Assessing Schmallenberg Virus Disease in Sardinia (Italy) After the First Epidemic Episode in 2012
Abstract
1. Introduction
2. Materials and Methods
2.1. Retrospective Study
2.2. Serological Investigation
2.3. Real-Time PCR (RT-PCR) Analysis
2.4. Isolation of SBV in Cell Cultures
2.5. Sanger Sequencing and Phylogenetic Analysis
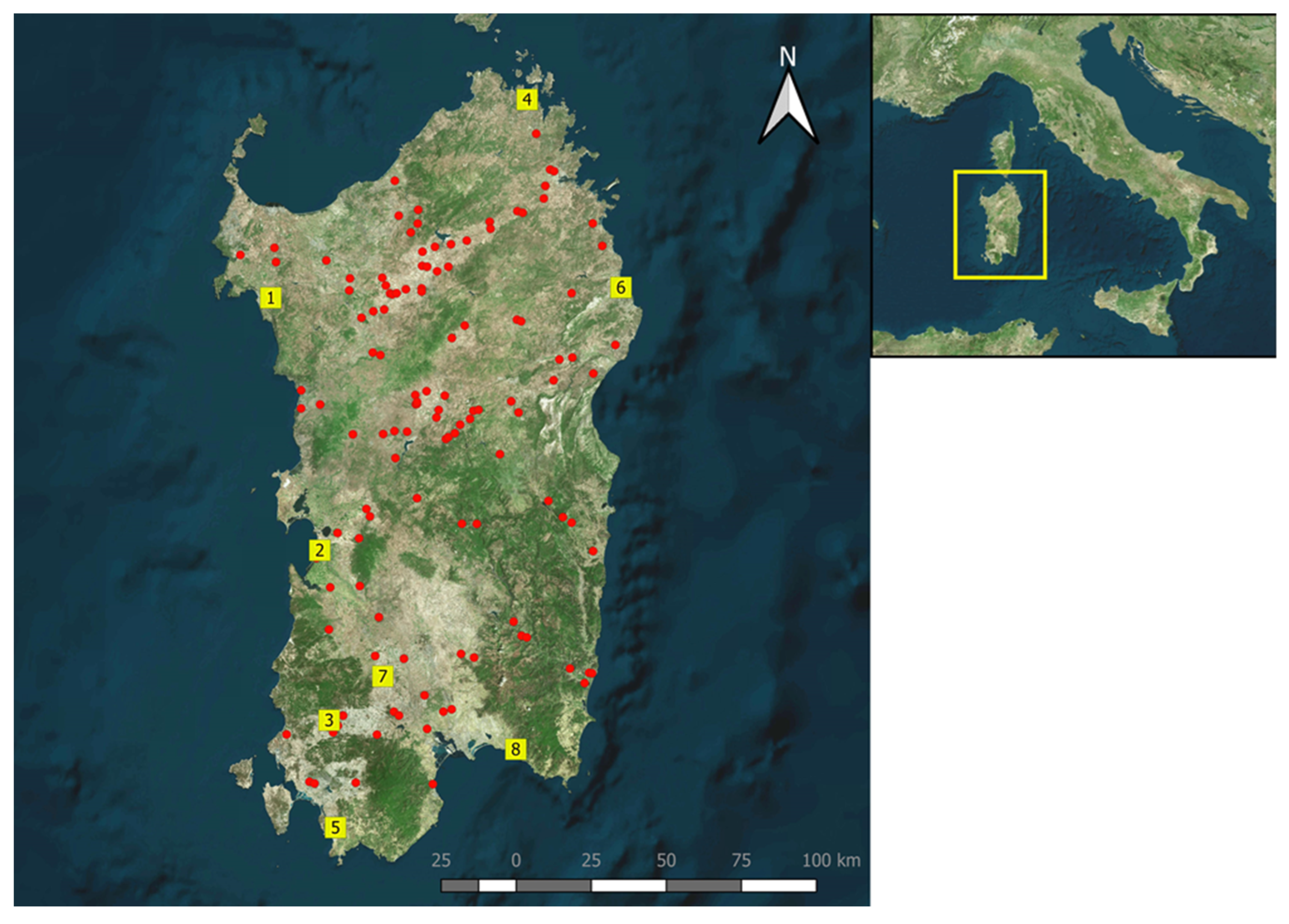
2.6. Entomological Survey
3. Results
3.1. Anatomo-Pathological Survey
3.2. Serological Survey
3.3. Isolation of SBV in Cell Cultures
3.4. Sequencing
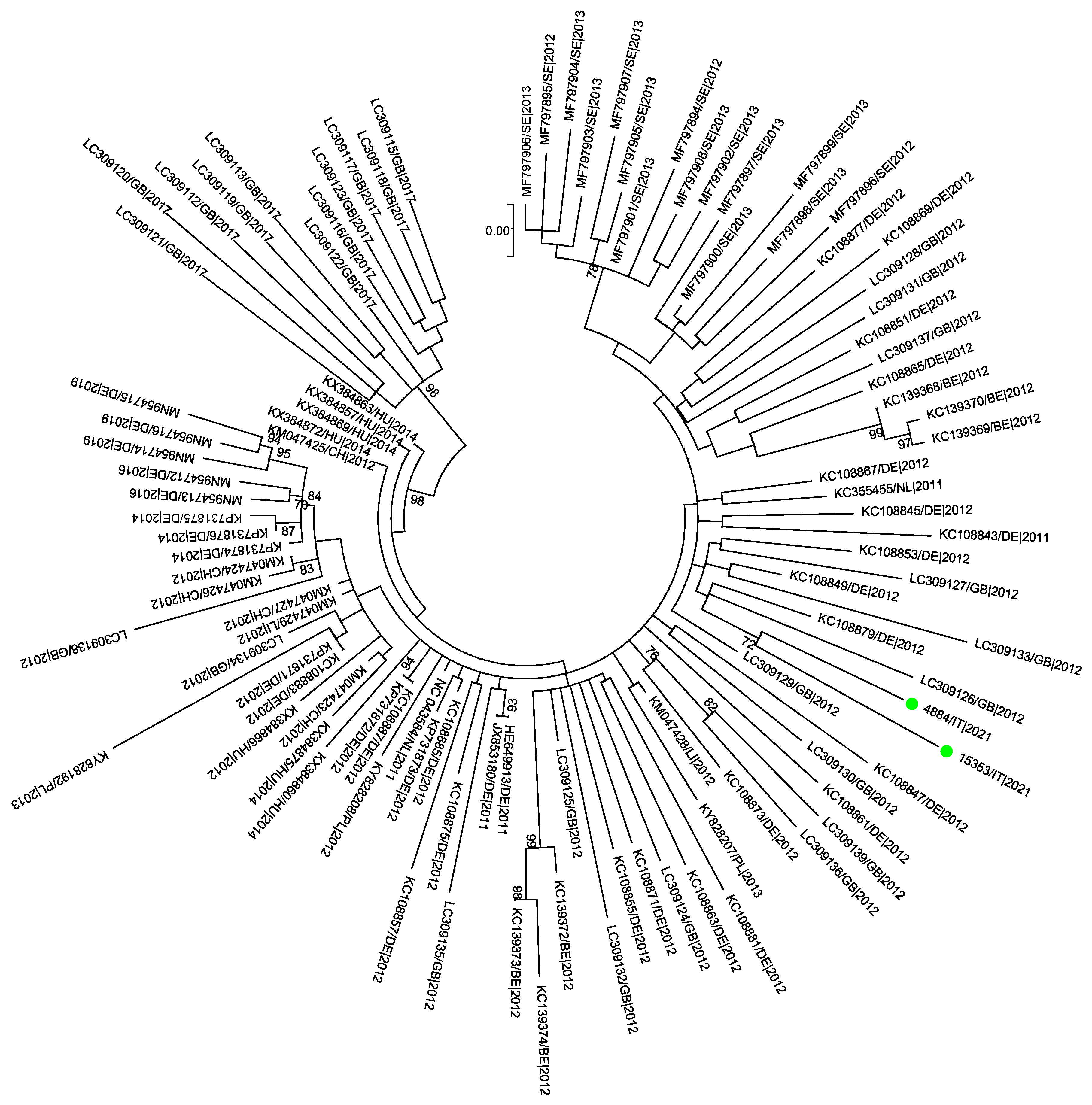
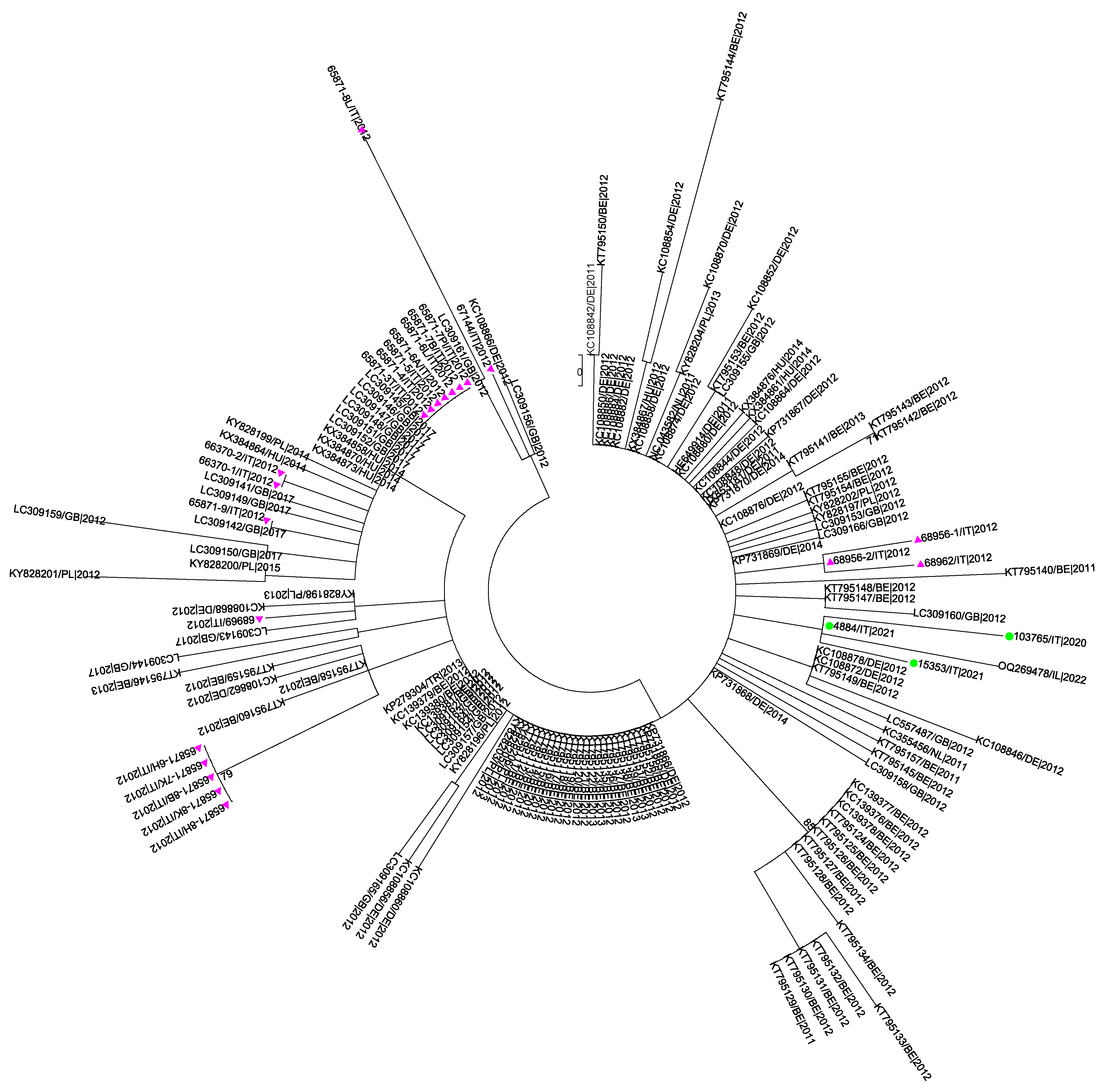
3.5. Phylogenetic Analysis
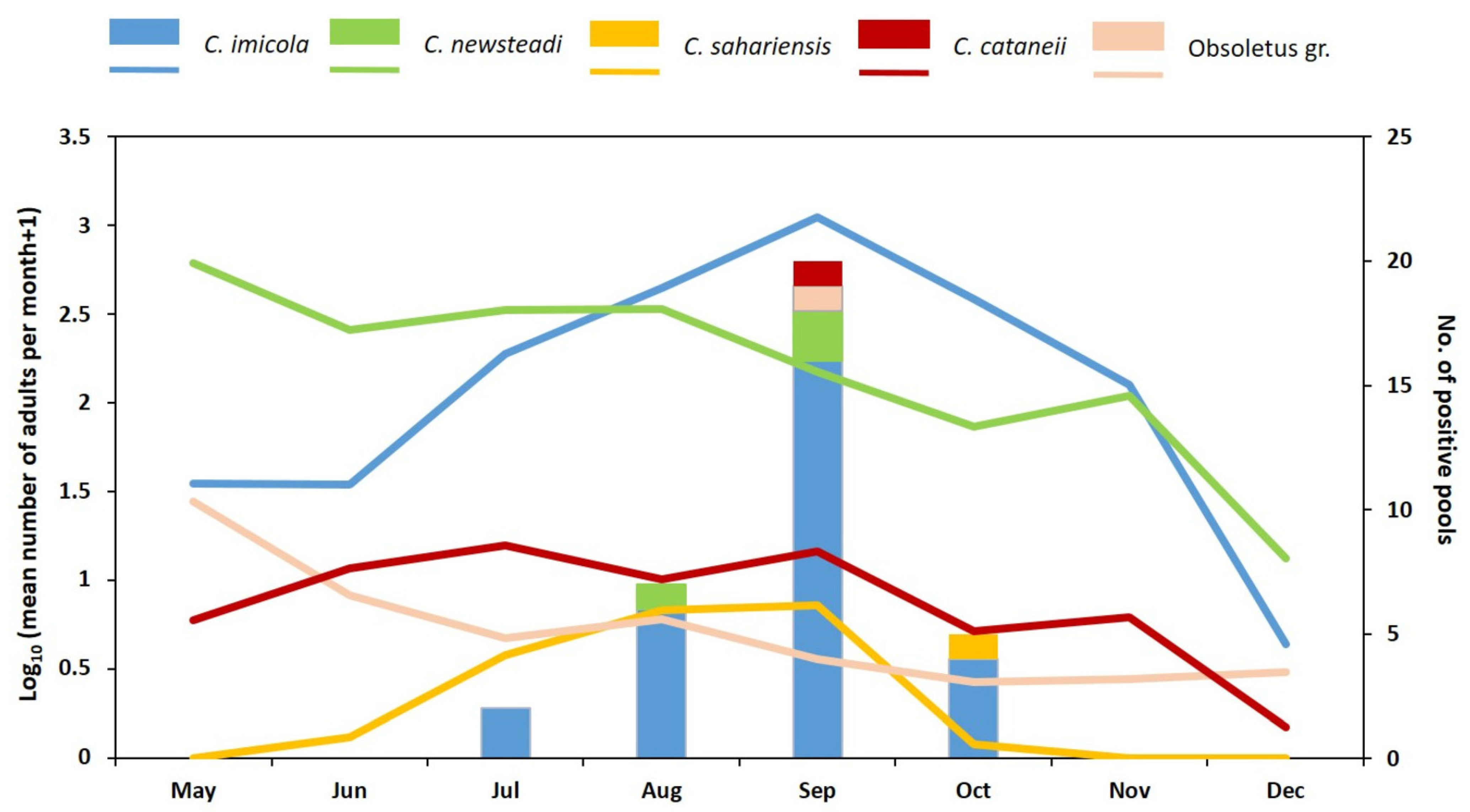
3.6. Entomological Survey
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hoffmann, B.; Scheuch, M.; Höper, D.; Jungblut, R.; Holsteg, M.; Schirrmeier, H.; Eschbaumer, M.; Goller, K.V.; Wernike, K.; Fischer, M.; et al. Novel orthobunyavirus in Cattle, Europe, 2011. Emerg. Infect. Dis. 2012, 18, 469–472. [Google Scholar] [CrossRef] [PubMed]
- van den Brom, R.; Luttikholt, S.J.; Lievaart-Peterson, K.; Peperkamp, N.H.; Mars, M.H.; van der Poel, W.H.; Vellema, P. Epizootic of ovine congenital malformations associated with Schmallenberg virus infection. Tijdschr. Diergeneeskd. 2012, 137, 106–111. [Google Scholar] [PubMed]
- Helmer, C.; Eibach, R.; Tegtmeyer, P.C.; Humann-Ziehank, E.; Ganter, M. Survey of Schmallenberg virus (SBV) infection in German goat flocks. Epidemiol. Infect. 2013, 141, 2335–2345. [Google Scholar] [CrossRef] [PubMed]
- Laloy, E.; Bréard, E.; Sailleau, C.; Viarouge, C.; Desprat, A.; Zientara, S.; Klein, F.; Hars, J.; Rossi, S. Schmallenberg virus infection among red deer, France, 2010–2012. Emerg. Infect. Dis. 2014, 20, 131–134. [Google Scholar] [CrossRef]
- Ferrara, G.; Wernike, K.; Iovane, G.; Pagnini, U.; Montagnaro, S. First evidence of Schmallenberg virus infection in southern Italy. BMC Vet. Res. 2023, 19, 95. [Google Scholar] [CrossRef]
- Herder, V.; Wohlsein, P.; Peters, M.; Hansmann, F.; Baumgärtner, W. Salient lesions in domestic ruminants infected with the emerging so-called Schmallenberg virus in Germany. Vet. Pathol. 2012, 49, 588–591. [Google Scholar] [CrossRef]
- Elbers, A.R.W.; Meiswinkel, R.; van Weezep, E.; Sloet van Oldruitenborgh-Oosterbaan, M.M.; Kooi, E.A. Schmallenberg virus detected by RT-PCR in Culicoides biting midges captured during the 2011 epidemic in The Netherlands. Emerg. Infect. Dis. 2013, 19, 106–109. [Google Scholar] [CrossRef]
- De Regge, N.; Deblauwe, I.; De Deken, R.; Vantieghem, P.; Madder, M.; Geysen, D.; Smeets, F.; Losson, B.; van den Berg, T.; Cay, A.B. Detection of Schmallenberg virus in different Culicoides spp. by real-time RT-PCR. Transbound. Emerg. Dis. 2012, 59, 471–475. [Google Scholar] [CrossRef]
- Goffredo, M.; Monaco, F.; Capelli, G.; Quaglia, M.; Federici, V.; Catalani, M.; Montarsi, F.; Polci, A.; Pinoni, C.; Calistri, P.; et al. Schmallenberg virus in Italy: A retrospective survey in Culicoides stored during the bluetongue Italian surveillance program. Prev. Vet. Med. 2013, 111, 230–236. [Google Scholar] [CrossRef]
- Ségard, A.; Gardès, L.; Jacquier, E.; Grillet, C.; Mathieu, B.; Rakotoarivony, I.; Setier-Rio, M.L.; Chavernac, D.; Cêtre-Sossah, C.; Balenghien, T.; et al. Schmallenberg virus in Culicoides Latreille (Diptera: Ceratopogonidae) populations in France during 2011-2012 outbreak. Transbound. Emerg. Dis. 2018, 65, e94–e103. [Google Scholar] [CrossRef]
- Kęsik-Maliszewska, J.; Larska, M.; Collins, Á.B.; Rola, J. Post-Epidemic Distribution of Schmallenberg Virus in Culicoides Arbovirus Vectors in Poland. Viruses 2019, 11, 447. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Elliott, R.M.; Brennan, B. Emerging phleboviruses. Curr. Opin. Virol. 2014, 5, 50–57. [Google Scholar] [CrossRef]
- Wernike, K.; Reimann, I.; Banyard, A.C.; Kraatz, F.; La Rocca, S.A.; Hoffmann, B.; McGowan, S.; Hechinger, S.; Choudhury, B.; Aebischer, A.; et al. High genetic variability of Schmallenberg virus M-segment leads to efficient immune escape from neutralizing antibodies. PLoS Pathog. 2021, 17, e1009247. [Google Scholar] [CrossRef] [PubMed]
- Wernike, K.; Beer, M. Misinterpretation of Schmallenberg virus sequence variations: The sample material makes the difference. Virus Genes 2019, 55, 123–126. [Google Scholar] [CrossRef] [PubMed]
- Muskens, J.; Smolenaars, A.J.; van der Poel, W.H.; Mars, M.H.; van Wuijckhuise, L.; Holzhauer, M.; van Weering, H.; Kock, P. Diarree en productiedaling op Nederlandse melkveebedrijven door het Schmallenbergvirus [Diarrhea and loss of production on Dutch dairy farms caused by the Schmallenberg virus]. Tijdschr. Voor Diergeneeskd. 2012, 137, 112–115. [Google Scholar]
- Veldhuis, A.; Mars, J.; Stegeman, A.; van Schaik, G. Changing surveillance objectives during the different phases of an emerging vector-borne disease outbreak: The Schmallenberg virus example. Prev. Vet. Med. 2019, 166, 21–27. [Google Scholar] [CrossRef]
- Wernike, K.; Beer, M. Re-circulation of Schmallenberg virus, Germany, 2019. Transbound. Emerg. Dis. 2020, 67, 2290–2295. [Google Scholar] [CrossRef]
- Balenghien, T.; Pagès, N.; Goffredo, M.; Carpenter, S.; Augot, D.; Jacquier, E.; Talavera, S.; Monaco, F.; Depaquit, J.; Grillet, C.; et al. The emergence of Schmallenberg virus across Culicoides communities and ecosystems in Europe. Prev. Vet. Med. 2014, 116, 360–369. [Google Scholar] [CrossRef]
- Sternberg, E.D.; Thomas, M.B. Local adaptation to temperature and the implications for vector-borne diseases. Trends Parasitol. 2014, 30, 115–122. [Google Scholar] [CrossRef]
- Bilk, S.; Schulze, C.; Fischer, M.; Beer, M.; Hlinak, A.; Hoffmann, B. Organ distribution of Schmallenberg virus RNA in malformed newborns. Vet. Microbiol. 2012, 159, 236–238. [Google Scholar] [CrossRef]
- Toussaint, J.F.; Sailleau, C.; Breard, E.; Zientara, S.; De Clercq, K. Bluetongue virus detection by two real-time RT-qPCRs targeting two different genomic segments. J. Virol. Methods 2007, 140, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Mansfield, K.L.; La Rocca, S.A.; Khatri, M.; Johnson, N.; Steinbach, F.; Fooks, A.R. Detection of Schmallenberg virus serum neutralising antibodies. J. Virol. Methods 2013, 188, 139–144. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Martin, D.P.; Varsani, A.; Roumagnac, P.; Botha, G.; Maslamoney, S.; Schwab, T.; Kelz, Z.; Kumar, V.; Murrell, B. RDP5: A Computer Program for Analyzing Recombination in, and Removing Signals of Recombination from, Nucleotide Sequence Datasets. Virus Evol. 2021, 7, 87. [Google Scholar] [CrossRef]
- Posada, D. JModelTest: Phylogenetic Model Averaging. Mol. Biol. Evol. 2008, 25, 1253–1256. [Google Scholar] [CrossRef]
- Schmidt, H.A.; Strimmer, K.; Vingron, M.; Von Haeseler, A. TREE-PUZZLE: Maximum Likelihood Phylogenetic Analysis Using Quartets and Parallel Computing. Bioinformatics 2002, 18, 502–504. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K.; Dudley, J. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Kremer, M. Contribution à l’étude du genre Culicoides Latreille particulièrement en France. Encycl. Entomol. A 1965, 39, 1–299. [Google Scholar]
- Delécolle, J.C. Nouvelle Contribution à L’étude Systématique et Iconographique des Espèces du Genre Culicoides (Diptera: Ceratopogonidae) du Nord–Est de la France. Ph.D. Thesis, Université Louis Pasteur de Strasbourg, Strasbourg, France, 1988. [Google Scholar]
- Glukhova, V.M. Culicoides (Diptera: Ceratopogonidae) of Russian and adjacent lands. Intern. J. Dipterolog. Res. 2005, 16, 3–75. [Google Scholar]
- Dyce, A.L. The recognition of nulliparous and parous Culicoides (Diptera: Ceratopogonidae) without dissection. J. Aust. Entomol. Soc. 1969, 8, 11–15. [Google Scholar] [CrossRef]
- Agerholm, J.S.; Arnbjerg, J.; Andersen, O. Familial chondrodysplasia in Holstein calves. J. Vet. Diagn. Investig. 2004, 16, 293–298. [Google Scholar]
- Lima Bezerra, J.J.; Lucena, R.B. Poisonings in ruminants by Cenostigma pyramidale (Tul.) Gagnon & G.P.Lewis (Fabaceae): A mini-review of teratogenic potential and phytochemical evidence. Toxicon 2024, 246, 107794. [Google Scholar] [CrossRef] [PubMed]
- Catalano, L.W., Jr.; Sever, J.L. The role of viruses as causes of congenital diseases. Annu Rev. Microbiol. 1971, 25, 255–282. [Google Scholar] [CrossRef]
- Wernike, K.; Beer, M. Schmallenberg Virus: A Novel Virus of Veterinary Importance. Adv. Virus Res. 2017, 99, 39–60. [Google Scholar] [CrossRef]
- Gache, K.; Zientara, S.; Collin, E.; Authié, E.; Dion, F.; Garin, E.; Zanella, G.; Calavas, D. Spatial and temporal patterns of Schmallenberg virus in France in 2016. Vet. Rec. 2018, 182, 575. [Google Scholar] [CrossRef] [PubMed]
- Sohier, C.; Deblauwe, I.; Van Loo, T.; Hanon, J.B.; Cay, A.B.; De Regge, N. Evidence of extensive renewed Schmallenberg virus circulation in Belgium during summer of 2016-increase in arthrogryposis-hydranencephaly cases expected. Transbound. Emerg. Dis. 2017, 64, 1015–1019. [Google Scholar] [CrossRef] [PubMed]
- Wernike, K.; Fischer, L.; Twietmeyer, S.; Beer, M. Extensive Schmallenberg virus circulation in Germany, 2023. Vet. Res. 2024, 55, 134. [Google Scholar] [CrossRef]
- Endalew, A.D.; Faburay, B.; Wilson, W.C.; Richt, J.A. Schmallenberg Disease-A Newly Emerged Culicoides-borne Viral Disease of Ruminants. Viruses 2019, 11, 1065. [Google Scholar] [CrossRef]
- Claine, F.; Coupeau, D.; Wiggers, L.; Muylkens, B.; Kirschvink, N. Schmallenberg virus infection of ruminants: Challenges and opportunities for veterinarians. Vet. Med. 2015, 6, 261–272. [Google Scholar] [CrossRef]
- Wernike, K.; Holsteg, M.; Sasserath, M.; Beer, M. Schmallenberg virus antibody development and decline in a naturally infected dairy cattle herd in Germany, 2011–2014. Vet. Microbiol. 2015, 181, 294–297. [Google Scholar] [CrossRef] [PubMed]
- Larska, M.; Polak, M.P.; Grochowska, M.; Lechowski, L.; Związek, J.S.; Zmudziński, J.F. First report of Schmallenberg virus infection in cattle and midges in Poland. Transbound. Emerg. Dis. 2013, 60, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Elbers, A.R.; Meiswinkel, R.; van Weezep, E.; Kooi, E.A.; van der Poel, W.H. Schmallenberg Virus in Culicoides Biting Midges in the Netherlands in 2012. Transbound. Emerg. Dis. 2015, 62, 339–342. [Google Scholar] [CrossRef] [PubMed]
- Goffredo, M.; Catalani, M.; Federici, V.; Portanti, O.; Marini, V.; Mancini, G.; Quaglia, M.; Santilli, A.; Teodori, L.; Savini, G. Vector species of Culicoides midges implicated in the 2012–2014 Bluetongue epidemics in Italy. Vet. Ital. 2015, 51, 131–138. [Google Scholar] [CrossRef]
- Foxi, C.; Delrio, G.; Falchi, G.; Marche, M.G.; Satta, G.; Ruiu, L. Role of different Culicoides vectors (Diptera: Ceratopogonidae) in bluetongue virus transmission and overwintering in Sardinia (Italy). Parasites Vectors 2016, 9, 440. [Google Scholar] [CrossRef]
- Boorman, J. Culicoides (Diptera: Ceratopogonidae) from Cyprus. Cah. ORSTOM Serv. Entomol. Med. Parasitol. 1974, 12, 7–13. [Google Scholar]
- Foxi, C.; Delrio, G. Larval habitats and seasonal abundance of Culicoides biting midges found in association with sheep in northern Sardinia, Italy. Med. Vet. Entomol. 2010, 24, 199–209. [Google Scholar] [CrossRef]
- Foxi, C.; Pinna, M.; Monteys, V.S.I.; Delrio, G. An updated checklist of the Culicoides Latreille (Diptera: Ceratopogonidae) of Sardinia (Italy), and seasonality in proven and potential vectors for bluetongue virus (BTV). Proc. Entomol. Soc. Wash. 2011, 113, 403–416. [Google Scholar] [CrossRef]
- Kieffer, J.J. Ceratopogonines recueillis au Sahara constantinois. Arch. Inst. Pasteur D’algérie 1923, 1, 654–683. [Google Scholar]
- Boorman, J. Culicoides (Diptera: Ceratopogonidae) of the Arabian peninsula with notes on their medical and veterinary importance. Fauna Saudi Arab. 1989, 10, 160–224. [Google Scholar]
- Veronesi, E.; Henstock, M.; Gubbins, S.; Batten, C.; Manley, R.; Barber, J.; Hoffmann, B.; Beer, M.; Attoui, H.; Mertens, P.P.; et al. Implicating Culicoides biting midges as vectors of Schmallenberg virus using semi-quantitative RT-PCR. PLoS ONE 2013, 8, e57747. [Google Scholar] [CrossRef] [PubMed]
- Sick, F.; Zeiske, S.; Beer, M.; Wernike, K. Characterization of a natural ‘dead-end’ variant of Schmallenberg virus. J. Gen. Virol. 2024, 105, 002005. [Google Scholar] [CrossRef]
- McGowan, S.L.; La Rocca, S.A.; Grierson, S.S.; Dastjerdi, A.; Choudhury, B.; Steinbach, F. Incursion of Schmallenberg virus into Great Britain in 2011 and emergence of variant sequences in 2016. Vet. J. 2018, 234, 77–84. [Google Scholar] [CrossRef]
- Barry, G.; Varela, M.; Ratinier, M.; Blomström, A.L.; Caporale, M.; Seehusen, F.; Hahn, K.; Schnettler, E.; Baumgärtner, W.; Kohl, A.; et al. NSs protein of Schmallenberg virus counteracts the antiviral response of the cell by inhibiting its transcriptional machinery. J. Gen. Virol. 2014, 95 Pt 8, 1640–1646. [Google Scholar] [CrossRef]
- Kraatz, F.; Wernike, K.; Hechinger, S.; König, P.; Granzow, H.; Reimann, I.; Beer, M. Deletion mutants of Schmallenberg virus are avirulent and protect from virus challenge. J. Virol. 2015, 89, 1825–1837. [Google Scholar] [CrossRef] [PubMed]
- Varela, M.; Pinto, R.M.; Caporale, M.; Piras, I.M.; Taggart, A.; Seehusen, F.; Hahn, K.; Janowicz, A.; de Souza, W.M.; Baumgärtner, W.; et al. Mutations in the Schmallenberg Virus Gc Glycoprotein Facilitate Cellular Protein Synthesis Shutoff and Restore Pathogenicity of NSs Deletion Mutants in Mice. J. Virol. 2016, 90, 5440–5450. [Google Scholar] [CrossRef]
- Fischer, M.; Hoffmann, B.; Goller, K.V.; Höper, D.; Wernike, K.; Beer, M. A mutation ‘hot spot’ in the Schmallenberg virus M segment. J. Gen. Virol. 2013, 94 Pt 6, 1161–1167. [Google Scholar] [CrossRef]
| Years | Species | No. Examined Farms | No. Farms with Malformed Newborns | No. Farms with RT-PCR Positive Newborns |
|---|---|---|---|---|
| 2013–2015 | Sheep | 91 | 35 | 0 |
| Cattle | 20 | 5 | 0 | |
| Goat | 12 | 1 | 1 | |
| 2016–2018 | Sheep | 49 | 12 | 1 |
| Cattle | 4 | 0 | 0 | |
| Goat | 4 | 0 | 0 | |
| 2019–2021 | Sheep | 9 | 4 | 3 |
| Cattle | 2 | 0 | 0 | |
| Goat | 2 | 0 | 0 | |
| 2022–2023 | Sheep | 11 | 8 | 1 |
| Cattle | 7 | 0 | 0 | |
| Goat | 2 | 1 | 0 |
| Year | Host | No. Tested Samples (% Positive) | No. Tested Farms (% Positive) |
|---|---|---|---|
| 2013 | sheep | 7348 (33.89) | 206 (79.61) |
| cattle | 1735 (69.74) | 51 (92.16) | |
| goat | 957 (29.15) | 42 (85.71) | |
| 2014 | sheep | 3516 (27.93) | 65 (87.69) |
| cattle | 1007 (90.37) | 18 (88.89) | |
| goat | 98 (19.39) | 4 (N.A.) | |
| 2015 | sheep | 1623 (7.33) | 47 (46.81) |
| cattle | 418 (25.84) | 17 (76.47) | |
| goat | 380 (10.00) | 2 (N.A.) | |
| 2016 | sheep | 611 (7.36) | 32 (28.13) |
| cattle | 613 (22.68) | 12 (91.67) | |
| goat | 421 (8.31) | 9 (N.A.) | |
| 2017 | sheep | 109 (10.09) | 23 (34.78) |
| cattle | 124 (46.77) | 14 (64.29) | |
| goat | 1 (0.00) | 1 (N.A.) | |
| 2018 | sheep | 118 (7.63) | 19 (26.32) |
| cattle | 104 (42.31) | 19 (68.42) | |
| goat | 48 (0.00) | 2 (N.A.) | |
| 2019 | sheep | 13 (7.69) | 3 (N.A.) |
| cattle | 55 (67.27) | 6 (N.A.) | |
| goat | 20 (15.00) | 2 (N.A.) | |
| 2020 | sheep | 30 (13.33) | 5 (N.A.) |
| cattle | 103 (36.89) | 12 (83.33) | |
| goat | 17 (11.76) | 2 (N.A.) | |
| 2021 | sheep | 140 (48.57) | 11 (54.55) |
| cattle | 102 (90.20) | 15 (100) | |
| 2022 | sheep | 34 (8.82) | 7 (N.A.) |
| cattle | 34 (64.71) | 6 (N.A.) | |
| goat | 2 (N.A.) | 1 (N.A.) | |
| 2023 | sheep | 79 (2.53) | 3 (N.A.) |
| cattle | 23 (95.65) | 4 (N.A.) |
| Year | No. Sheep/% Positive | C.I. 95% |
|---|---|---|
| 2022 | 311/16.40 | 12.28–20.52 |
| 2024 | 339/21.53 | 17.15–25.91 |
| Species | Municipalities | |||||||
|---|---|---|---|---|---|---|---|---|
| Alghero (25) | Arborea (21) | Iglesias (31) | Palau (32) | Quartu S.E. (30) | S.A. Arresi (29) | Siniscola (30) | Villacidro (28) | |
| C. imicola | 1012 | 1449 | 8765 | 45,279 | 2266 | 1202 | 11,297 | 3391 |
| C. newsteadi | 1631 | 358 | 1847 | 36,238 | 106 | 391 | 5318 | 1326 |
| C. circumscriptus | 79 | 13 | 340 | 432 | 14 | 1710 | 245 | 236 |
| C. jumineri | 71 | 82 | 103 | 430 | 8 | 165 | 102 | 1662 |
| C. cataneii | 227 | 7 | 177 | 477 | 13 | 899 | 59 | 96 |
| Obsoletus gr. | 79 | 27 | 318 | 377 | 125 | 2 | 91 | 21 |
| C. kibunensis | 28 | 0 | 36 | 270 | 31 | 4 | 22 | 2 |
| C. punctatus | 37 | 41 | 67 | 198 | 6 | 28 | 192 | 293 |
| C. sahariensis | 22 | 1 | 85 | 168 | 2 | 160 | 14 | 49 |
| C. pulicaris | 333 | 0 | 4 | 82 | 10 | 0 | 3 | 7 |
| C. univittatus | 0 | 0 | 31 | 154 | 7 | 240 | 0 | 2 |
| C. paolae | 23 | 4 | 34 | 10 | 13 | 185 | 17 | 33 |
| C. festivipennis | 32 | 0 | 13 | 33 | 2 | 0 | 1 | 0 |
| C. puncticollis | 27 | 0 | 1 | 4 | 0 | 29 | 4 | 0 |
| C. maritimus | 0 | 3 | 0 | 0 | 0 | 43 | 3 | 0 |
| C. parroti | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| C. subfagineus | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| C. brunnicans | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| C. lupicaris | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total | 3605 | 1985 | 11,822 | 84,152 | 2603 | 5058 | 17,368 | 7118 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Foxi, C.; Pintus, D.; Zinellu, S.; Macciocu, S.; Angioi, P.P.; Sechi, A.M.; Fiori, M.S.; Ladu, A.; Puggioni, G.; Denti, S.; et al. Assessing Schmallenberg Virus Disease in Sardinia (Italy) After the First Epidemic Episode in 2012. Pathogens 2025, 14, 349. https://doi.org/10.3390/pathogens14040349
Foxi C, Pintus D, Zinellu S, Macciocu S, Angioi PP, Sechi AM, Fiori MS, Ladu A, Puggioni G, Denti S, et al. Assessing Schmallenberg Virus Disease in Sardinia (Italy) After the First Epidemic Episode in 2012. Pathogens. 2025; 14(4):349. https://doi.org/10.3390/pathogens14040349
Chicago/Turabian StyleFoxi, Cipriano, Davide Pintus, Susanna Zinellu, Simonetta Macciocu, Pier Paolo Angioi, Anna Maria Sechi, Mariangela Stefania Fiori, Anna Ladu, Graziella Puggioni, Stefano Denti, and et al. 2025. "Assessing Schmallenberg Virus Disease in Sardinia (Italy) After the First Epidemic Episode in 2012" Pathogens 14, no. 4: 349. https://doi.org/10.3390/pathogens14040349
APA StyleFoxi, C., Pintus, D., Zinellu, S., Macciocu, S., Angioi, P. P., Sechi, A. M., Fiori, M. S., Ladu, A., Puggioni, G., Denti, S., Sanna, M. L., Madrau, M. P., Satta, G., Oggiano, A., Ligios, C., & Dei Giudici, S. (2025). Assessing Schmallenberg Virus Disease in Sardinia (Italy) After the First Epidemic Episode in 2012. Pathogens, 14(4), 349. https://doi.org/10.3390/pathogens14040349






