Patterns of the Circulation of Influenza in a Targeted Jordanian Subpopulation from November 2021 to April 2023
Abstract
1. Introduction
2. Materials and Methods
2.1. Cohort Description
2.2. Nucleic Acid Extraction and Multiplex Respiratory Panel
2.3. Subtype Identification and Analysis of Influenza Viruses
2.4. Statistical Analysis
3. Results
3.1. Demographic of Study Participants
3.2. Clinical Characteristics of Study Participants
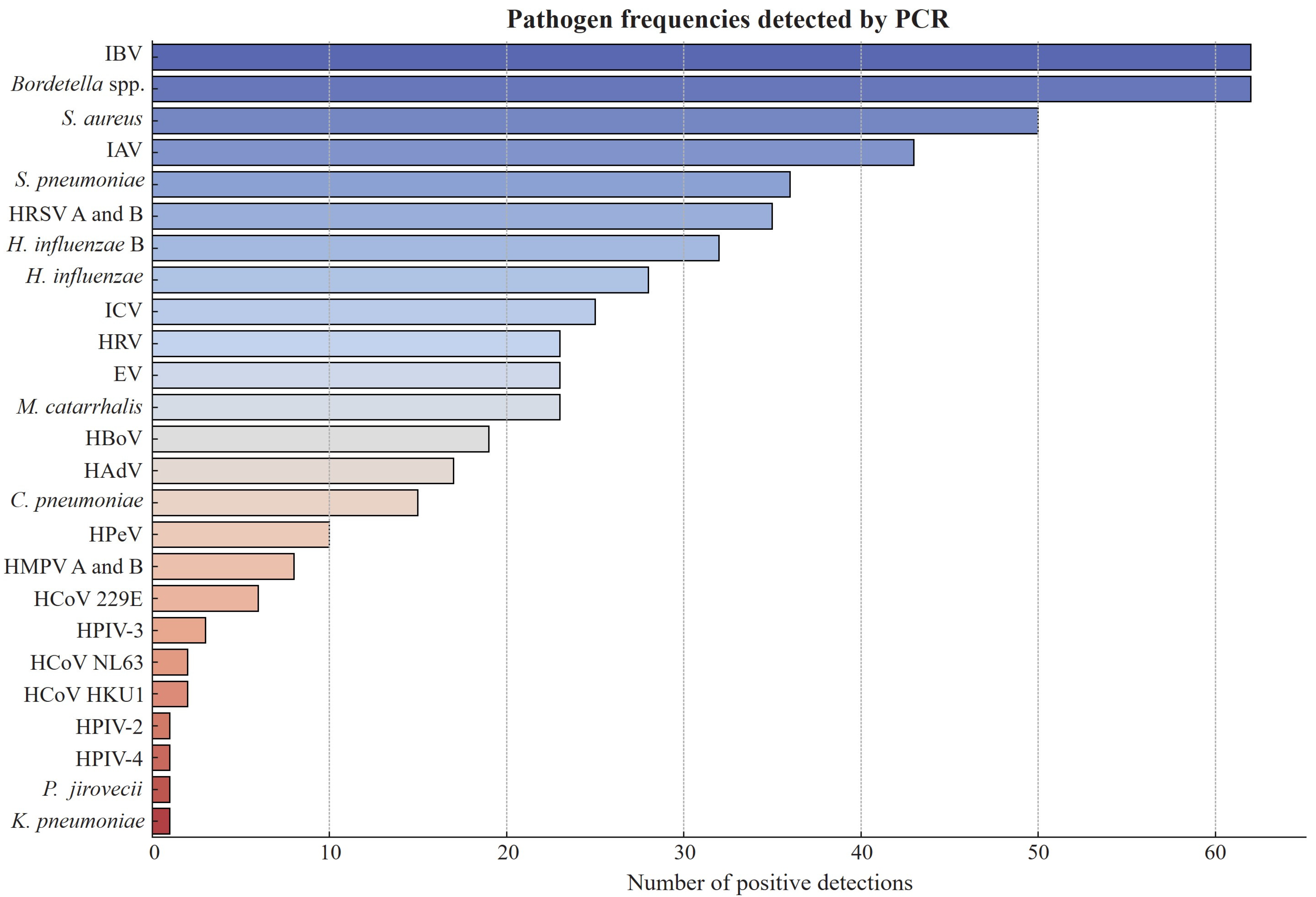
3.3. Distribution of Infections and Coinfections Among Study Participants
3.4. Coinfections with Influenza Viruses
3.5. Subtype Distribution of Influenza A and B Viruses
4. Discussion
5. Study Limitations
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Thompson, M.G.; Levine, M.Z.; Bino, S.; Hunt, D.R.; Al-Sanouri, T.M.; Simões, E.A.F.; Porter, R.M.; Biggs, H.M.; Gresh, L.; Simaku, A.; et al. Underdetection of Laboratory-Confirmed Influenza-Associated Hospital Admissions among Infants: A Multicentre, Prospective Study HHS Public Access. Lancet Child Adolesc. Health 2019, 3, 781–794. [Google Scholar] [CrossRef] [PubMed]
- Rolsma, S.L.; Rankin, D.A.; Haddadin, Z.; Hamdan, L.; Rahman, H.K.; Faouri, S.; Shehabi, A.; Williams, J.V.; Khuri-Bulos, N.; Halasa, N.B.; et al. Assessing the Epidemiology and Seasonality of Influenza among Children under Two Hospitalized in Amman, Jordan, 2010–2013. Influ. Other Respir. Viruses 2021, 15, 284–292. [Google Scholar] [CrossRef] [PubMed]
- Abu-Helalah, M.; Al-Shatnawi, S.F.; Lubad, M.A.; Al-Zayadneh, E.; Al-Hanaktah, M.; Harahsheh, M.; AL-Iede, M.; Nafi, O.; Yousef, R.; Almaaitah, I.; et al. The Epidemiology and Health Burdens of Influenza Infections Amongst Hospitalized Children Under 5 Years of Age in Jordan: A National Multi-Center Cross-Sectional Study. Vaccines 2025, 13, 12. [Google Scholar] [CrossRef] [PubMed]
- Hamdan, O.; Amarin, J.Z.; Potter, M.; Haddadin, Z.; Yanis, A.; Shawareb, Y.; Khuri-Bulos, N.; Haddadin, R.; Halasa, N.B.; Spieker, A.J. Seasonal Influenza Vaccination: Attitudes and Practices of Healthcare Providers in Jordan. PLoS ONE 2024, 19, e0314224. [Google Scholar] [CrossRef]
- Ababneh, M.; Jaber, M.; Rababa’h, A.; Ababneh, F. Seasonal Influenza Vaccination among Older Adults in Jordan: Prevalence, Knowledge, and Attitudes. Hum. Vaccin. Immunother. 2020, 16, 2252–2256. [Google Scholar] [CrossRef]
- Al Khatib, H.A.; Al Thani, A.A.; Gallouzi, I.; Yassine, H.M. Epidemiological and Genetic Characterization of PH1N1 and H3N2 Influenza Viruses Circulated in MENA Region during 2009–2017. BMC Infect. Dis. 2019, 19, 314. [Google Scholar] [CrossRef]
- Abdalla, O.; Mohammed, M.; Hakawi, A.M.; Aljifri, A.; Abdalla, M.; Eltigani, S.; Mujib, S.A.; Assiri, A. Hospital-Based Surveillance of Influenza A(H1N1)Pdm09 Virus in Saudi Arabia, 2010–2016. Ann. Saudi Med. 2020, 40, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Althaqafi, A.; Farahat, F.; Alsaedi, A.; Alshamrani, M.; Alsaeed, M.S.; AlhajHussein, B.; El-Kafrawy, S.A.; Azhar, E.I. Molecular Detection of Influenza A and B Viruses in Four Consecutive Influenza Seasons 2015–16 to 2018–19 in a Tertiary Center in Western Saudi Arabia. J. Epidemiol. Glob. Health 2021, 11, 208–215. [Google Scholar] [CrossRef]
- Abo Shama, N.M.; Mahmoud, S.H.; Bagato, O.; AbdElsalam, E.T.; Alkhazindar, M.; Kandeil, A.; McKenzie, P.P.; Webby, R.J.; Ali, M.A.; Kayali, G.; et al. Incidence and Neutralizing Antibody Seroprevalence of Influenza B Virus in Egypt: Results of a Community-Based Cohort Study. PLoS ONE 2022, 17, e0269321. [Google Scholar] [CrossRef]
- Gomaa, M.R.; Badra, R.; El Rifay, A.S.; Kandeil, A.; Kamel, M.N.; Abo Shama, N.M.; El-Shesheny, R.; Barakat, A.B.; Ali, M.A.; Kayali, G. Incidence and Seroprevalence of Seasonal Influenza a Viruses in Egypt: Results of a Community-Based Cohort Study. Influ. Other Respir. Viruses 2022, 16, 749–755. [Google Scholar] [CrossRef]
- Farah, Z.; El Naja, H.A.; Tempia, S.; Saleh, N.; Abubakar, A.; Maison, P.; Ghosn, N. Estimation of the Influenza-Associated Respiratory Hospitalization Burden Using Sentinel Surveillance Data, Lebanon, 2015–2020. Influ. Other Respir. Viruses 2023, 17, e13138. [Google Scholar] [CrossRef]
- Fahim, M.; Roshdy, W.H.; Deghedy, O.; Kamel, R.; Naguib, A.; Showky, S.; Elguindy, N.; Abdel Fattah, M.; Afifi, S.; Mohsen, A.; et al. Epidemiology, Disease Severity and Outcome of Severe Acute Respiratory Syndrome Coronavirus 2 and Influenza Viruses Coinfection Seen at Egypt Integrated Acute Respiratory Infections Surveillance, 2020–2022. Can. J. Infect. Dis. Med. Microbiol. 2022, 2022, 7497500. [Google Scholar] [CrossRef]
- Khan, A.; AlBalwi, M.A.; AlAbdulkareem, I.; AlMasoud, A.; AlAsiri, A.; AlHarbi, W.; AlSehile, F.; El-Saed, A.; Balkhy, H.H. Atypical Influenza A(H1N1)Pdm09 Strains Caused an Influenza Virus Outbreak in Saudi Arabia during the 2009–2011 Pandemic Season. J. Infect. Public Health 2019, 12, 557–567. [Google Scholar] [CrossRef]
- Khasawneh, A.I.; Himsawi, N.M.; Abu-Raideh, J.A.; Sammour, A.; Safieh, H.A.; Obeidat, A.; Azab, M.; Tarifi, A.A.; Al Khawaldeh, A.; Al-Momani, H.; et al. Prevalence of SARS-COV-2 and Other Respiratory Pathogens among a Jordanian Subpopulation during Delta-to-Omicron Transition: Winter 2021/2022. PLoS ONE 2023, 18, e0283804. [Google Scholar] [CrossRef] [PubMed]
- Khasawneh, A.I.; Himsawi, N.; Sammour, A.; Safieh, H.A.; Burayzat, S.; Al-Momani, H.; Alotaibi, M.R.; Al Shboul, S.; Saleh, T. Molecular Characterization of Human Respiratory Syncytial Virus Strains Circulating among Hospitalized Children in Jordan. BMC Infect. Dis. 2024, 24, 1347. [Google Scholar] [CrossRef] [PubMed]
- Bancej, C.; Rahal, A.; Lee, L.; Buckrell, S.; Schmidt, K.; Nathalie Bastien, N. National FluWatch Mid-Season Report, 2021–2022: Sporadic Influenza Activity Returns. Can. Commun. Dis. Rep. 2022, 48, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Influenza Surveillance Report: Key Updates for Week 52, Ending December 31, 2022. Weekly US, 6 January 2023.
- Pendrey, C.G.A.; Strachan, J.; Peck, H.; Aziz, A.; Moselen, J.; Moss, R.; Rahaman, M.R.; Barr, I.G.; Subbarao, K.; Sullivan, S.G. The Re-Emergence of Influenza Following the COVID-19 Pandemic in Victoria, Australia, 2021 to 2022. Eurosurveillance 2023, 28, 2300118. [Google Scholar] [CrossRef]
- Nott, R.; Fuller, T.L.; Brasil, P.; Nielsen-Saines, K. Out-of-Season Influenza during a COVID-19 Void in the State of Rio de Janeiro, Brazil: Temperature Matters. Vaccines 2022, 10, 821. [Google Scholar] [CrossRef]
- Chow, E.J.; Uyeki, T.M.; Chu, H.Y. The Effects of the COVID-19 Pandemic on Community Respiratory Virus Activity. Nat. Rev. Microbiol. 2022, 21, 195. [Google Scholar] [CrossRef]
- Moghoofei, M.; Monavari, S.H.; Mostafaei, S.; Hadifar, S.; Ghasemi, A.; Babaei, F.; Kavosi, H.; Tavakoli, A.; Javanmard, D.; Esghaei, M.; et al. Prevalence of Influenza A Infection in the Middle-East: A Systematic Review and Meta-Analysis. Clin. Respir. J. 2018, 12, 1787–1801. [Google Scholar] [CrossRef]
- Perofsky, A.C.; Huddleston, J.; Hansen, C.L.; Barnes, J.R.; Rowe, T.; Xu, X.; Kondor, R.; Wentworth, D.E.; Lewis, N.; Whittaker, L.; et al. Antigenic Drift and Subtype Interference Shape A(H3N2) Epidemic Dynamics in the United States. Elife 2024, 13, RP91849. [Google Scholar] [CrossRef]
- Jayadas, T.T.P.; Jeewandara, C.; Senadheera, B.; Kuruppu, H.; Wickramanayake, R.; Chathurangika, P.H.; Senatilleke, N.; Warnakulasuriya, N.; Bary, F.; Wijewickrama, A.; et al. Genomic Surveillance and Evolutionary Dynamics of Influenza a Virus in Sri Lanka. Virol. J. 2024, 21, 304. [Google Scholar] [CrossRef] [PubMed]
- Haddara, A.; Houry, Z.; Zahreddine, N.; Atallah, M.; Boutros, C.F.; Tannous, J.; Sadaka, C.; Wehbe, S.; Kadi, T.; Ibrahim, A.; et al. Characteristics of Medically Attended Influenza Infection across Age Groups before the COVID-19 Pandemic in Lebanon. J. Infect. Public. Health 2024, 17, 102521. [Google Scholar] [CrossRef] [PubMed]
- Dandachi, I.; Alrezaihi, A.; Amin, D.; AlRagi, N.; Alhatlani, B.; Binjomah, A.; Aleisa, K.; Dong, X.; Hiscox, J.A.; Aljabr, W. Molecular Surveillance of Influenza A Virus in Saudi Arabia: Whole-Genome Sequencing and Metagenomic Approaches. Microbiol. Spectr. 2024, 12, e0066524. [Google Scholar] [CrossRef] [PubMed]
- Cane, J.; Sanderson, N.; Barnett, S.; Vaughan, A.; Pott, M.; Kapel, N.; Morgan, M.; Jesuthasan, G.; Samuel, R.; Ehsaan, M.; et al. Nanopore Sequencing of Influenza A and B in Oxfordshire and the United Kingdom, 2022–2023. J. Infect. 2024, 88, 106164. [Google Scholar] [CrossRef]
- Brcko, I.C.; Souza, V.C.d.; Ribeiro, G.; Lima, A.R.J.; Martins, A.J.; Barros, C.R.d.S.; de Carvalho, E.; Pereira, J.S.; de Lima, L.P.O.; Viala, V.L.; et al. Comprehensive Molecular Epidemiology of Influenza Viruses in Brazil: Insights from a Nationwide Analysis. Virus Evol. 2024, 11, veae102. [Google Scholar] [CrossRef]
- Chung, J.R.; Price, A.M.; Zimmerman, R.K.; Moehling Geffel, K.; House, S.L.; Curley, T.; Wernli, K.J.; Phillips, C.H.; Martin, E.T.; Vaughn, I.A.; et al. Influenza Vaccine Effectiveness against Medically Attended Outpatient Illness, United States, 2023–2024 Season. Clin. Infect. Dis. 2025, ciae658. [Google Scholar] [CrossRef]
- Klein, E.Y.; Monteforte, B.; Gupta, A.; Jiang, W.; May, L.; Hsieh, Y.H.; Dugas, A. The Frequency of Influenza and Bacterial Coinfection: A Systematic Review and Meta-analysis. Influ. Other Respir. Viruses 2016, 10, 394. [Google Scholar] [CrossRef]
- Morris, D.E.; Cleary, D.W.; Clarke, S.C. Secondary Bacterial Infections Associated with Influenza Pandemics. Front. Microbiol. 2017, 8, 1041. [Google Scholar] [CrossRef]
- Kırca, F.; Aydoğan, S.; Gozalan, A.; Güler, E.; Uyan Erten, A.Z.; Özşen Uygur, A.S.; Doğan, A.; Dinc, B. Impact of Non-Pharmaceutical Interventions on Circulating Respiratory Viruses during the COVID-19 Pandemic in Turkey. Ann. Saudi Med. 2023, 43, 143–153. [Google Scholar] [CrossRef]
- Kampenusa, I.; Niedre-Otomere, B.; Trofimova, J.; Pole, I.; Pakarna, G.; Savicka, O.; Nikisins, S. Circulation and Codetections of Influenza Virus, SARS-CoV-2, Respiratory Syncytial Virus, Rhinovirus, Adenovirus, Bocavirus, and Other Respiratory Viruses During 2022–2023 Season in Latvia. Viruses 2024, 16, 1650. [Google Scholar] [CrossRef] [PubMed]
- Farzi, R.; Pirbonyeh, N.; Kadivar, M.R.; Moattari, A. Prevalence of Influenza Viruses A and B, Adenovirus, Respiratory Syncytial Virus, and Human Metapneumonia Viruses among Children with Acute Respiratory Tract Infection. Adv. Virol. 2024, 2024, 7613948. [Google Scholar] [CrossRef] [PubMed]
- Grohskopf, L.A.; Blanton, L.H.; Ferdinands, J.M.; Chung, J.R.; Broder, K.R.; Talbot, H.K.; Morgan, R.L.; Fry, A.M. Prevention and Control of Seasonal Influenza with Vaccines: Recommendations of the Advisory Committee on Immunization Practices—United States, 2022–2023 Influenza Season. MMWR Recomm. Rep. 2022, 71, 1–28. [Google Scholar] [CrossRef] [PubMed]
- Zalloum, W.A.; Elayeh, E.R.; Ali, B.A.H.; Zalloum, N. Perception, Knowledge and Attitude towards Influenza Vaccine during COVID-19 Pandemic in Jordanian Population. Eur. J. Integr. Med. 2022, 49, 102100. [Google Scholar] [CrossRef]


| Variable | N = 458 (%) | IAV N = 43 (%) | IBV N = 62 (%) | ICV N = 25 (%) | p Value |
|---|---|---|---|---|---|
| Gender Male Female | 262 (57.2) 196 (42.8) | 33 (76.7) 10 (23.3) | 40 (64.5) 22 (35.5) | 13 (52.0) 12 (48.0) | 0.107 |
| Age 0–5 6–18 19–30 31–50 >50 | 87 (19.0) 80 (17.5) 122 (26.6) 129 (28.2) 40 (8.7) | 4 (9.3) 2 (4.7) 28 (65.1) 8 (18.6) 1 (2.3) | 2 (3.2) 19 (30.6) 15 (24.2) 19 (30.6) 7 (11.4) | 18 (72.0) 4 (16.0) 2 (8.0) 1 (4.0) 0 | <0.001 |
| Residential city Amman Zarqa Irbid Others | 278 (60.7) 101 (22.1) 63 (13.8) 7 (1.5) | 7 (16.3) 31 (72.1) 2 (4.7) 3 (6.9) | 37 (59.6) 17 (27.4) 7 (11.4) 1 (1.6) | 23 (92.0) 0 1 (4.0) 1 (4.0) | <0.001 |
| Educational level Preschool Primary school Bachelor’s Master’s PhD | 91 (19.9) 177 (38.6) 178 (38.9) 7 (1.5) 5 (1.1) | 6 (14.0) 20 (46.5) 17 (39.5) 0 0 | 2 (3.2) 33 (53.1) 24 (38.7) 0 3 | 19 (76.0) 4 (16.0) 2 (8.0) 0 0 | <0.001 NA |
| Variable | N = 458 (%) | IAV N = 43 (%) | IBV N = 62 (%) | ICV N = 25 (%) | p Value |
|---|---|---|---|---|---|
| Symptoms Cough Nasal discharge Sore throat Headache Myalgia Fever Chills Difficulty breathing Nausea Vomiting Diarrhea | 362 (79.0) 291 (63.5) 290 (63.3) 272 (59.4) 252 (55.0) 247 (53.9) 217 (47.4) 214 (46.7) 115 (25.1) 115 (25.1) 48 (10.5) | 38 (88.4) 25 (58.1) 33 (76.7) 28 (65.1) 28 (65.1) 24 (55.8) 24 (55.8) 25 (58.1) 6 (14.0) 6 (14.0) 1 (2.3) | 52 (83.9) 45 (72.6) 47 (75.8) 43 (69.4) 32 (51.6) 31 (50.0) 32 (51.6) 25 (40.3) 12 (19.4) 12 (19.4) 7 (11.3) | 25 (100) 18 (72.0) 13 (52.0) 4 (16.0) 4 (16.0) 12 (48.0) 4 (16.0) 19 (76.0) 12 (48.0) 12 (48.0) 5 (20.0) | <0.001 |
| Health status Healthy Hypertension Allergy Asthma Diabetes Heart disease Other illnesses | 367 (80.1) 26 (5.7) 21 (4.6) 19 (4.1) 15 (3.3) 5 (1.1) 5 (1.1) | 35 (81.4) 1 (2.3) 3 (6.9) 2 (4.7) 0 0 2 (4.7) | 49 (79.0) 4 (6.5) 3 (4.8) 1 (1.6) 1 (1.6) 0 4 (6.5) | 24 (96.0) 0 0 1 (4.0) 0 0 0 | NA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khasawneh, A.I.; Himsawi, N.M.; Abu-Raideh, J.A.; Sammour, A.; Safieh, H.A.; Al Qudah, M.; Obeidat, A.; Alotaibi, M.R.; Al-Momani, H.; Khasawneh, R.; et al. Patterns of the Circulation of Influenza in a Targeted Jordanian Subpopulation from November 2021 to April 2023. Pathogens 2025, 14, 365. https://doi.org/10.3390/pathogens14040365
Khasawneh AI, Himsawi NM, Abu-Raideh JA, Sammour A, Safieh HA, Al Qudah M, Obeidat A, Alotaibi MR, Al-Momani H, Khasawneh R, et al. Patterns of the Circulation of Influenza in a Targeted Jordanian Subpopulation from November 2021 to April 2023. Pathogens. 2025; 14(4):365. https://doi.org/10.3390/pathogens14040365
Chicago/Turabian StyleKhasawneh, Ashraf I., Nisreen M. Himsawi, Jumana A. Abu-Raideh, Ashraf Sammour, Hazem Abu Safieh, Mohammad Al Qudah, Ali Obeidat, Moureq R. Alotaibi, Hafez Al-Momani, Rame Khasawneh, and et al. 2025. "Patterns of the Circulation of Influenza in a Targeted Jordanian Subpopulation from November 2021 to April 2023" Pathogens 14, no. 4: 365. https://doi.org/10.3390/pathogens14040365
APA StyleKhasawneh, A. I., Himsawi, N. M., Abu-Raideh, J. A., Sammour, A., Safieh, H. A., Al Qudah, M., Obeidat, A., Alotaibi, M. R., Al-Momani, H., Khasawneh, R., Al Shboul, S., & Saleh, T. (2025). Patterns of the Circulation of Influenza in a Targeted Jordanian Subpopulation from November 2021 to April 2023. Pathogens, 14(4), 365. https://doi.org/10.3390/pathogens14040365






