N.P.; Boldyrev, N.D.; Svyatchenko, S.V.; Danilenko, A.V.; Goncharova, N.I.; Shadrinova, K.N.; Danilenko, E.I.; Onkhonova, G.S.; Kosenko, M.N.; et al. An Investigation of Severe Influenza Cases in Russia during the 2022–2023 Epidemic Season and an Analysis of HA-D222G/N Polymorphism in Newly Emerged and Dominant Clade 6B.1A.5a.2a A(H1N1)Pdm09 Viruses. Pathogens 2023, 13, 1. [Google Scholar] [CrossRef]
- Wolf, R.M.; Antoon, J.W. Influenza in Children and Adolescents: Epidemiology, Management, and Prevention. Pediatr. Rev. 2023, 44, 605–617. [Google Scholar] [CrossRef]
- Gaitonde, D.Y.; Moore, F.C.; Morgan, M.K. Influenza: Diagnosis and Treatment. Am. Fam. Physician 2019, 100, 751–758. [Google Scholar]
- Lewandowski, K.; Xu, Y.; Pullan, S.T.; Lumley, S.F.; Foster, D.; Sanderson, N.; Vaughan, A.; Morgan, M.; Bright, N.; Kavanagh, J.; et al. Metagenomic Nanopore Sequencing of Influenza Virus Direct from Clinical Respiratory Samples. J. Clin. Microbiol. 2019, 58, e00963-19. [Google Scholar] [CrossRef]
- Chung, H.-Y.; Jian, M.-J.; Chang, C.-K.; Lin, J.-C.; Yeh, K.-M.; Chen, C.-W.; Chiu, S.-K.; Wang, Y.-H.; Liao, S.-J.; Li, S.-Y.; et al. Novel Dual Multiplex Real-Time RT-PCR Assays for the Rapid Detection of SARS-CoV-2, Influenza A/B, and Respiratory Syncytial Virus Using the BD MAX Open System. Emerg. Microbes Infect. 2021, 10, 161–166. [Google Scholar] [CrossRef]
- Sadeghi, P.; Sohrabi, H.; Hejazi, M.; Jahanban-Esfahlan, A.; Baradaran, B.; Tohidast, M.; Majidi, M.R.; Mokhtarzadeh, A.; Tavangar, S.M.; de la Guardia, M. Lateral Flow Assays (LFA) as an Alternative Medical Diagnosis Method for Detection of Virus Species: The Intertwine of Nanotechnology with Sensing Strategies. TrAC Trends Anal. Chem. 2021, 145, 116460. [Google Scholar] [CrossRef]
- Allwinn, R.; Preiser, W.; Rabenau, H.; Buxbaum, S.; Stürmer, M.; Doerr, H.W. Laboratory Diagnosis of Influenza–Virology or Serology? Med. Microbiol. Immunol. 2002, 191, 157–160. [Google Scholar] [CrossRef]
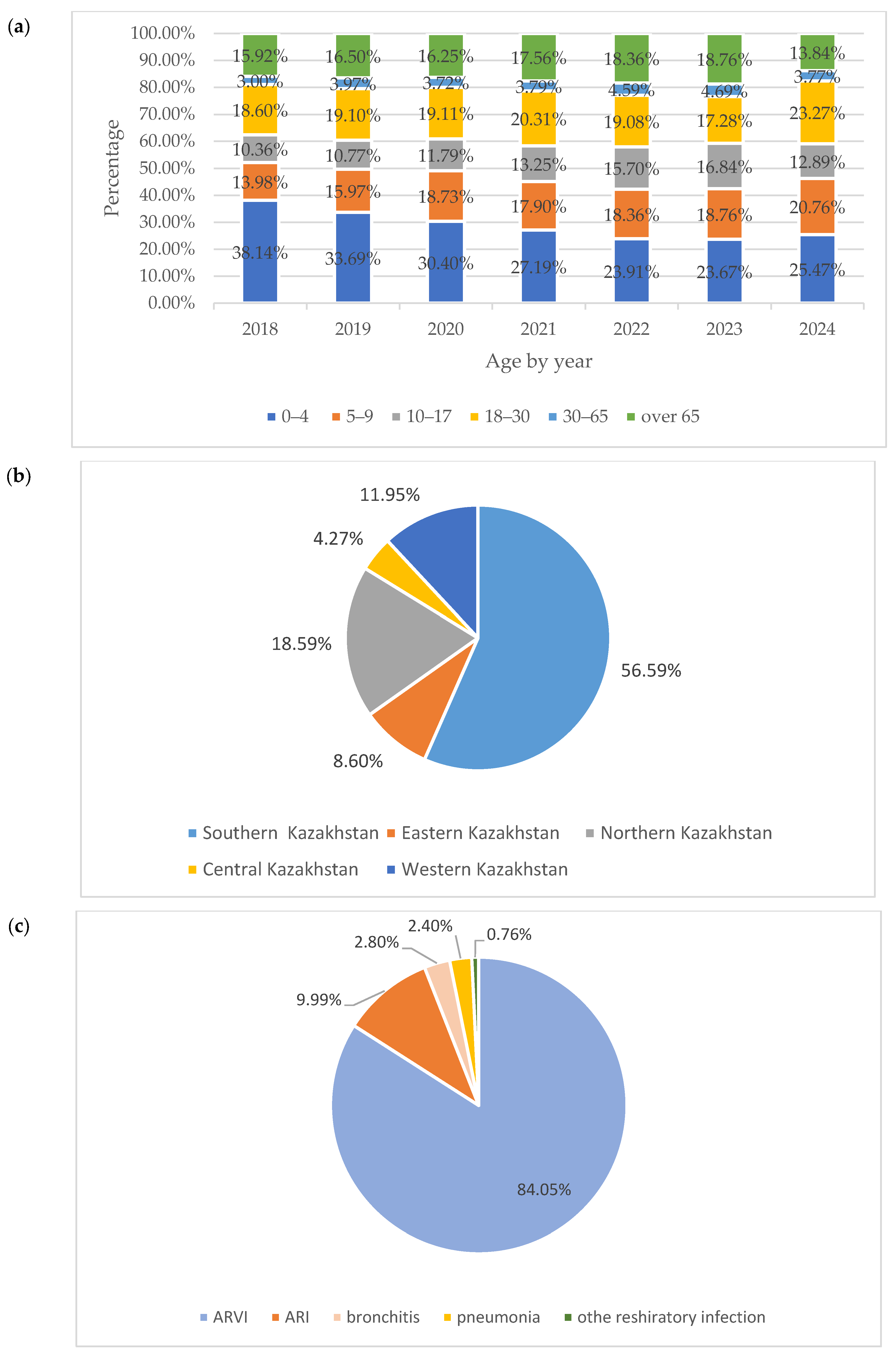
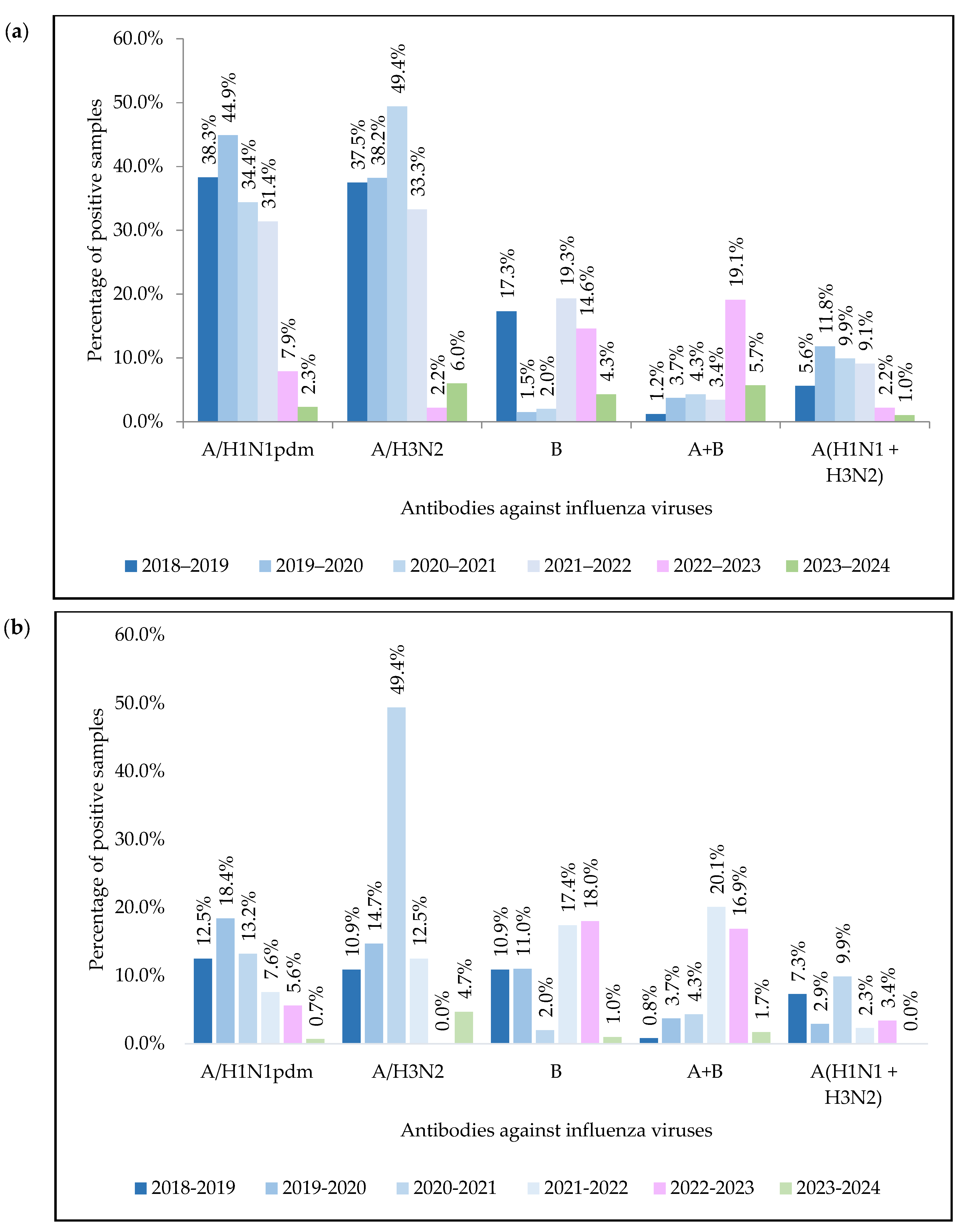
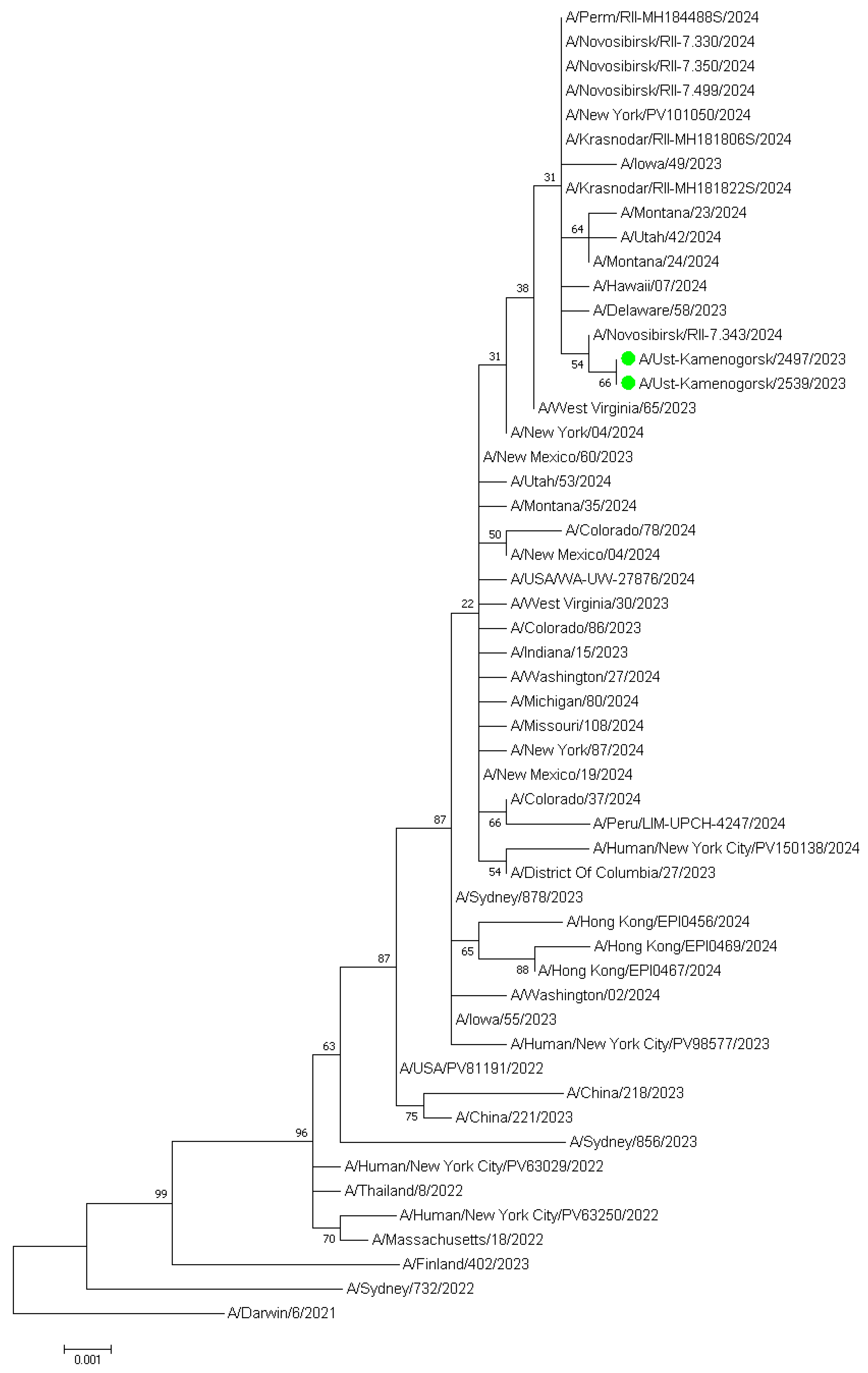
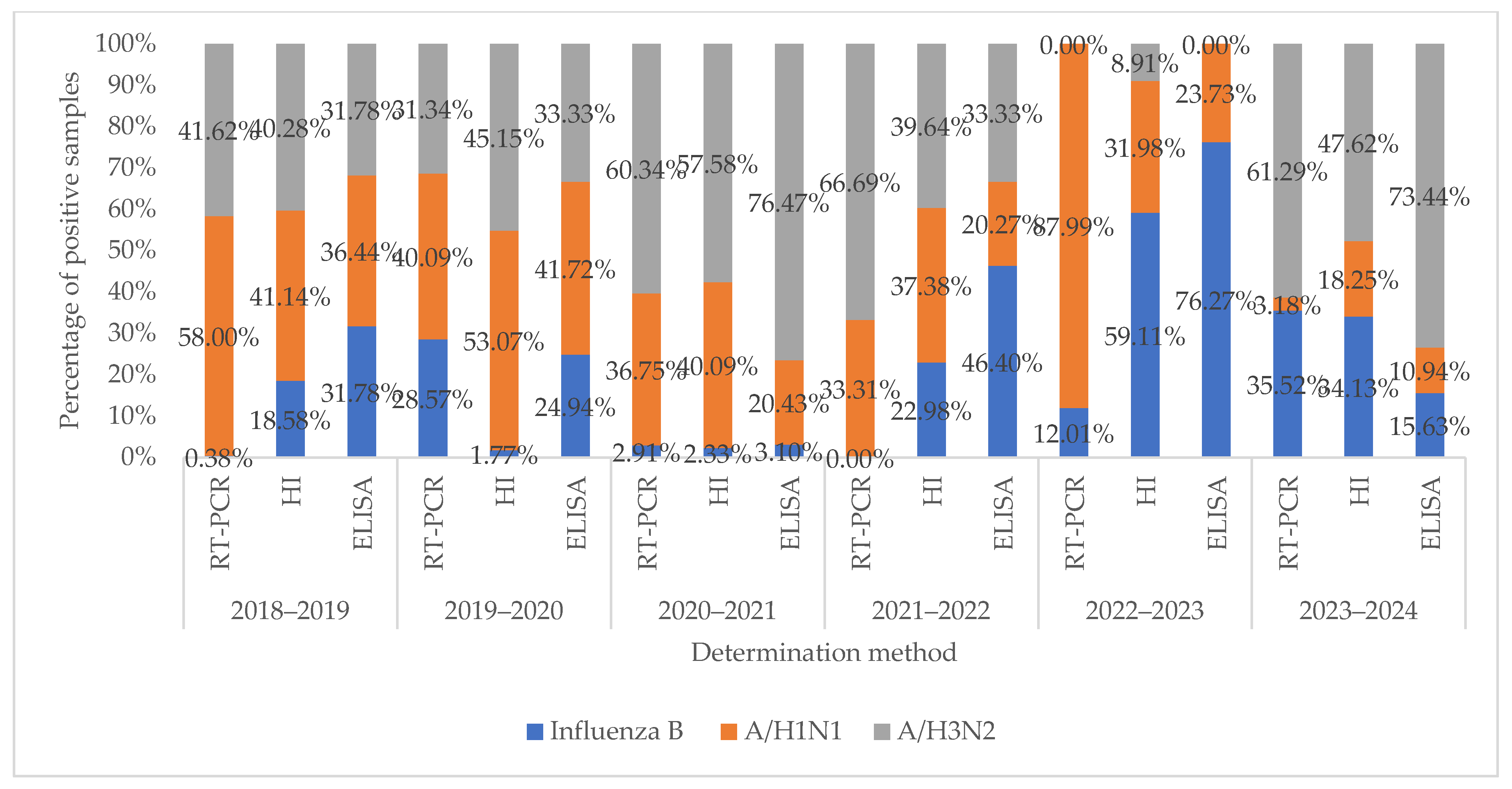
| Virus | Number of PCR-Positive Virus Samples by Year | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 2017–2018 | 2018–2019 | 2019–2020 | 2020–2021 | 2021–2022 | 2022–2023 | 2023–2024 | Total | p-Value | |
| Number of tested samples | 1602 | 1309 | 806 | 581 | 414 | 469 | 318 | 5499 | 0.0591 |
| Adenovirus (hAdv) | 14 (0.87) 1 | 13 (0.99) | 1 (0.12) | 1 (0.17) | 1 (0.24) | 2 (0.43) | 1 (0.31) | 33 (0.60) | 0.0809 |
| Bocavirus (hBov) | 2 (0.12) | 1 (0.08) | 2 (0.25) | 2 (0.34) | 2 (0.48) | 1 (0.21) | 1 (0.31) | 11 (0.20) | 0.0543 |
| Coronavirus (hCov) | 10 (0.62) | 8 (0.61) | 4 (0.50) | 0 | 0 | 2 (0.43) | 1 (0.31) | 25 (0.45) | 0.0742 |
| Metapneumovirus (hMpv) | 11 (0.69) | 10 (0.76) | 1 (0.12) | 0 | 0 | 1 (0.21) | 0 | 23 (0.42) | 0.0923 |
| Parainfluenza (hPiv) 1/3 | 10 (0.62) | 9 (0.69) | 2 (0.25) | 2 (0.34) | 2 (0.48) | 2 (0.43) | 2 (0.63) | 29 (0.53) | 0.0660 |
| Parainfluenza (hPiv) 2/4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Respiratory syncytial virus (hRSv) | 201 (12.55) | 214 (16.35) | 22 (2.73) | 14 (2.41) | 13 (3.14) | 15 (3.20) | 10 (3.14) | 489 (8.89) | 0.0842 |
| Rhinovirus (hRv) | 68 (4.24) | 41 (3.13) | 7 (0.87) | 7 (1.20) | 7 (1.69) | 8 (1.71) | 4 (1.26) | 142 (2.58) | 0.0782 |
| ARVI in total | 316 (19.73) | 296 (22.61) | 39 (4.84) | 26 (4.48) | 25 (6.04) | 31 (6.61) | 19 (5.97) | 752 (13.68) | 0.0804 |
| Influenza A non-detected | 216 (13.48) | 15 (1.15) | 13 (1.61) | 0 | 0 | 70 (14.93) | 26 (8.18) | 340 (6.18) | 0.0885 |
| Influenza B | 0 (0.00) | 2 (0.15) | 10 (1.24) | 2 (0.34) | 0 | 3 (0.64) | 11 (3.46) | 28 (0.51) | 0.0753 |
| A(H1N1)pdm09 | 213 (13.30) | 301 (22.99) | 14 (1.74) | 25 (4.30) | 22 (5.31) | 22 (4.69) | 1 (0.31) | 598 (10.87) | 0.0871 |
| A(H3N2) | 0 (0.00) | 216 (16.50) | 11 (1.36) | 41 (7.06) | 44 (10.63) | 0 | 19 (5.97) | 331 (6.02) | 0.0983 |
| Influenza in total | 429 (26.78) | 534 (40.79) | 48 (5.96) | 68 (11.70) | 66 (15.94) | 95 (20.26) | 57 (17.92) | 1297 (23.59) | 0.0738 |
| A total of positive samples | 745 (46.50) | 830 (63.41) | 87 (10.79) | 94 (16.18) | 91 (21.98) | 126 (26.87) | 76 (23.90) | 2049 (37.26) | 0.0729 |
| Age Group (Years) | Sample Size | Numbers of Participants That Are Immune to Influenza Viruses | |||||||
|---|---|---|---|---|---|---|---|---|---|
| A(H1N1)pdm | A(H3N2) | Type B | Total | ||||||
| HI | ELISA | HI | ELISA | HI | ELISA | HI | ELISA | ||
| 0–4 | 64 (4.21) 1 | 17 (26.56) | 11 (17.19) | 33 (51.56) | 19 (29.69) | 0 | 0 | 50 (78.13) | 30 (46.88) |
| 5–9 | 29 (1.91) | 5 (17.24) | 4 (13.79) | 5 (17.24) | 4 (13.79) | 5 (17.24) | 8 (27.59) | 15 (51.72) | 16 (55.17) |
| 10–17 | 36 (2.37) | 19 (52.78) | 10 (27.78) | 12 (33.33) | 7 (19.44) | 8 (22.22) | 7 (19.44) | 39 (108.33) | 24 (66.67) |
| 18–30 | 175 (11.51) | 87 (49.71) | 52 (29.71) | 50 (28.57) | 28 (16.00) | 34 (19.43) | 47 (26.86) | 171 (97.71) | 127 (72.57) |
| 30–65 | 627 (41.22) | 234 (37.32) | 138 (22.01) | 320 (51.04) | 176 (28.07) | 126 (20.10) | 161 (25.68) | 680 (108.45) | 475 (75.76) |
| ≥65 | 590 (38.79) | 219 (37.12) | 130 (22.03) | 215 (36.44) | 124 (21.02) | 55 (9.32) | 71 (12.03) | 489 (82.88) | 325 (55.08) |
| Total | 1521 | 581 (38.20) | 345 (22.68) | 635 (41.75) | 358 (23.54) | 228 (14.99) | 294 (19.33) | 1444 (94.94) | 997 (65.55) |
| p-value | 0.0779 | 0.0863 | 0.0812 | 0.0830 | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Glebova, T.; Klivleyeva, N.; Baimukhametova, A.; Lukmanova, G.; Saktaganov, N.; Ongarbayeva, N.; Baimakhanova, B.; Kassymova, G.; Sagatova, M.; Rachimbayeva, A.; et al. Acute Respiratory and Influenza Viruses Circulating in Kazakhstan During 2018–2024. Pathogens 2025, 14, 493. https://doi.org/10.3390/pathogens14050493
Glebova T, Klivleyeva N, Baimukhametova A, Lukmanova G, Saktaganov N, Ongarbayeva N, Baimakhanova B, Kassymova G, Sagatova M, Rachimbayeva A, et al. Acute Respiratory and Influenza Viruses Circulating in Kazakhstan During 2018–2024. Pathogens. 2025; 14(5):493. https://doi.org/10.3390/pathogens14050493
Chicago/Turabian StyleGlebova, Tatyana, Nailya Klivleyeva, Assem Baimukhametova, Galina Lukmanova, Nurbol Saktaganov, Nuray Ongarbayeva, Baiken Baimakhanova, Gulmira Kassymova, Madisha Sagatova, Almagul Rachimbayeva, and et al. 2025. "Acute Respiratory and Influenza Viruses Circulating in Kazakhstan During 2018–2024" Pathogens 14, no. 5: 493. https://doi.org/10.3390/pathogens14050493
APA StyleGlebova, T., Klivleyeva, N., Baimukhametova, A., Lukmanova, G., Saktaganov, N., Ongarbayeva, N., Baimakhanova, B., Kassymova, G., Sagatova, M., Rachimbayeva, A., Zhanuzakova, N., Naidenova, T., Rakhmonova, N., & Webby, R. (2025). Acute Respiratory and Influenza Viruses Circulating in Kazakhstan During 2018–2024. Pathogens, 14(5), 493. https://doi.org/10.3390/pathogens14050493






