Reassortment Dynamics: Phylogeography and Evolution of H4N9 Influenza Viruses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sequence Preprocessing
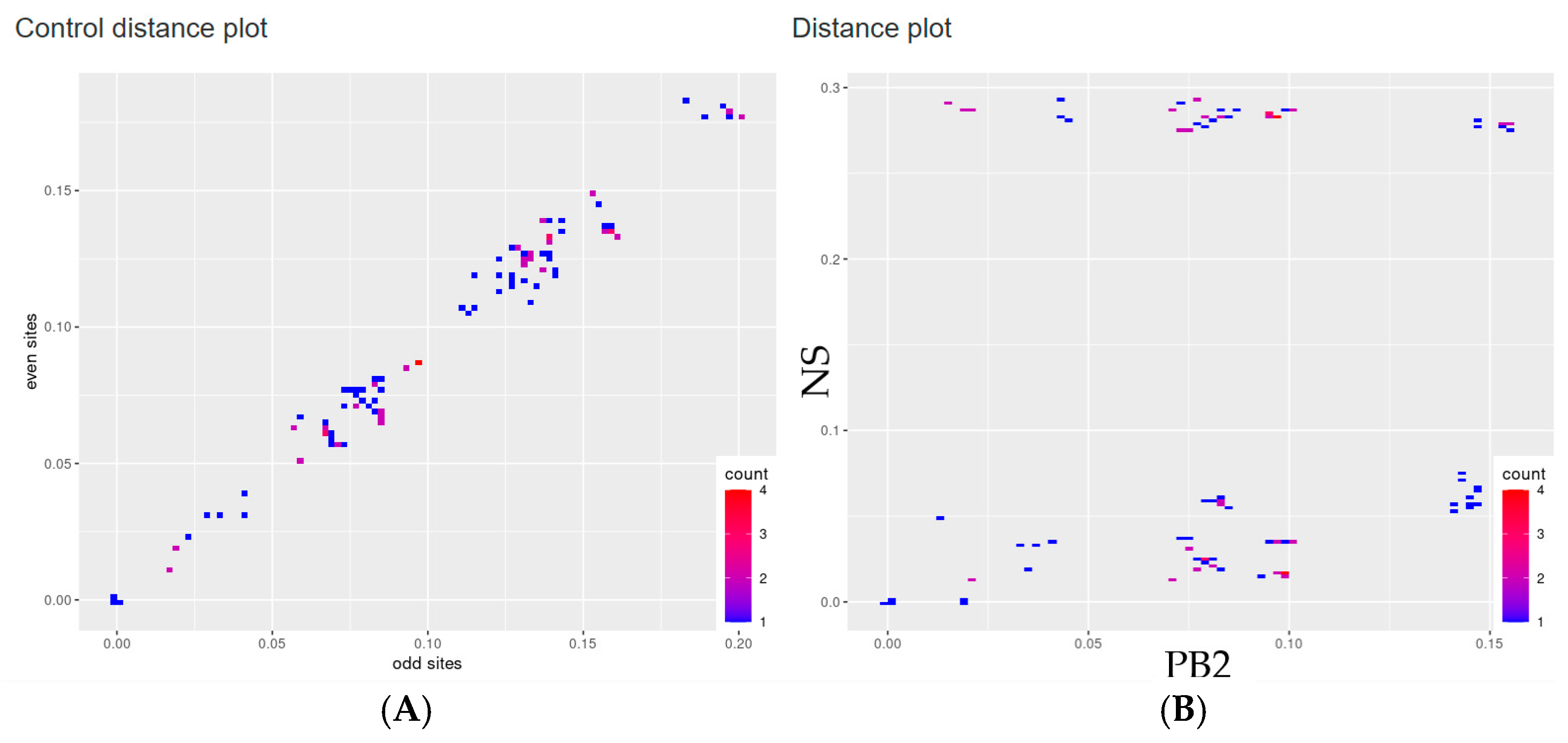
2.2. Recombination and Reassortment Analysis
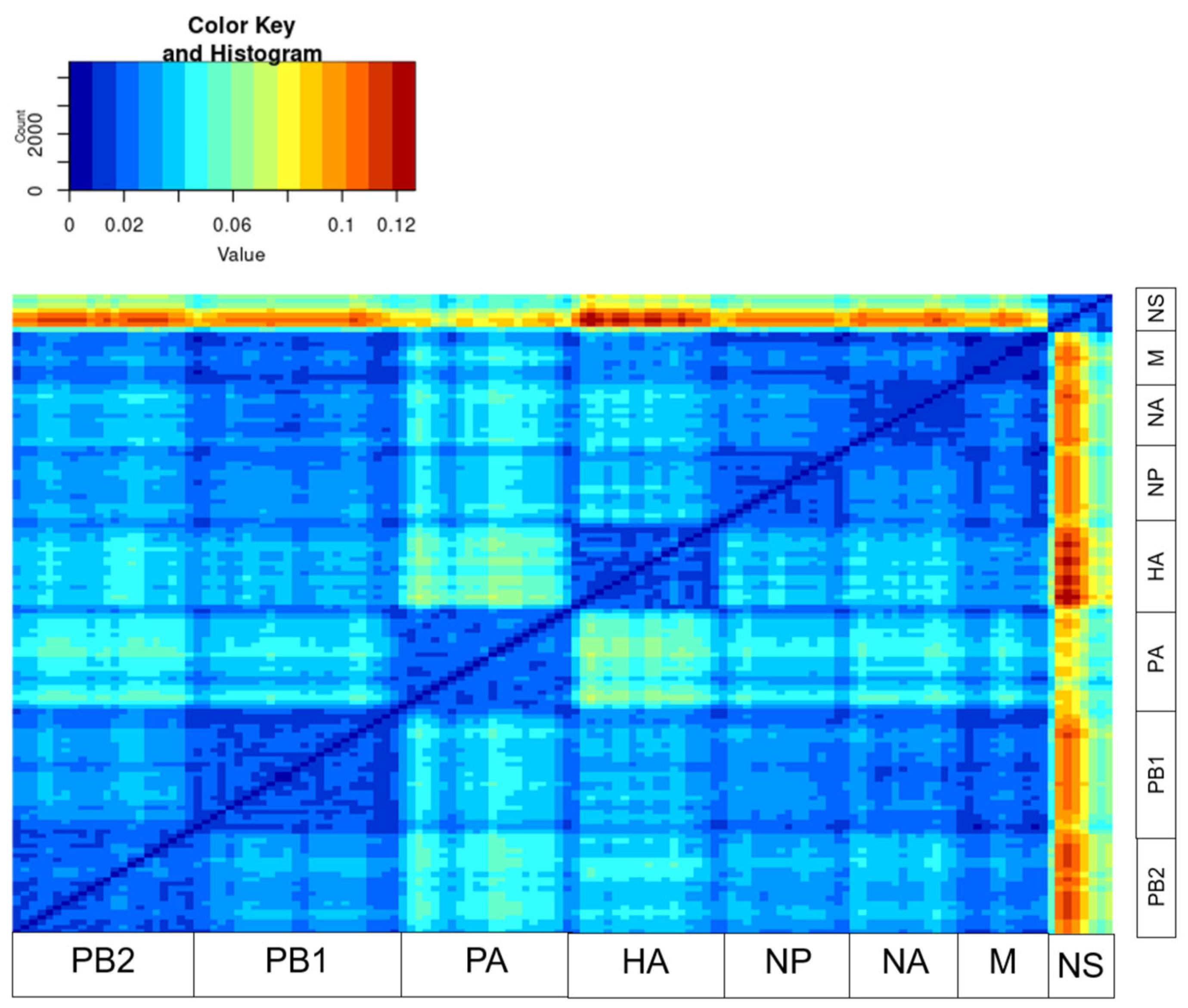
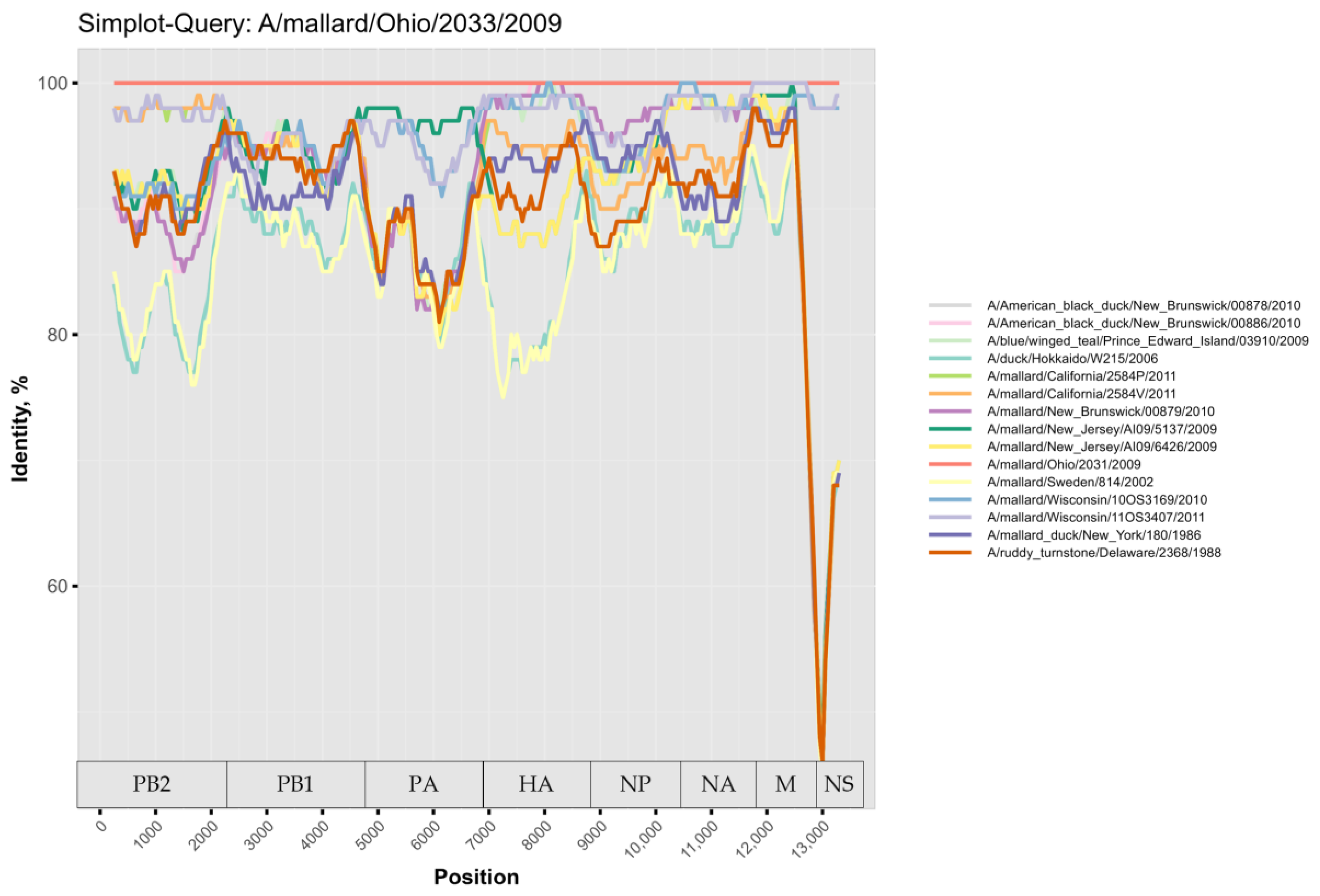
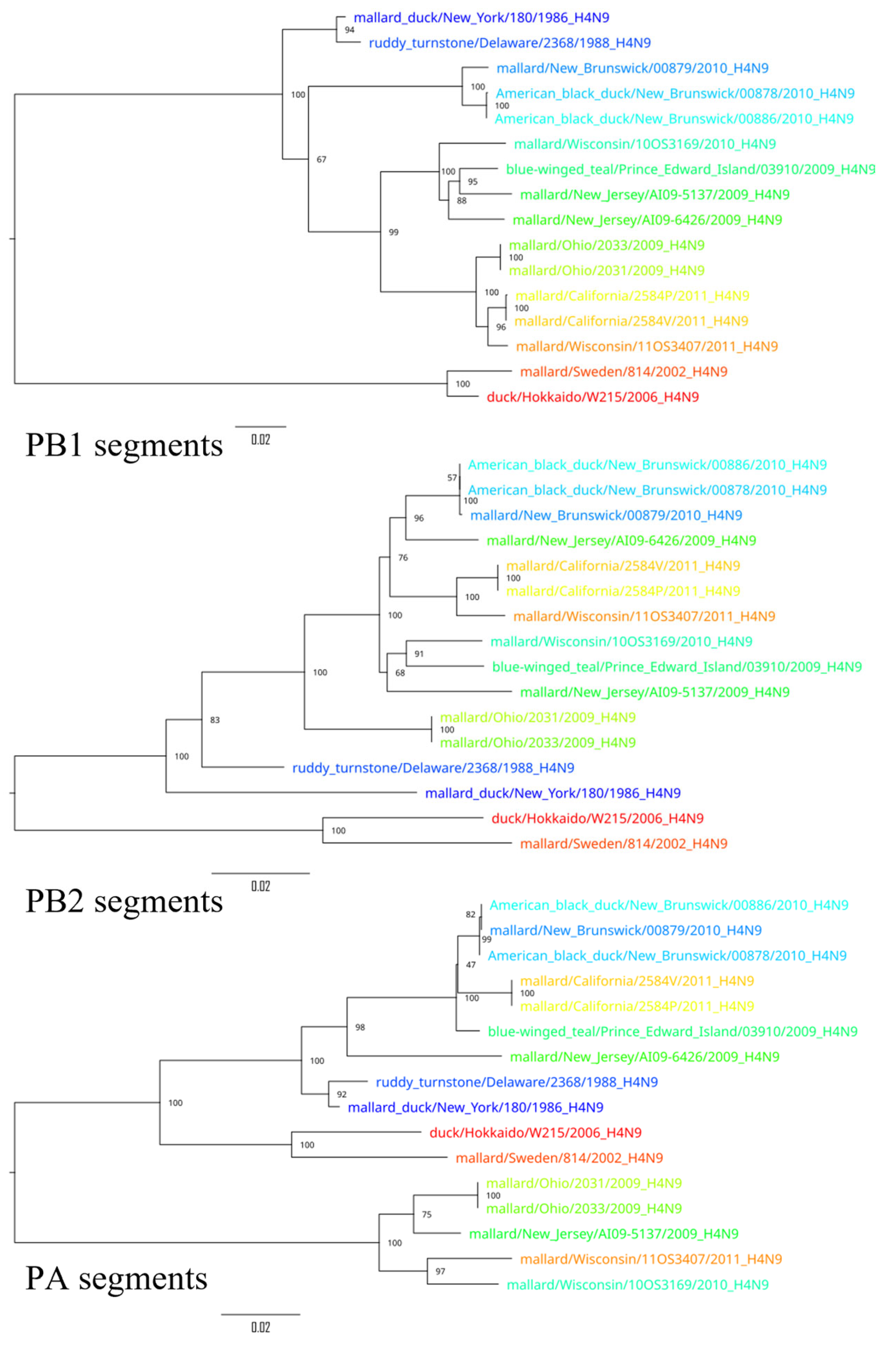
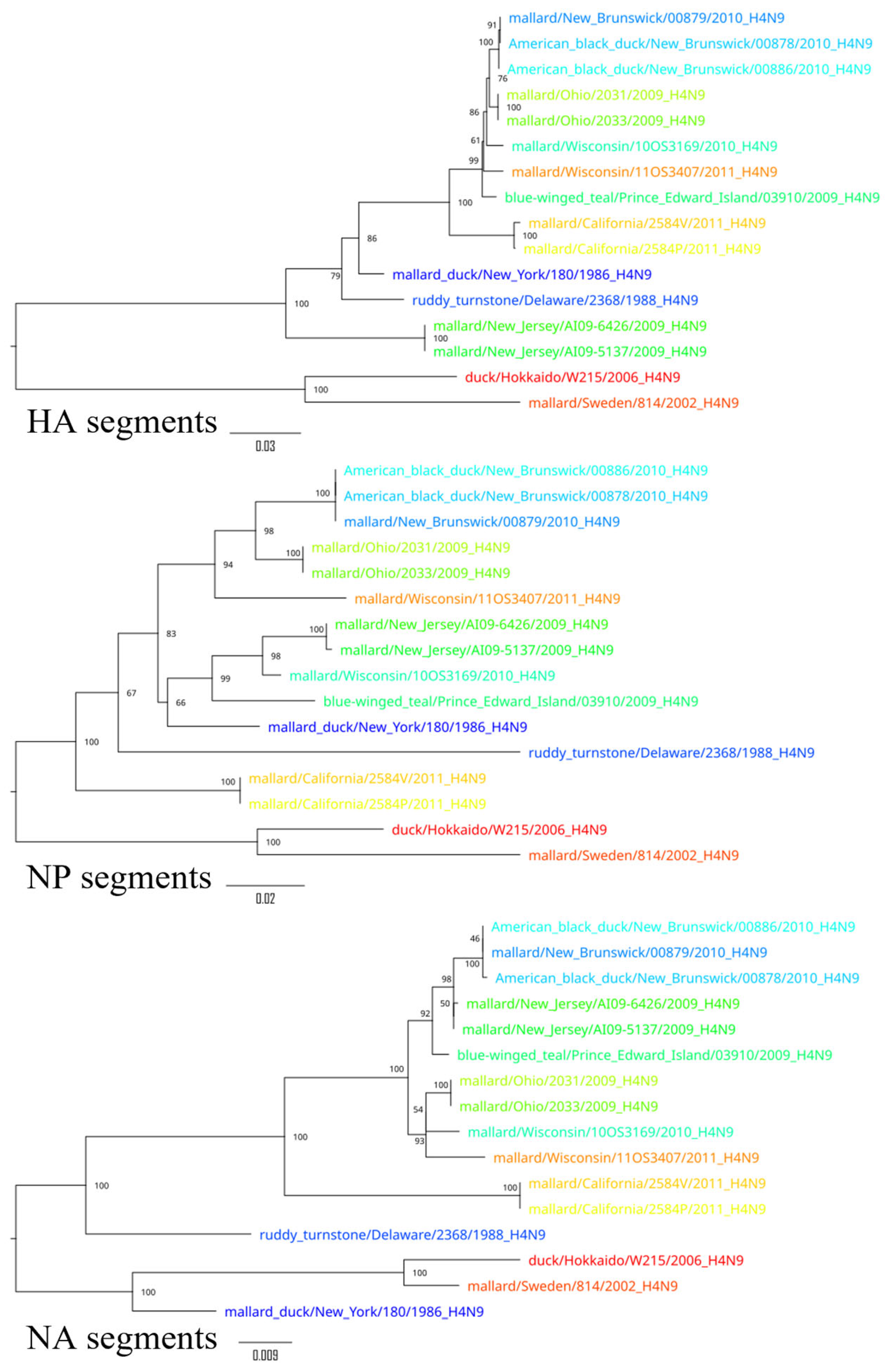
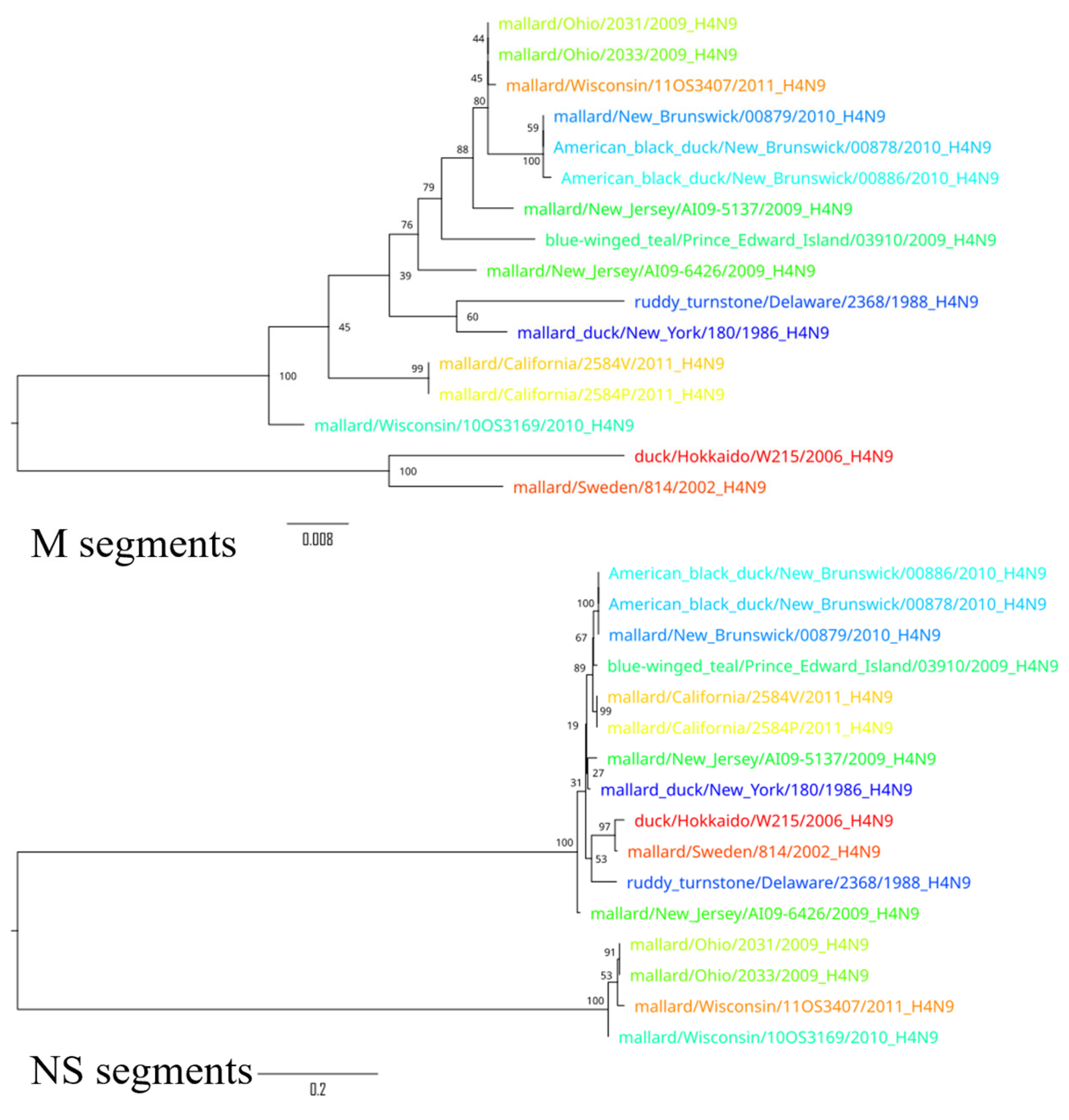
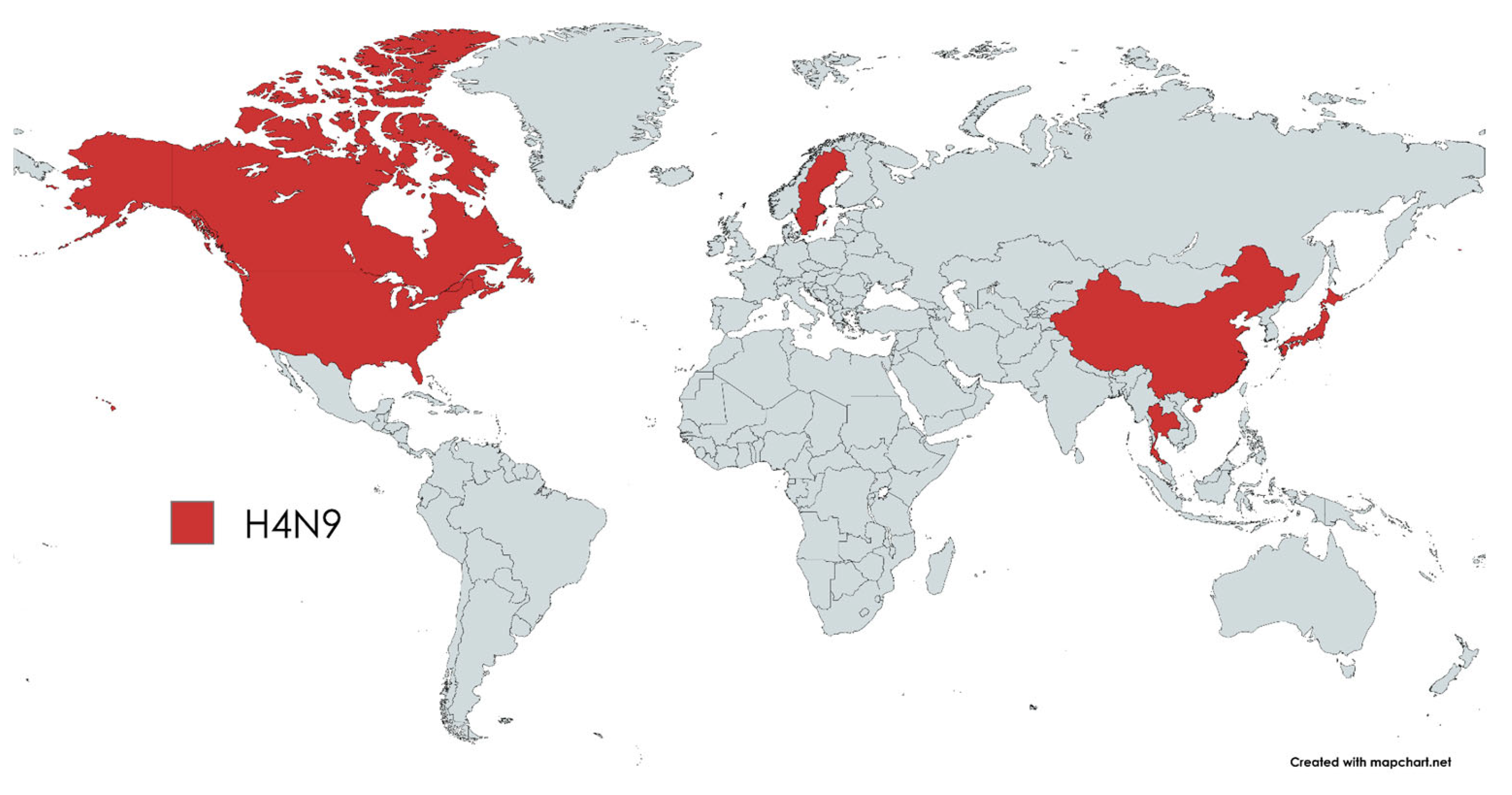
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| LPAI | Low-pathogenic influenza A |
| IAVs | Influenza A viruses |
| LBM | Live bird market |
| NCBI | National Center for Biotechnology Information |
| VSC | Viral Segment Concatenator |
| PDCP | Pairwise distance correspondence plot |
| PDDM | Pairwise distance deviation matrix |
References
- Ganti, K.; Bagga, A.; Carnaccini, S.; Ferreri, L.M.; Geiger, G.; Caceres, C.J.; Seibert, B.; Li, Y.; Wang, L.; Kwon, T.; et al. Influenza A virus reassortment in mammals gives rise to genetically distinct within-host subpopulations. Nat. Commun. 2022, 13, 6846. [Google Scholar] [CrossRef] [PubMed]
- Monto, A.S.; Fukuda, K. Lessons from influenza pandemics of the last 100 years. Clin. Infect. Dis. 2020, 70, 951–957. [Google Scholar] [CrossRef] [PubMed]
- Taubenberger, J.K.; Morens, D.M. 1918 Influenza: The mother of all pandemics. Rev. Biomed. 2006, 17, 69–79. [Google Scholar] [CrossRef]
- Viboud, C.; Miller, M.; Olson, D.R.; Osterholm, M.; Simonsen, L. Preliminary Estimates of Mortality and Years of Life Lost Associated with the 2009 A/H1N1 Pandemic in the US and Comparison with Past Influenza Seasons. PLoS Curr. 2010, 2, RRN1153. [Google Scholar] [CrossRef]
- Nuwarda, R.F.; Alharbi, A.A.; Kayser, V. An Overview of Influenza Viruses and Vaccines. Vaccines 2021, 9, 1032. [Google Scholar] [CrossRef]
- McDonald, S.M.; Nelson, M.I.; Turner, P.E.; Patton, J.T. Reassortment in segmented RNA viruses: Mechanisms and outcomes. Nat. Rev. Microbiol. 2016, 14, 448–460. [Google Scholar] [CrossRef]
- Steel, J.; Lowen, A.C. Influenza a Virus Reassortment; Current Topics in Microbiology and Immunology; Springer: Cham, Switzerland, 2014; Volume 385, pp. 377–401. [Google Scholar] [CrossRef]
- Mena, I.; Nelson, M.I.; Quezada-Monroy, F.; Jayeeta Dutta, J.; Cortes-Fernández, R.; Lara-Puente, J.H.; Castro-Peralta, F.; Cunha, L.F.; Trovão, N.S.; Lozano-Dubernard, B.; et al. Origins of the 2009 H1N1 influenza pandemic in swine in Mexico. eLife 2016, 5, e16777. [Google Scholar] [CrossRef]
- Krug, R.M.; Yuan, W.; Noah, D.L.; Latham, A.G. Intracellular warfare between human influenza viruses and human cells: The roles of the viral NS1 protein. Virology 2003, 309, 181–189. [Google Scholar] [CrossRef]
- Fereidouni, S.; Starick, E.; Karamendin, K.; Di Genova, C.; Scott, S.D.; Khan, Y.; Harder, T.; Kydyrmanov, A. Genetic characterization of a new candidate hemagglutinin subtype of influenza A viruses. Emerg. Microbes Infect. 2023, 12, 2225645. [Google Scholar] [CrossRef]
- Cumulative Number of Confirmed Human Cases of Avian Influenza A(H5N1) Reported to WHO, 2003–2014. Available online: https://cdn.who.int/media/docs/default-source/influenza/h5n1-human-case-cumulative-table/cumulative-number-of-confirmed-human-cases-for-avian-influenza-a(h5n1)-reported-to-who--2003-2023.pdf?sfvrsn=74bc4d1_1&download=true (accessed on 10 September 2024).
- Guarner, J.; Falcón-Escobedo, R. Comparison of the Pathology Caused by H1N1, H5N1, and H3N2 Influenza Viruses. Arch. Med Res. 2009, 40, 655–661. [Google Scholar] [CrossRef]
- Feoktistova, S.G.; Ivanova, A.O.; Degtyarev, E.P.; Smirnova, D.I.; Volchkov, P.Y.; Deviatkin, A.A. Phylogenetic Insights into H7Nx Influenza Viruses: Uncovering Reassortment Patterns and Geographic Variability. Viruses 2024, 16, 1656. [Google Scholar] [CrossRef] [PubMed]
- Webster, R.G.; Bean, W.J.; Gorman, O.T.; Chambers, T.M.; Kawaoka, Y. Evolution and ecology of influenza A viruses. Microbiol. Rev. 1992, 56, 152–179. [Google Scholar] [CrossRef] [PubMed]
- Ramey, A.M.; Reeves, A.B.; Drexler, J.Z.; Ackerman, J.T.; De La Cruz, S.; Lang, A.S.; Leyson, C.; Link, P.; Prosser, D.J.; Robertson, G.J.; et al. Influenza A viruses remain infectious for more than seven months in northern wetlands of North America: IAVs in wetlands. Proc. R. Soc. B Biol. Sci. 2020, 287, 20201680. [Google Scholar] [CrossRef] [PubMed]
- Poulson, R.L.; Reeves, A.B.; Ahlstrom, C.A.; Scott, L.C.; Hubbard, L.E.; Fojtik, A.; Carter, D.L.; Stallknecht, D.E.; Ramey, A.M. Infectivity of Wild-Bird Origin Influenza A Viruses in Minnesota Wetlands across Seasons. Pathogens 2024, 13, 406. [Google Scholar] [CrossRef]
- Liu, J.; Liang, Z.; Sun, W.; Hua, W.; Huang, S.; Wen, F. The H4 subtype of avian influenza virus: A review of its historical evolution, global distribution, adaptive mutations and receptor binding properties. Poult. Sci. 2024, 103, 103913. [Google Scholar] [CrossRef]
- Wisedchanwet, T.; Wongphatcharachai, M.; Boonyapisitsopa, S.; Bunpapong, N.; Kitikoon, P.; Amonsin, A. Genetic characterization of avian influenza subtype H4N6 and H4N9 from live bird market, Thailand. Virol. J. 2011, 8, 131. [Google Scholar] [CrossRef]
- Li, C.; Chen, H. Enhancement of Influenza Virus Transmission by Gene Reassortment; Current Topics in Microbiology and Immunology; Springer: Cham, Switzerland, 2014; Volume 385, pp. 185–204. [Google Scholar] [CrossRef]
- Reeth, K.V.; Vincent, A.L. Influenza Viruses. In Diseases of Swine; Wiley Online Library: Hoboken, NJ, USA, 2019; pp. 576–593. [Google Scholar] [CrossRef]
- Morales, A.C., Jr.; Hilt, D.A.; Williams, S.M.; Pantin-Jackwood, M.J.; Suarez, D.; Spackman, E.; Stallknecht, D.E.; Jackwood, M.W. Biologic Characterization of H4, H6, and H9 Type Low Pathogenicity Avian Influenza Viruses from Wild Birds in Chickens and Turkeys. Avian Dis. 2009, 53, 552–562. [Google Scholar] [CrossRef]
- Ivanova, A.; Volchkov, P.; Deviatkin, A. Concatenation of segmented viral genomes for reassortment analysis. bioRxiv 2024. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Vakulenko, Y.; Deviatkin, A.; Drexler, J.F.; Lukashev, A. Modular Evolution of Coronavirus Genomes. Viruses 2021, 13, 1270. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Adisasmito, W.B.; Almuhairi, S.; Behravesh, C.B.; Bilivogui, P.; Bukachi, S.A.; Casas, N.; Becerra, N.C.; Charron, D.F.; Chaudhary, A.; Zanelle, J.R.C.; et al. One Health: A new definition for a sustainable and healthy future. PLOS Pathog. 2022, 18, e1010537. [Google Scholar] [CrossRef]
- Monne, I.; Fusaro, A.; Nelson, M.I.; Bonfanti, L.; Mulatti, P.; Hughes, J.; Murcia, P.R.; Schivo, A.; Valastro, V.; Moreno, A.; et al. Emergence of a Highly Pathogenic Avian Influenza Virus from a Low-Pathogenic Progenitor. J. Virol. 2014, 88, 4375–4388. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Cui, P.; Song, Y.; Qi, Y.; Li, X.; Qi, W.; Xu, C.; Jiao, P.; Liao, M. PB2 segment promotes high-pathogenicity of H5N1 avian influenza viruses in mice. Front. Microbiol. 2015, 6, 73. [Google Scholar] [CrossRef]
- Hohensee, L.; Scheibner, D.; Schäfer, A.; Shelton, H.; Mettenleiter, T.C.; Breithaupt, A.; Dorhoi, A.; Abdelwhab, E.M.; Blohm, U. The role of PB1-F2 in adaptation of high pathogenicity avian influenza virus H7N7 in chickens. Veter. Res. 2024, 55, 5. [Google Scholar] [CrossRef]
- Gao, H.; Sun, Y.; Hu, J.; Qi, L.; Wang, J.; Xiong, X.; Wang, Y.; He, Q.; Lin, Y.; Kong, W.; et al. The contribution of PA-X to the virulence of pandemic 2009 H1N1 and highly pathogenic H5N1 avian influenza viruses. Sci. Rep. 2015, 5, 8262. [Google Scholar] [CrossRef]
- Wasilenko, J.L.; Sarmento, L.; Pantin-Jackwood, M.J. A single substitution in amino acid 184 of the NP protein alters the replication and pathogenicity of H5N1 avian influenza viruses in chickens. Arch. Virol. 2009, 154, 969–979. [Google Scholar] [CrossRef]
- Calderon, B.M.; Danzy, S.; Delima, G.K.; Jacobs, N.T.; Ganti, K.; Hockman, M.R.; Conn, G.L.; Lowen, A.C.; Steel, J. Dysregulation ofMsegment gene expression contributes to influenza A virus host restriction. PLoS Pathog. 2019, 15, e1007892. [Google Scholar] [CrossRef]
- Petersen, H.; Wang, Z.; Lenz, E.; Pleschka, S.; Rautenschlein, S. Reassortment of NS Segments Modifies Highly Pathogenic Avian Influenza Virus Interaction with Avian Hosts and Host Cells. J. Virol. 2013, 87, 5362–5371. [Google Scholar] [CrossRef]
- Marangon, S.; Capua, I.; Rossi, E.; Ferre’, N.; Pozza, M.D.; Bonfanti, L.; Mannelli, A. The control of avian influenza in areas at risk: The Italian experience 1997–2003. In Avian Influenza Prevention and Control. Wageningen; Schrijver, R., Koch, G., Eds.; Springer: Dordrecht, The Netherlands, 2005; pp. 33–39. [Google Scholar] [CrossRef]
- Banks, J.; Speidel, E.S.; Moore, E.; Plowright, L.; Piccirillo, A.; Capua, I.; Cordioli, P.; Fioretti, A.; Alexander, D.J. Changes in the haemagglutinin and the neuraminidase genes prior to the emergence of highly pathogenic H7N1 avian influenza viruses in Italy. Arch. Virol. 2001, 146, 963–973. [Google Scholar] [CrossRef]
- Worobey, M.; Han, G.Z.; Rambaut, A. A synchronized global sweep of the internal genes of modern avian influenza virus. Nature 2014, 508, 254–257. [Google Scholar] [CrossRef] [PubMed]
- Dugan, V.G.; Chen, R.; Spiro, D.J.; Sengamalay, N.; Zaborsky, J.; Ghedin, E.; Nolting, J.; Swayne, D.E.; Runstadler, J.A.; Happ, G.M.; et al. The Evolutionary Genetics and Emergence of Avian Influenza Viruses in Wild Birds. PLoS Pathog. 2008, 4, e1000076. [Google Scholar] [CrossRef]
- Hao, W.; Wang, L.; Li, S. Roles of the non-structural proteins of influenza a virus. Pathogens 2020, 9, 812. [Google Scholar] [CrossRef] [PubMed]
- Abdelwhab, E.M.; Veits, J.; Mettenleiter, T.C. Avian influenza virus NS1. Virulence 2013, 4, 583–588. [Google Scholar] [CrossRef]
- Henderson, D.A. The eradication of smallpox–An overview of the past, present, and future. Vaccine 2011, 29, D7–D9. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bobrova, N.A.; Lisenenkova, E.D.; Avsievich, E.S.; Mityaeva, O.N.; Volchkov, P.Y.; Deviatkin, A.A. Reassortment Dynamics: Phylogeography and Evolution of H4N9 Influenza Viruses. Pathogens 2025, 14, 469. https://doi.org/10.3390/pathogens14050469
Bobrova NA, Lisenenkova ED, Avsievich ES, Mityaeva ON, Volchkov PY, Deviatkin AA. Reassortment Dynamics: Phylogeography and Evolution of H4N9 Influenza Viruses. Pathogens. 2025; 14(5):469. https://doi.org/10.3390/pathogens14050469
Chicago/Turabian StyleBobrova, Nataliia A., Ekaterina D. Lisenenkova, Ekaterina S. Avsievich, Olga N. Mityaeva, Pavel Yu Volchkov, and Andrey A. Deviatkin. 2025. "Reassortment Dynamics: Phylogeography and Evolution of H4N9 Influenza Viruses" Pathogens 14, no. 5: 469. https://doi.org/10.3390/pathogens14050469
APA StyleBobrova, N. A., Lisenenkova, E. D., Avsievich, E. S., Mityaeva, O. N., Volchkov, P. Y., & Deviatkin, A. A. (2025). Reassortment Dynamics: Phylogeography and Evolution of H4N9 Influenza Viruses. Pathogens, 14(5), 469. https://doi.org/10.3390/pathogens14050469





