Investigating Correlation Between Gut Microbiota and Rheumatoid Arthritis Subtypes by Mendelian Randomization
Abstract
1. Introduction
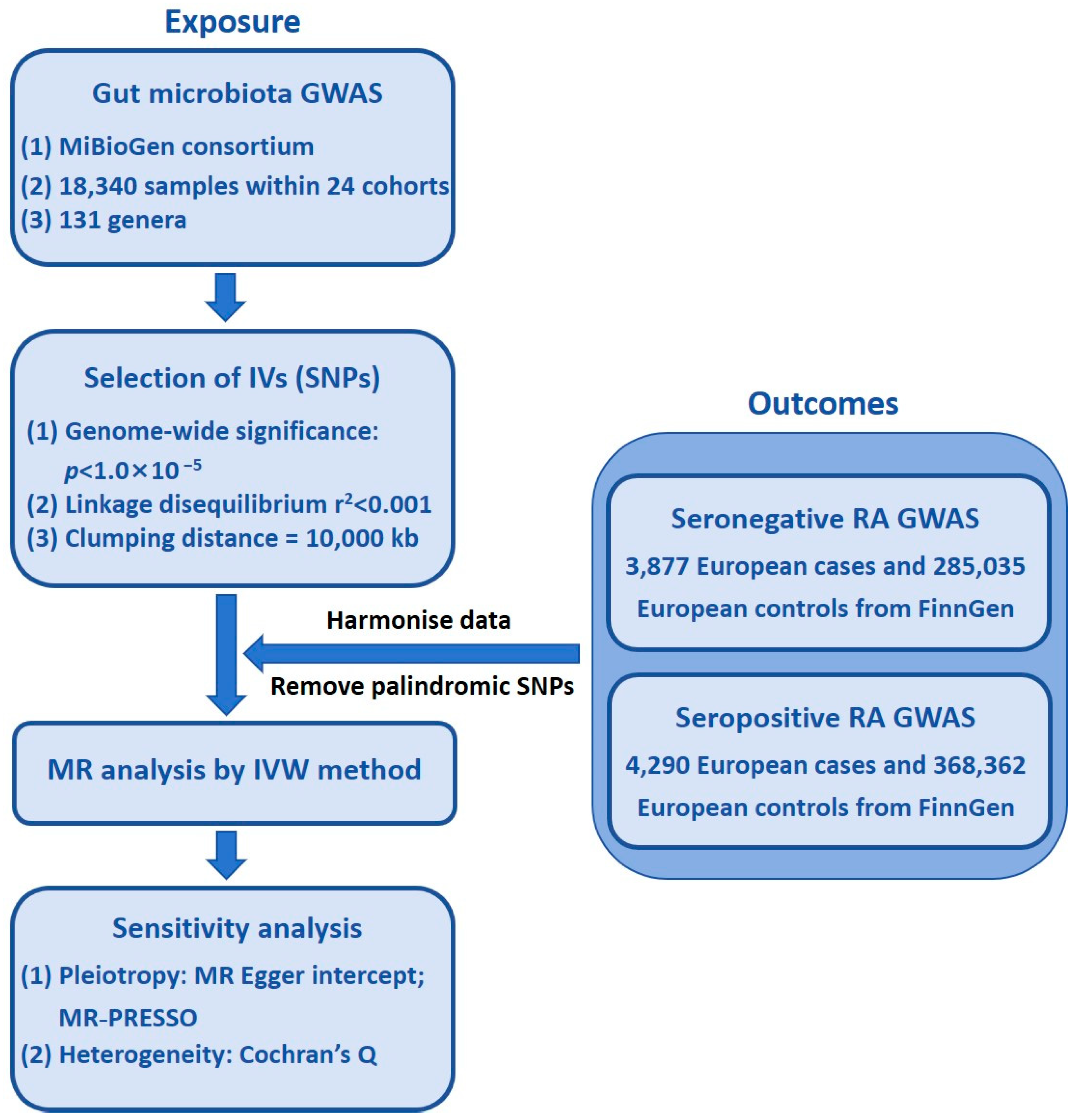
2. Materials and Methods
2.1. Study Design
- (1)
- The SNPs selected as IVs exhibit strong associations with gut microbiota;
- (2)
- The IVs are independent of confounders that could introduce bias;
- (3)
- The IVs influence the outcome solely through the exposure, ensuring no direct effects on RA beyond their association with GM.
2.2. Ethics Statement
2.3. Study Population and Data Sources
2.4. Exposure Data
2.5. Outcome Data
2.6. Selection of IVs
2.7. Statistical Analysis
- (a)
- Handling of continuous variables
- (b)
- MR models and SNP weighting
- IVW analysis (fixed or random effects, depending on heterogeneity): considered the primary MR estimate;
- Weighted median analysis: provides a consistent estimate even if up to 50% of the instruments are invalid;
- MR-Egger regression: used to assess pleiotropy and provide bias-corrected causal estimates;
- Simple mode and weighted mode analyses: offer additional robustness checks.
- (c)
- Handling of heterogeneity and pleiotropy
- (d)
- Sensitivity analyses
- MR-Egger intercept test: used to detect potential directional pleiotropy [24];
- Leave-one-out analysis: performed to evaluate whether any single SNP disproportionately influenced the results.
3. Study Results
3.1. IVs in GM Taxa
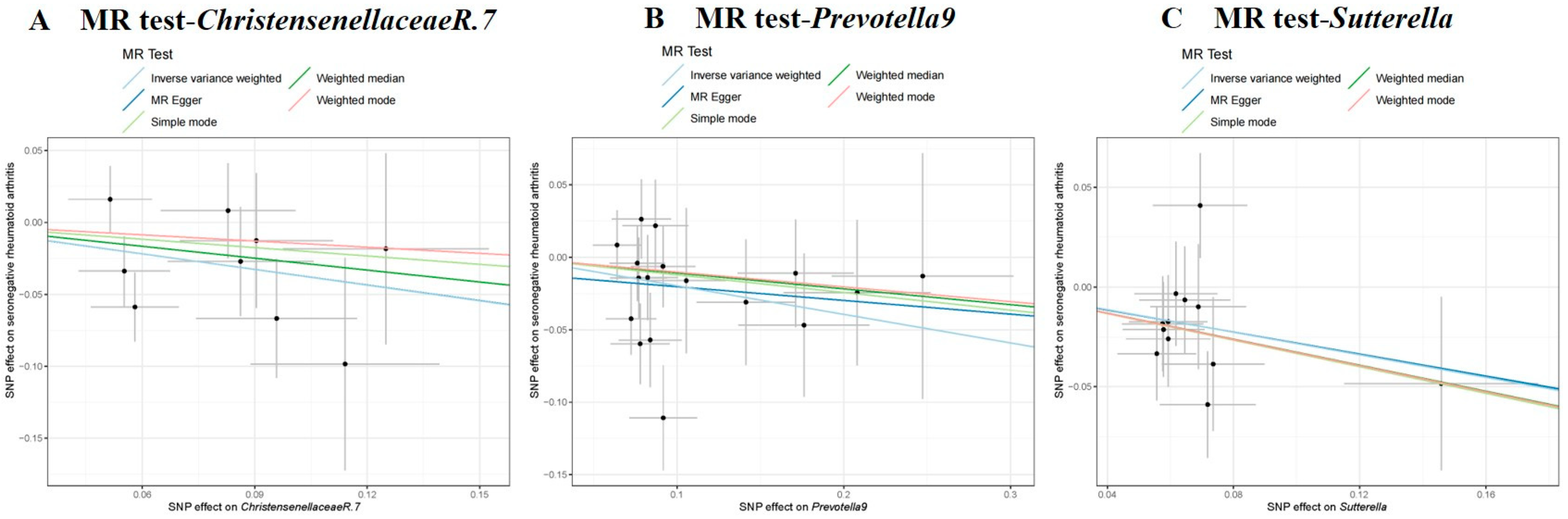
3.2. Causal Effect of GM on Seronegative RA
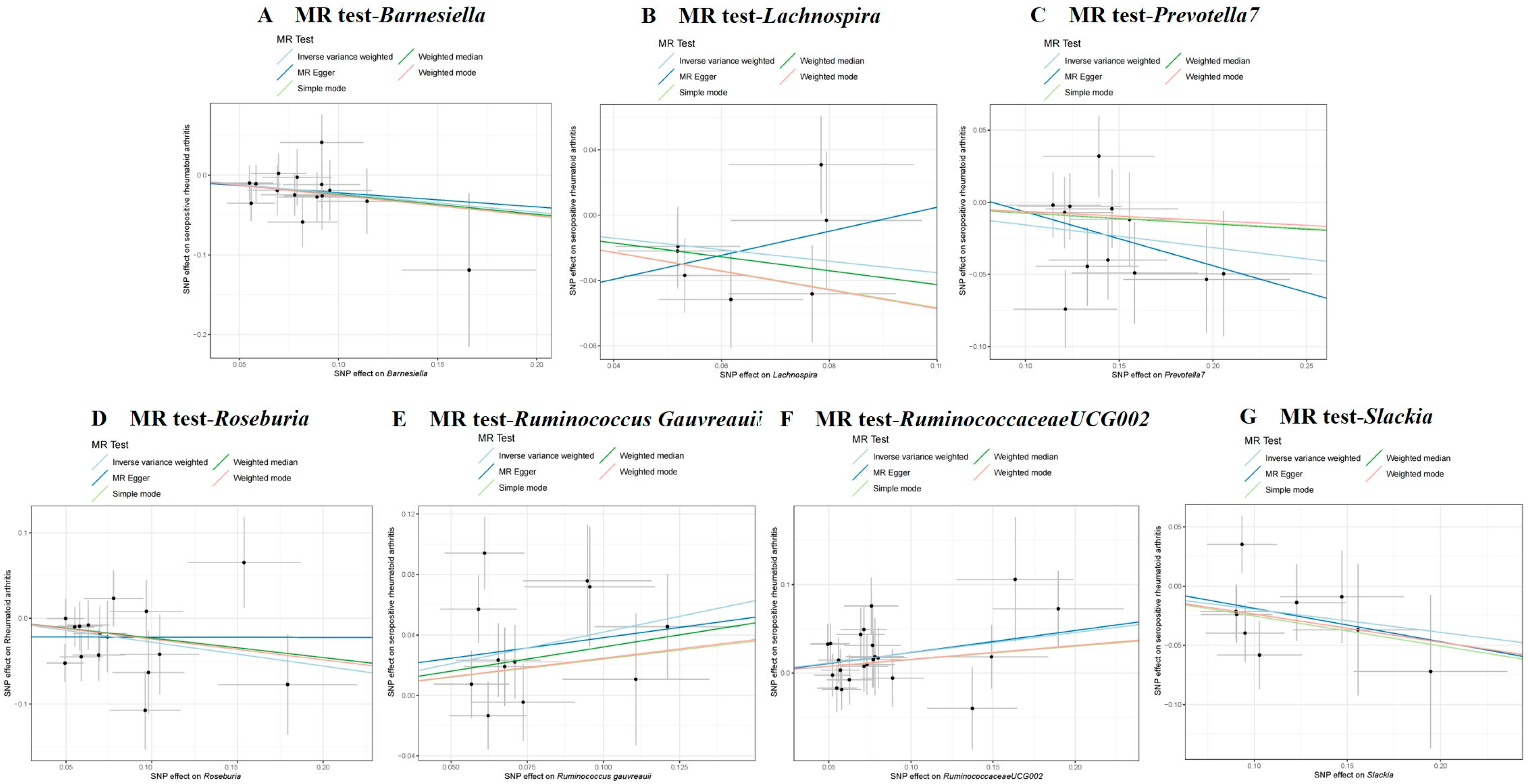
3.3. Causal Effect of GM on Seropositive RA
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Smolen, J.S.; Aletaha, D.; Barton, A.; Burmester, G.R.; Emery, P.; Firestein, G.S.; Kavanaugh, A.; McInnes, I.B.; Solomon, D.H.; Strand, V.; et al. Rheumatoid arthritis. Nat. Rev. Dis. Primers. 2018, 4, 18001. [Google Scholar] [CrossRef] [PubMed]
- Malmström, V.; Catrina, A.I.; Klareskog, L. The immunopathogenesis of seropositive rheumatoid arthritis: From triggering to targeting. Nat. Rev. Immunol. 2017, 17, 60–75. [Google Scholar] [CrossRef] [PubMed]
- Aletaha, D.; Alasti, F.; Smolen, J.S. Rheumatoid factor, not antibodies against citrullinated proteins, is associated with baseline disease activity in rheumatoid arthritis clinical trials. Arthritis Res. Ther. 2015, 17, 229. [Google Scholar] [CrossRef] [PubMed]
- Van Gaalen, F.A.; van Aken, J.; Huizinga, T.W.; Schreuder, G.M.T.; Breedveld, F.C.; Zanelli, E.; Van Venrooij, W.J.; Verweij, C.L.; Toes, R.E.; De Vries, R.R. Association between HLA class II genes and autoantibodies to cyclic citrullinated peptides (CCPs) influences the severity of rheumatoid arthritis. Arthritis Rheum. 2004, 50, 2113–2121. [Google Scholar] [CrossRef]
- Gonzalez, A.; Icen, M.; Kremers, H.M.; Crowson, C.S.; Davis, J.M.; Therneau, T.M.; Roger, V.L.; Gabriel, S.E. Mortality trends in rheumatoid arthritis: The role of rheumatoid factor. J. Rheumatol. 2008, 35, 1009–1014. [Google Scholar]
- Dong, X.; Zheng, Z.; Zhai, Y.; Zheng, Y.; Ding, J.; Jiang, J.; Zhu, P. ACPA mediates the interplay between innate and adaptive immunity in rheumatoid arthritis. Autoimmun. Rev. 2018, 17, 845–853. [Google Scholar] [CrossRef]
- De Stefano, L.; D’Onofrio, B.; Gandolfo, S.; Bozzalla Cassione, E.; Mauro, D.; Manzo, A.; Ciccia, F.; Bugatti, S. Seronegative rheumatoid arthritis: One year in review 2023. Clin. Exp. Rheumatol. 2023, 41, 554–564. [Google Scholar] [CrossRef]
- O’Hara, A.M.; Shanahan, F. The gut flora as a forgotten organ. EMBO Rep. 2006, 7, 688–693. [Google Scholar] [CrossRef]
- Zaiss, M.M.; Joyce Wu, H.J.; Mauro, D.; Schett, G.; Ciccia, F. The gut-joint axis in rheumatoid arthritis. Nat. Rev. Rheumatol. 2021, 17, 224–237. [Google Scholar] [CrossRef]
- Holers, V.M.; Demoruelle, M.K.; Kuhn, K.A.; Buckner, J.H.; Robinson, W.H.; Okamoto, Y.; Norris, J.M.; Deane, K.D. Rheumatoid arthritis and the mucosal origins hypothesis: Protection turns to destruction. Nat. Rev. Rheumatol. 2018, 14, 542–557. [Google Scholar] [CrossRef]
- Wells, P.M.; Adebayo, A.S.; Bowyer, R.C.E.; Freidin, M.B.; Finckh, A.; Strowig, T.; Lesker, T.R.; Alpizar-Rodriguez, D.; Gilbert, B.; Kirkham, B.; et al. Associations between gut microbiota and genetic risk for rheumatoid arthritis in the absence of disease: A cross-sectional study. Lancet Rheumatol. 2020, 2, e418–e427. [Google Scholar] [CrossRef] [PubMed]
- Rogier, R.; Ederveen, T.H.A.; Boekhorst, J.; Wopereis, H.; Scher, J.U.; Manasson, J.; Frambach, S.J.; Knol, J.; Garssen, J.; van der Kraan, P.M.; et al. Aberrant intestinal microbiota due to IL-1 receptor antagonist deficiency promotes IL-17- and TLR4-dependent arthritis. Microbiome 2017, 5, 63. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, D.; Jia, H.; Feng, Q.; Wang, D.; Liang, D.; Wu, X.; Li, J.; Tang, L.; Li, Y.; et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat. Med. 2015, 21, 895–905. [Google Scholar] [CrossRef] [PubMed]
- Weith, M.; Beyer, A. The next step in Mendelian randomization. Elife 2023, 12, e86416. [Google Scholar] [CrossRef]
- Davey Smith, G.; Hemani, G. Mendelian randomization: Genetic anchors for causal inference in epidemiological studies. Hum. Mol. Genet. 2014, 23, R89–R98. [Google Scholar] [CrossRef]
- Wang, J.; Kurilshikov, A.; Radjabzadeh, D.; Turpin, W.; Croitoru, K.; Bonder, M.J.; Jackson, M.A.; Medina-Gomez, C.; Frost, F.; Homuth, G.; et al. Meta-analysis of human genome-microbiome association studies: The MiBioGen consortium initiative. Microbiome 2018, 6, 101. [Google Scholar] [CrossRef]
- Kurilshikov, A.; Medina-Gomez, C.; Bacigalupe, R.; Radjabzadeh, D.; Wang, J.; Demirkan, A.; Le Roy, C.I.; Raygoza Garay, J.A.; Finnicum, C.T.; Liu, X.; et al. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat. Genet. 2021, 53, 156–165. [Google Scholar] [CrossRef]
- Ji, X.; Yang, Q.; Zhu, X.L.; Xu, L.; Guo, J.Y.; Rong, Y.; Cai, Y.L. Association between gut microbiota and endometriosis: A two-sample Mendelian randomization study. Front. Microbiol. 2023, 14, 1188458. [Google Scholar] [CrossRef]
- Li, P.; Wang, H.; Guo, L.; Gou, X.; Chen, G.; Lin, D.; Fan, D.; Guo, X.; Liu, Z. Association between gut microbiota and preeclampsia-eclampsia: A two-sample Mendelian randomization study. BMC Med. 2022, 20, 443. [Google Scholar] [CrossRef]
- Sanna, S.; van Zuydam, N.R.; Mahajan, A.; Kurilshikov, A.; Vich Vila, A.; Võsa, U.; Mujagic, Z.; Masclee, A.A.; Jonkers, D.M.; Oosting, M.; et al. Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat. Genet. 2019, 51, 600–605. [Google Scholar] [CrossRef]
- Verbanck, M.; Chen, C.Y.; Neale, B.; Do, R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 2018, 50, 693–698. [Google Scholar] [CrossRef] [PubMed]
- Pierce, B.L.; Ahsan, H.; Vanderweele, T.J. Power and instrument strength requirements for Mendelian randomization studies using multiple genetic variants. Int. J. Epidemiol. 2011, 40, 740–752. [Google Scholar] [CrossRef] [PubMed]
- Burgess, S.; Dudbridge, F.; Thompson, S.G. Combining information on multiple instrumental variables in Mendelian randomization: Comparison of allele score and summarized data methods. Stat. Med. 2016, 35, 1880–1906. [Google Scholar] [CrossRef] [PubMed]
- Bowden, J.; Davey Smith, G.; Burgess, S. Mendelian randomization with invalid instruments: Effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 2015, 44, 512–525. [Google Scholar] [CrossRef]
- Tett, A.; Pasolli, E.; Masetti, G.; Ercolini, D.; Segata, N. Prevotella diversity, niches and interactions with the human host. Nat. Rev. Microbiol. 2021, 19, 585–599. [Google Scholar] [CrossRef]
- Alpizar-Rodriguez, D.; Lesker, T.R.; Gronow, A.; Gilbert, B.; Raemy, E.; Lamacchia, C.; Gabay, C.; Finckh, A.; Strowig, T. Prevotella copri in individuals at risk for rheumatoid arthritis. Ann. Rheum. Dis. 2019, 78, 590–593. [Google Scholar] [CrossRef]
- Kishikawa, T.; Maeda, Y.; Nii, T.; Motooka, D.; Matsumoto, Y.; Matsushita, M.; Matsuoka, H.; Yoshimura, M.; Kawada, S.; Teshigawara, S.; et al. Metagenome-wide association study of gut microbiome revealed novel aetiology of rheumatoid arthritis in the Japanese population. Ann. Rheum. Dis. 2020, 79, 103–111. [Google Scholar] [CrossRef]
- Lee, J.Y.; Mannaa, M.; Kim, Y.; Kim, G.T.; Seo, Y.S. Comparative analysis of fecal microbiota composition between Rheumatoid arthritis and osteoarthritis patients. Genes 2019, 10, 748. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, B.; Li, S.; Yang, L.; Zhu, D.; Wang, Y.; Wang, H.; Wang, T.; Shi, B.; Gai, Z.; et al. Detection and characterization of bacterial nucleic acids in culture-negative synovial tissue and fluid samples from rheumatoid arthritis or osteoarthritis patients. Sci. Rep. 2018, 8, 14305. [Google Scholar] [CrossRef]
- Maeda, Y.; Kurakawa, T.; Umemoto, E.; Motooka, D.; Ito, Y.; Gotoh, K.; Hirota, K.; Matsushita, M.; Furuta, Y.; Narazaki, M.; et al. Dysbiosis contributes to arthritis development via activation of autoreactive T cells in the intestine. Arthritis Rheumatol. 2016, 68, 2646–2661. [Google Scholar] [CrossRef]
- Pianta, A.; Arvikar, S.; Strle, K.; Drouin, E.E.; Wang, Q.; Costello, C.E.; Steere, A.C. Evidence of the immune relevance of Prevotella copri, a gut microbe, in patients with Rheumatoid arthritis. Arthritis Rheumatol. 2017, 69, 964–975. [Google Scholar] [CrossRef] [PubMed]
- Marietta, E.V.; Murray, J.A.; Luckey, D.H.; Jeraldo, P.R.; Lamba, A.; Patel, R.; Luthra, H.S.; Mangalam, A.; Taneja, V. Suppression of inflammatory arthritis by human gut-derived Prevotella histicola in humanized mice. Arthritis Rheumatol. 2016, 68, 2878–2888. [Google Scholar] [CrossRef]
- Sun, Y.; Chen, Q.; Lin, P.; Xu, R.; He, D.; Ji, W.; Bian, Y.; Shen, Y.; Li, Q.; Liu, C.; et al. Characteristics of gut microbiota in patients with Rheumatoid arthritis in Shanghai, China. Front. Cell Infect. Microbiol. 2019, 9, 369. [Google Scholar] [CrossRef] [PubMed]
- Yun, H.; Wang, X.; Wei, C.; Liu, Q.; Li, X.; Li, N.; Zhang, G.; Cui, D.; Liu, R. Alterations of the intestinal microbiome and metabolome in women with rheumatoid arthritis. Clin. Exp. Med. 2023, 23, 4695–4706. [Google Scholar] [CrossRef] [PubMed]
- Essex, M.; Rios Rodriguez, V.; Rademacher, J.; Proft, F.; Löber, U.; Markó, L.; Pleyer, U.; Strowig, T.; Marchand, J.; Kirwan, J.A.; et al. Shared and Distinct Gut Microbiota in Spondyloarthritis, Acute Anterior Uveitis, and Crohn’s Disease. Arthritis Rheumatol. 2024, 76, 48–58. [Google Scholar] [CrossRef]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; Van Der Veeken, J.; Deroos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhou, J.; Zhang, X.; Mao, R.; Zhang, C. Mendelian randomization supports causality between gut microbiota and chronic hepatitis B. Front. Microbiol. 2023, 14, 1243811. [Google Scholar] [CrossRef]
- Wu, R.; Wang, D.; Cheng, L.; Su, R.; Li, B.; Fan, C.; Gao, C.; Wang, C. Impaired immune tolerance mediated by reduced Tfr cells in rheumatoid arthritis linked to gut microbiota dysbiosis and altered metabolites. Arthritis Res. Ther. 2024, 26, 21. [Google Scholar] [CrossRef]
- Wu, X.; Liu, J.; Xiao, L.; Lu, A.; Zhang, G. Alterations of gut microbiome in rheumatoid arthritis. Osteoarthr. Cartil. 2017, 25, S287–S288. [Google Scholar] [CrossRef]
| Traits (Outcome) | Taxonomies | GM | Significant Results for MR Analysis of the Samples | Analysis of Sensitivity and Potential Pleiotropy Detection | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MR Method | No. SNP | Beta | SE | p Value | OR | 95 %CI | Method | Q | p | Egger-Intercept | p | MR-PRESSO | |||
| Seronegative RA | Genus | Prevotella9 | IVW | 17 | −0.197 | 0.083 | 0.017 | 0.821 | 0.699–0.965 | IVW | 17.652 | 0.345 | −0.011 | 0.648 | 0.417 |
| MR Egger | 17.400 | 0.295 | |||||||||||||
| Genus | Sutterella | IVW | 12 | −0.283 | 0.114 | 0.013 | 0.753 | 0.602–0.943 | IVW | 9.066 | 0.616 | −5.07 × 104 | 0.988 | 0.619 | |
| MR Egger | 9.066 | 0.526 | |||||||||||||
| Genus | Christensenellaceae R.7 | IVW | 9 | −0.363 | 0.153 | 0.018 | 0.696 | 0.515–0.940 | IVW | 7.987 | 0.435 | −1.53 × 104 | 0.997 | 0.454 | |
| MR Egger | 7.987 | 0.334 | |||||||||||||
| Seropositive RA | Genus | RuminococcaceaeUCG002 | IVW | 22 | 0.230 | 0.083 | 0.006 | 1.258 | 1.070–1.480 | IVW | 16.643 | 0.733 | −0.002 | 0.926 | 0.744 |
| MR Egger | 16.634 | 0.677 | |||||||||||||
| Genus | Ruminococcus gauvreauii | IVW | 12 | 0.418 | 0.137 | 0.002 | 1.519 | 1.161–1.988 | IVW | 18.500 | 0.071 | 0.011 | 0.812 | 0.105 | |
| MR Egger | 18.390 | 0.049 | |||||||||||||
| Genus | Lachnospira | IVW | 7 | −0.352 | 0.170 | 0.038 | 0.703 | 0.504–0.981 | IVW | 6.359 | 0.384 | −0.069 | 0.272 | 0.410 | |
| MR Egger | 4.839 | 0.436 | |||||||||||||
| Genus | Slackia | IVW | 9 | −0.195 | 0.093 | 0.036 | 0.823 | 0.685–0.987 | IVW | 8.207 | 0.413 | 0.009 | 0.855 | 0.453 | |
| MR Egger | 8.165 | 0.318 | |||||||||||||
| Genus | Roseburia | IVW | 16 | −0.278 | 0.112 | 0.013 | 0.757 | 0.608–0.943 | IVW | 16.201 | 0.369 | −0.022 | 0.383 | 0.369 | |
| MR Egger | 15.314 | 0.357 | |||||||||||||
| Genus | Barnesiella | IVW | 15 | −0.232 | 0.101 | 0.021 | 0.793 | 0.651–0.966 | IVW | 7.513 | 0.913 | −0.004 | 0.901 | 0.906 | |
| MR Egger | 7.497 | 0.875 | |||||||||||||
| Genus | Prevotella7 | IVW | 12 | −0.157 | 0.060 | 0.009 | 0.855 | 0.760–0.962 | IVW | 11.882 | 0.373 | 0.031 | 0.575 | 0.405 | |
| MR Egger | 11.497 | 0.320 | |||||||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, J.; Peng, Y.; Tian, R.; Yu, H.; Hu, H.; Huang, Q.; Xu, Y.; Liu, L.; Pan, H. Investigating Correlation Between Gut Microbiota and Rheumatoid Arthritis Subtypes by Mendelian Randomization. Pathogens 2025, 14, 385. https://doi.org/10.3390/pathogens14040385
Wu J, Peng Y, Tian R, Yu H, Hu H, Huang Q, Xu Y, Liu L, Pan H. Investigating Correlation Between Gut Microbiota and Rheumatoid Arthritis Subtypes by Mendelian Randomization. Pathogens. 2025; 14(4):385. https://doi.org/10.3390/pathogens14040385
Chicago/Turabian StyleWu, Jiaqi, Yao Peng, Ruimin Tian, Hao Yu, Huating Hu, Qingchun Huang, Youhua Xu, Liang Liu, and Hudan Pan. 2025. "Investigating Correlation Between Gut Microbiota and Rheumatoid Arthritis Subtypes by Mendelian Randomization" Pathogens 14, no. 4: 385. https://doi.org/10.3390/pathogens14040385
APA StyleWu, J., Peng, Y., Tian, R., Yu, H., Hu, H., Huang, Q., Xu, Y., Liu, L., & Pan, H. (2025). Investigating Correlation Between Gut Microbiota and Rheumatoid Arthritis Subtypes by Mendelian Randomization. Pathogens, 14(4), 385. https://doi.org/10.3390/pathogens14040385







