Immunogenicity of Trypanosoma cruzi Multi-Epitope Recombinant Protein as an Antigen Candidate for Chagas Disease Vaccine in Humans
Abstract
1. Introduction
2. Materials and Methods
2.1. Trypanosoma cruzi Epitope Prediction from Databases
2.2. Epitope Validation
2.3. Trypanosoma cruzi-Epitopes Conservation
2.4. Docking of T. cruzi Epitopes to HLA Class I Molecules
2.5. In Silico Design of the Multi-Epitope Recombinant Protein
2.6. Expression of the Multi-Epitope Protein
2.7. Multi-Epitope Recombinant Protein Validation
2.8. Data Analysis
2.9. Ethical Considerations
3. Results
3.1. Identification of T. cruzi Epitopes to HLA-A*02:01
3.2. Epitopes Induced IFN-γ in PBMC from Chagasic Patients
3.3. Epitopes Conserved in Multiples DTUs of T. cruzi
3.4. Promiscuous Epitopes and Population Coverage
3.5. Protein Expression of Multi-Epitope in E. coli
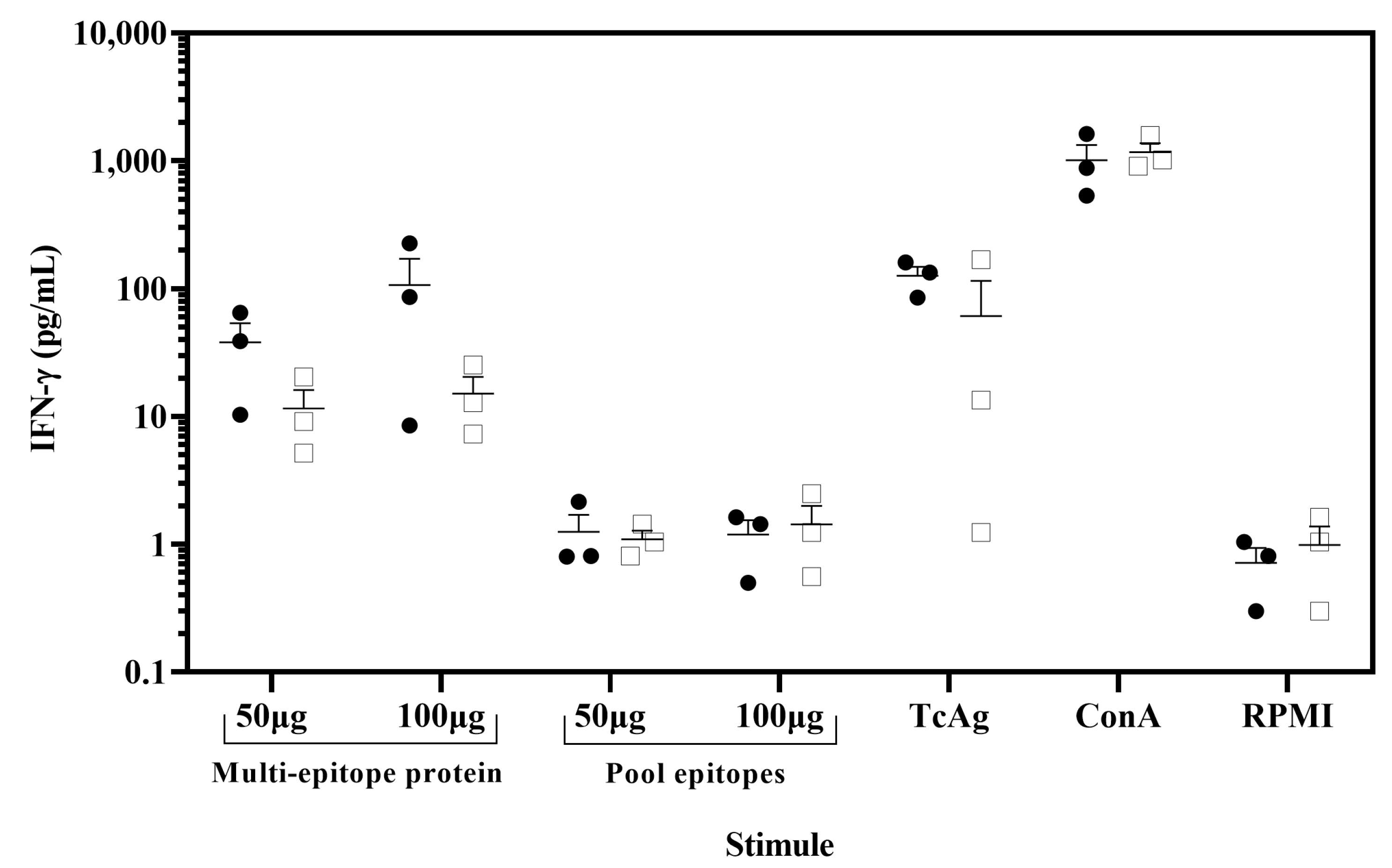
3.6. Immunogenicity of the Multi-Epitope Protein in Chagasic Patients
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- IHME. Chagas Disease—Level 3 Cause. Institute for Health Metrics and Evaluation. Available online: https://www.healthdata.org/research-analysis/diseases-injuries-risks/factsheets/2021-chagas-disease-level-3-disease (accessed on 1 August 2024).
- Bern, C. Chagas’ Disease. N. Engl. J. Med. 2015, 373, 1882. [Google Scholar] [CrossRef] [PubMed]
- Perez-Molina, J.A.; Crespillo-Andujar, C.; Bosch-Nicolau, P.; Molina, I. Trypanocidal treatment of Chagas disease. Enfermedades Infecc. Y Microbiol. Clin. Engl. Ed. 2020, 39, 458–470. [Google Scholar] [CrossRef]
- Robinson, J.; Halliwell, J.A.; Hayhurst, J.D.; Flicek, P.; Parham, P.; Marsh, S.G. The IPD and IMGT/HLA database: Allele variant databases. Nucleic Acids Res. 2015, 43, D423–D431. [Google Scholar] [CrossRef] [PubMed]
- Maranon, C.; Thomas, M.C.; Planelles, L.; Lopez, M.C. The immunization of A2/K(b) transgenic mice with the KMP11-HSP70 fusion protein induces CTL response against human cells expressing the T. cruzi KMP11 antigen: Identification of A2-restricted epitopes. Mol. Immunol. 2001, 38, 279–287. [Google Scholar] [CrossRef]
- Wizel, B.; Palmieri, M.; Mendoza, C.; Arana, B.; Sidney, J.; Sette, A.; Tarleton, R. Human infection with Trypanosoma cruzi induces parasite antigen-specific cytotoxic T lymphocyte responses. J. Clin. Investig. 1998, 102, 1062–1071. [Google Scholar] [CrossRef]
- Laucella, S.A.; Postan, M.; Martin, D.; Hubby Fralish, B.; Albareda, M.C.; Alvarez, M.G.; Lococo, B.; Barbieri, G.; Viotti, R.J.; Tarleton, R.L. Frequency of interferon- gamma -producing T cells specific for Trypanosoma cruzi inversely correlates with disease severity in chronic human Chagas disease. J. Infect. Dis. 2004, 189, 909–918. [Google Scholar] [CrossRef]
- Egui, A.; Thomas, M.C.; Morell, M.; Maranon, C.; Carrilero, B.; Segovia, M.; Puerta, C.J.; Pinazo, M.J.; Rosas, F.; Gascon, J.; et al. Trypanosoma cruzi paraflagellar rod proteins 2 and 3 contain immunodominant CD8(+) T-cell epitopes that are recognized by cytotoxic T cells from Chagas disease patients. Mol. Immunol. 2012, 52, 289–298. [Google Scholar] [CrossRef]
- Maranon, C.; Egui, A.; Carrilero, B.; Thomas, M.C.; Pinazo, M.J.; Gascon, J.; Segovia, M.; Lopez, M.C. Identification of HLA-A *02:01-restricted CTL epitopes in Trypanosoma cruzi heat shock protein-70 recognized by Chagas disease patients. Microbes Infect. 2011, 13, 1025–1032. [Google Scholar] [CrossRef]
- Fonseca, S.G.; Moins-Teisserenc, H.; Clave, E.; Ianni, B.; Nunes, V.L.; Mady, C.; Iwai, L.K.; Sette, A.; Sidney, J.; Marin, M.L.; et al. Identification of multiple HLA-A*0201-restricted cruzipain and FL-160 CD8+ epitopes recognized by T cells from chronically Trypanosoma cruzi-infected patients. Microbes Infect. 2005, 7, 688–697. [Google Scholar] [CrossRef]
- Al Zamane, S.; Nobel, F.A.; Jebin, R.A.; Amin, M.B.; Somadder, P.D.; Antora, N.J.; Hossain, M.I.; Islam, M.J.; Ahmed, K.; Moni, M.A. Development of an in silico multi-epitope vaccine against SARS-COV-2 by precised immune-informatics approaches. Inform. Med. Unlocked 2021, 27, 100781. [Google Scholar] [CrossRef]
- Pandey, R.K.; Ojha, R.; Aathmanathan, V.S.; Krishnan, M.; Prajapati, V.K. Immunoinformatics approaches to design a novel multi-epitope subunit vaccine against HIV infection. Vaccine 2018, 36, 2262–2272. [Google Scholar] [CrossRef] [PubMed]
- Tahir Ul Qamar, M.; Ahmad, S.; Fatima, I.; Ahmad, F.; Shahid, F.; Naz, A.; Abbasi, S.W.; Khan, A.; Mirza, M.U.; Ashfaq, U.A.; et al. Designing multi-epitope vaccine against Staphylococcus aureus by employing subtractive proteomics, reverse vaccinology and immuno-informatics approaches. Comput. Biol. Med. 2021, 132, 104389. [Google Scholar] [CrossRef] [PubMed]
- Maharaj, L.; Adeleke, V.T.; Fatoba, A.J.; Adeniyi, A.A.; Tshilwane, S.I.; Adeleke, M.A.; Maharaj, R.; Okpeku, M. Immunoinformatics approach for multi-epitope vaccine design against P. falciparum malaria. Infect. Genet. Evol. 2021, 92, 104875. [Google Scholar] [CrossRef] [PubMed]
- Lage, D.P.; Ribeiro, P.A.F.; Dias, D.S.; Mendonca, D.V.C.; Ramos, F.F.; Carvalho, L.M.; de Oliveira, D.; Steiner, B.T.; Martins, V.T.; Perin, L.; et al. A candidate vaccine for human visceral leishmaniasis based on a specific T cell epitope-containing chimeric protein protects mice against Leishmania infantum infection. npj Vaccines 2020, 5, 75. [Google Scholar] [CrossRef]
- Khatoon, N.; Ojha, R.; Mishra, A.; Prajapati, V.K. Examination of antigenic proteins of Trypanosoma cruzi to fabricate an epitope-based subunit vaccine by exploiting epitope mapping mechanism. Vaccine 2018, 36, 6290–6300. [Google Scholar] [CrossRef]
- Michel-Todo, L.; Reche, P.A.; Bigey, P.; Pinazo, M.J.; Gascon, J.; Alonso-Padilla, J. In silico Design of an Epitope-Based Vaccine Ensemble for Chagas Disease. Front. Immunol. 2019, 10, 2698. [Google Scholar] [CrossRef]
- Rawal, K.; Sinha, R.; Abbasi, B.A.; Chaudhary, A.; Nath, S.K.; Kumari, P.; Preeti, P.; Saraf, D.; Singh, S.; Mishra, K.; et al. Identification of vaccine targets in pathogens and design of a vaccine using computational approaches. Sci. Rep. 2021, 11, 17626. [Google Scholar] [CrossRef]
- Gonzalez-Lopez, C.; Chen, W.H.; Alfaro-Chacon, A.; Villanueva-Lizama, L.E.; Rosado-Vallado, M.; Ramirez-Sierra, M.J.; Teh-Poot, C.F.; Pollet, J.; Asojo, O.; Jones, K.M.; et al. A novel multi-epitope recombinant protein elicits an antigen-specific CD8+ T cells response in Trypanosoma cruzi-infected mice. Vaccine 2022, 40, 6445–6449. [Google Scholar] [CrossRef]
- Teh-Poot, C.; Tzec-Arjona, E.; Martinez-Vega, P.; Ramirez-Sierra, M.J.; Rosado-Vallado, M.; Dumonteil, E. From genome screening to creation of vaccine against Trypanosoma cruzi by use of immunoinformatics. J. Infect. Dis. 2015, 211, 258–266. [Google Scholar] [CrossRef]
- Barquera, R.; Hernandez-Zaragoza, D.I.; Bravo-Acevedo, A.; Arrieta-Bolanos, E.; Clayton, S.; Acuna-Alonzo, V.; Martinez-Alvarez, J.C.; Lopez-Gil, C.; Adalid-Sainz, C.; Vega-Martinez, M.D.R.; et al. The immunogenetic diversity of the HLA system in Mexico correlates with underlying population genetic structure. Hum. Immunol. 2020, 81, 461–474. [Google Scholar] [CrossRef]
- Single, R.M.; Meyer, D.; Nunes, K.; Francisco, R.S.; Hunemeier, T.; Maiers, M.; Hurley, C.K.; Bedoya, G.; Gallo, C.; Hurtado, A.M.; et al. Demographic history and selection at HLA loci in Native Americans. PLoS ONE 2020, 15, e0241282. [Google Scholar] [CrossRef]
- Andreatta, M.; Nielsen, M. Gapped sequence alignment using artificial neural networks: Application to the MHC class I system. Bioinformatics 2016, 32, 511–517. [Google Scholar] [CrossRef] [PubMed]
- Reche, P.A.; Reinherz, E.L. Prediction of peptide-MHC binding using profiles. Methods Mol. Biol. 2007, 409, 185–200. [Google Scholar] [CrossRef] [PubMed]
- Rammensee, H.; Bachmann, J.; Emmerich, N.P.; Bachor, O.A.; Stevanovic, S. SYFPEITHI: Database for MHC ligands and peptide motifs. Immunogenetics 1999, 50, 213–219. [Google Scholar]
- Parker, K.C.; Bednarek, M.A.; Coligan, J.E. Scheme for ranking potential HLA-A2 binding peptides based on independent binding of individual peptide side-chains. J. Immunol. 1994, 152, 163–175. [Google Scholar] [CrossRef]
- Singh, H.; Raghava, G.P. ProPred1: Prediction of promiscuous MHC Class-I binding sites. Bioinformatics 2003, 19, 1009–1014. [Google Scholar] [CrossRef]
- Lata, S.; Bhasin, M.; Raghava, G.P. Application of machine learning techniques in predicting MHC binders. Methods Mol. Biol. 2007, 409, 201–215. [Google Scholar] [CrossRef]
- Schueler-Furman, O.; Altuvia, Y.; Sette, A.; Margalit, H. Structure-based prediction of binding peptides to MHC class I molecules: Application to a broad range of MHC alleles. Protein Sci. 2000, 9, 1838–1846. [Google Scholar] [CrossRef]
- Kim, Y.; Ponomarenko, J.; Zhu, Z.; Tamang, D.; Wang, P.; Greenbaum, J.; Lundegaard, C.; Sette, A.; Lund, O.; Bourne, P.E.; et al. Immune epitope database analysis resource. Nucleic Acids Res. 2012, 40, W525–W530. [Google Scholar] [CrossRef]
- Bhasin, M.; Raghava, G.P. Prediction of CTL epitopes using QM, SVM and ANN techniques. Vaccine 2004, 22, 3195–3204. [Google Scholar] [CrossRef]
- Hattotuwagama, C.K.; Guan, P.; Doytchinova, I.A.; Zygouri, C.; Flower, D.R. Quantitative online prediction of peptide binding to the major histocompatibility complex. J. Mol. Graph. Model. 2004, 22, 195–207. [Google Scholar] [CrossRef] [PubMed]
- Doytchinova, I.A.; Guan, P.; Flower, D.R. EpiJen: A server for multistep T cell epitope prediction. BMC Bioinform. 2006, 7, 131. [Google Scholar] [CrossRef]
- Larsen, M.V.; Lundegaard, C.; Lamberth, K.; Buus, S.; Lund, O.; Nielsen, M. Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinform. 2007, 8, 424. [Google Scholar] [CrossRef]
- Stranzl, T.; Larsen, M.V.; Lundegaard, C.; Nielsen, M. NetCTLpan: Pan-specific MHC class I pathway epitope predictions. Immunogenetics 2010, 62, 357–368. [Google Scholar] [CrossRef]
- Karosiene, E.; Lundegaard, C.; Lund, O.; Nielsen, M. NetMHCcons: A consensus method for the major histocompatibility complex class I predictions. Immunogenetics 2012, 64, 177–186. [Google Scholar] [CrossRef]
- Erup Larsen, M.; Kloverpris, H.; Stryhn, A.; Koofhethile, C.K.; Sims, S.; Ndung’u, T.; Goulder, P.; Buus, S.; Nielsen, M. HLArestrictor-a tool for patient-specific predictions of HLA restriction elements and optimal epitopes within peptides. Immunogenetics 2011, 63, 43–55. [Google Scholar] [CrossRef]
- Teh-Poot, C.; Dumonteil, E. Mining Trypanosoma cruzi Genome Sequences for Antigen Discovery and Vaccine Development. Methods Mol. Biol. 2019, 1955, 23–34. [Google Scholar] [CrossRef]
- Singh, H.; Raghava, G.P. BLAST-based structural annotation of protein residues using Protein Data Bank. Biol. Direct 2016, 11, 4. [Google Scholar] [CrossRef]
- Villanueva-Lizama, L.E.; Cruz-Chan, J.V.; Aguilar-Cetina, A.D.C.; Herrera-Sanchez, L.F.; Rodriguez-Perez, J.M.; Rosado-Vallado, M.E.; Ramirez-Sierra, M.J.; Ortega-Lopez, J.; Jones, K.; Hotez, P.; et al. Trypanosoma cruzi vaccine candidate antigens Tc24 and TSA-1 recall memory immune response associated with HLA-A and -B supertypes in Chagasic chronic patients from Mexico. PLoS Negl. Trop. Dis. 2018, 12, e0006240. [Google Scholar] [CrossRef]
- Aslett, M.; Aurrecoechea, C.; Berriman, M.; Brestelli, J.; Brunk, B.P.; Carrington, M.; Depledge, D.P.; Fischer, S.; Gajria, B.; Gao, X.; et al. TriTrypDB: A functional genomic resource for the Trypanosomatidae. Nucleic Acids Res. 2010, 38, D457–D462. [Google Scholar] [CrossRef]
- Kurcinski, M.; Badaczewska-Dawid, A.; Kolinski, M.; Kolinski, A.; Kmiecik, S. Flexible docking of peptides to proteins using CABS-dock. Protein Sci. 2020, 29, 211–222. [Google Scholar] [CrossRef] [PubMed]
- Kurcinski, M.; Jamroz, M.; Blaszczyk, M.; Kolinski, A.; Kmiecik, S. CABS-dock web server for the flexible docking of peptides to proteins without prior knowledge of the binding site. Nucleic Acids Res. 2015, 43, W419–W424. [Google Scholar] [CrossRef] [PubMed]
- Mashiach, E.; Schneidman-Duhovny, D.; Andrusier, N.; Nussinov, R.; Wolfson, H.J. FireDock: A web server for fast interaction refinement in molecular docking. Nucleic Acids Res. 2008, 36, W229–W232. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Sidney, J.; Peters, B.; Frahm, N.; Brander, C.; Sette, A. HLA class I supertypes: A revised and updated classification. BMC Immunol. 2008, 9, 1. [Google Scholar] [CrossRef]
- Diaz, A.A.; Tomba, E.; Lennarson, R.; Richard, R.; Bagajewicz, M.J.; Harrison, R.G. Prediction of protein solubility in Escherichia coli using logistic regression. Biotechnol. Bioeng. 2010, 105, 374–383. [Google Scholar] [CrossRef]
- Magnan, C.N.; Randall, A.; Baldi, P. SOLpro: Accurate sequence-based prediction of protein solubility. Bioinformatics 2009, 25, 2200–2207. [Google Scholar] [CrossRef]
- Hebditch, M.; Carballo-Amador, M.A.; Charonis, S.; Curtis, R.; Warwicker, J. Protein-Sol: A web tool for predicting protein solubility from sequence. Bioinformatics 2017, 33, 3098–3100. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.e.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Springer: Berlin/Heidelberg, Germany, 2005; pp. 571–607. [Google Scholar] [CrossRef]
- Doytchinova, I.A.; Flower, D.R. VaxiJen: A server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform. 2007, 8, 4. [Google Scholar] [CrossRef]
- Dimitrov, I.; Bangov, I.; Flower, D.R.; Doytchinova, I. AllerTOP v.2--a server for in silico prediction of allergens. J. Mol. Model. 2014, 20, 2278. [Google Scholar] [CrossRef]
- Saha, S.; Raghava, G.P. AlgPred: Prediction of allergenic proteins and mapping of IgE epitopes. Nucleic Acids Res. 2006, 34, W202–W209. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Wang, S.; Zhao, F.; Xu, J. Protein threading using context-specific alignment potential. Bioinformatics 2013, 29, i257–i265. [Google Scholar] [CrossRef] [PubMed]
- Bui, H.H.; Sidney, J.; Dinh, K.; Southwood, S.; Newman, M.J.; Sette, A. Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC Bioinform. 2006, 7, 153. [Google Scholar] [CrossRef]
- Dumonteil, E.; Herrera, C. The Case for the Development of a Chagas Disease Vaccine: Why? How? When? Trop. Med. Infect. Dis. 2021, 6, 16. [Google Scholar] [CrossRef]
- Bae, J.; Smith, R.; Daley, J.; Mimura, N.; Tai, Y.T.; Anderson, K.C.; Munshi, N.C. Myeloma-specific multiple peptides able to generate cytotoxic T lymphocytes: A potential therapeutic application in multiple myeloma and other plasma cell disorders. Clin. Cancer Res. 2012, 18, 4850–4860. [Google Scholar] [CrossRef]
- Mahajan, B.; Berzofsky, J.A.; Boykins, R.A.; Majam, V.; Zheng, H.; Chattopadhyay, R.; de la Vega, P.; Moch, J.K.; Haynes, J.D.; Belyakov, I.M.; et al. Multiple antigen peptide vaccines against Plasmodium falciparum malaria. Infect. Immun. 2010, 78, 4613–4624. [Google Scholar] [CrossRef]
- Pishraft Sabet, L.; Taheri, T.; Memarnejadian, A.; Mokhtari Azad, T.; Asgari, F.; Rahimnia, R.; Alavian, S.M.; Rafati, S.; Samimi Rad, K. Immunogenicity of multi-epitope DNA and peptide vaccine candidates based on Core, E2, NS3 and NS5B HCV epitopes in BALB/c mice. Hepat. Mon. 2014, 14, e22215. [Google Scholar] [CrossRef]
- Sanchez Alberti, A.; Bivona, A.E.; Cerny, N.; Schulze, K.; Weissmann, S.; Ebensen, T.; Morales, C.; Padilla, A.M.; Cazorla, S.I.; Tarleton, R.L.; et al. Engineered trivalent immunogen adjuvanted with a STING agonist confers protection against Trypanosoma cruzi infection. npj Vaccines 2017, 2, 9. [Google Scholar] [CrossRef]
- Pacini, M.F.; Perdomini, A.; Bulfoni Balbi, C.; Dinatale, B.; Herrera, F.E.; Perez, A.R.; Marcipar, I. The high identity of the Trypanosoma cruzi Group-I of trans-sialidases points them as promising vaccine immunogens. Proteins 2023, 91, 1444–1460. [Google Scholar] [CrossRef]
- Islam, S.I.; Sanjida, S.; Ahmed, S.S.; Almehmadi, M.; Allahyani, M.; Aljuaid, A.; Alsaiari, A.A.; Halawi, M. Core Proteomics and Immunoinformatic Approaches to Design a Multiepitope Reverse Vaccine Candidate against Chagas Disease. Vaccines 2022, 10, 1669. [Google Scholar] [CrossRef] [PubMed]
- Climaco, M.C.; de Figueiredo, L.A.; Lucas, R.C.; Pinheiro, G.R.G.; Dias Magalhaes, L.M.; Oliveira, A.L.G.; Almeida, R.M.; Barbosa, F.S.; Castanheira Bartholomeu, D.; Bueno, L.L.; et al. Development of chimeric protein as a multivalent vaccine for human Kinetoplastid infections: Chagas disease and leishmaniasis. Vaccine 2023, 41, 5400–5411. [Google Scholar] [CrossRef] [PubMed]
- de la Cruz, J.J.; Villanueva-Lizama, L.; Dzul-Huchim, V.; Ramirez-Sierra, M.J.; Martinez-Vega, P.; Rosado-Vallado, M.; Ortega-Lopez, J.; Flores-Pucheta, C.I.; Gillespie, P.; Zhan, B.; et al. Production of recombinant TSA-1 and evaluation of its potential for the immuno-therapeutic control of Trypanosoma cruzi infection in mice. Hum. Vaccin. Immunother. 2019, 15, 210–219. [Google Scholar] [CrossRef]
- Martinez-Campos, V.; Martinez-Vega, P.; Ramirez-Sierra, M.J.; Rosado-Vallado, M.; Seid, C.A.; Hudspeth, E.M.; Wei, J.; Liu, Z.; Kwityn, C.; Hammond, M.; et al. Expression, purification, immunogenicity, and protective efficacy of a recombinant Tc24 antigen as a vaccine against Trypanosoma cruzi infection in mice. Vaccine 2015, 33, 4505–4512. [Google Scholar] [CrossRef]
- Macaluso, G.; Grippi, F.; Di Bella, S.; Blanda, V.; Gucciardi, F.; Torina, A.; Guercio, A.; Cannella, V. A Review on the Immunological Response against Trypanosoma cruzi. Pathogens 2023, 12, 282. [Google Scholar] [CrossRef]
- Tarleton, R.L. CD8+ T cells in Trypanosoma cruzi infection. Semin. Immunopathol. 2015, 37, 233–238. [Google Scholar] [CrossRef]
- da Costa, K.M.; Marques da Fonseca, L.; Dos Reis, J.S.; Santos, M.; Previato, J.O.; Mendonca-Previato, L.; Freire-de-Lima, L. Trypanosoma cruzi trans-Sialidase as a Potential Vaccine Target Against Chagas Disease. Front. Cell. Infect. Microbiol. 2021, 11, 768450. [Google Scholar] [CrossRef]
- Garg, N.J. An Update on Vaccines Against Trypanosoma cruzi and Chagas Disease. Pathogens 2025, 14, 124. [Google Scholar] [CrossRef]
- Acosta-Serrano, A.; Almeida, I.C.; Freitas-Junior, L.H.; Yoshida, N.; Schenkman, S. The mucin-like glycoprotein super-family of Trypanosoma cruzi: Structure and biological roles. Mol. Biochem. Parasitol. 2001, 114, 143–150. [Google Scholar] [CrossRef]
- Alvarez, M.G.; Postan, M.; Weatherly, D.B.; Albareda, M.C.; Sidney, J.; Sette, A.; Olivera, C.; Armenti, A.H.; Tarleton, R.L.; Laucella, S.A. HLA Class I-T cell epitopes from trans-sialidase proteins reveal functionally distinct subsets of CD8+ T cells in chronic Chagas disease. PLoS Negl. Trop. Dis. 2008, 2, e288. [Google Scholar] [CrossRef]
- Majeau, A.; Murphy, L.; Herrera, C.; Dumonteil, E. Assessing Trypanosoma cruzi Parasite Diversity through Comparative Genomics: Implications for Disease Epidemiology and Diagnostics. Pathogens 2021, 10, 212. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Peng, D.; Baptista, R.P.; Li, Y.; Kissinger, J.C.; Tarleton, R.L. Strain-specific genome evolution in Trypanosoma cruzi, the agent of Chagas disease. PLoS Pathog. 2021, 17, e1009254. [Google Scholar] [CrossRef] [PubMed]
- Santos, E.; McCabe, A.; Gonzalez-Galarza, F.F.; Jones, A.R.; Middleton, D. Allele Frequencies Net Database: Improvements for storage of individual genotypes and analysis of existing data. Hum. Immunol. 2016, 77, 238–248. [Google Scholar] [CrossRef] [PubMed]
- Ayo, C.M.; Dalalio, M.M.; Visentainer, J.E.; Reis, P.G.; Sippert, E.A.; Jarduli, L.R.; Alves, H.V.; Sell, A.M. Genetic susceptibility to Chagas disease: An overview about the infection and about the association between disease and the immune response genes. Biomed. Res. Int. 2013, 2013, 284729. [Google Scholar] [CrossRef]
- Zingales, B. Trypanosoma cruzi genetic diversity: Something new for something known about Chagas disease manifestations, serodiagnosis and drug sensitivity. Acta Trop. 2018, 184, 38–52. [Google Scholar] [CrossRef]
- Silvestrini, M.M.A.; Alessio, G.D.; Frias, B.E.D.; Sales Junior, P.A.; Araujo, M.S.S.; Silvestrini, C.M.A.; Brito Alvim de Melo, G.E.; Martins-Filho, O.A.; Teixeira-Carvalho, A.; Martins, H.R. New insights into Trypanosoma cruzi genetic diversity, and its influence on parasite biology and clinical outcomes. Front. Immunol. 2024, 15, 1342431. [Google Scholar] [CrossRef]
- Velasquez-Ortiz, N.; Herrera, G.; Hernandez, C.; Munoz, M.; Ramirez, J.D. Discrete typing units of Trypanosoma cruzi: Geographical and biological distribution in the Americas. Sci. Data 2022, 9, 360. [Google Scholar] [CrossRef]
- Zingales, B.; Andrade, S.G.; Briones, M.R.; Campbell, D.A.; Chiari, E.; Fernandes, O.; Guhl, F.; Lages-Silva, E.; Macedo, A.M.; Machado, C.R.; et al. A new consensus for Trypanosoma cruzi intraspecific nomenclature: Second revision meeting recommends TcI to TcVI. Mem. Inst. Oswaldo Cruz 2009, 104, 1051–1054. [Google Scholar] [CrossRef]
- Sarobe, P.; Lasarte, J.J.; Larrea, E.; Golvano, J.J.; Prieto, I.; Gullon, A.; Prieto, J.; Borras-Cuesta, F. Enhancement of peptide immunogenicity by insertion of a cathepsin B cleavage site between determinants recognized by B and T cells. Res. Immunol. 1993, 144, 257–262. [Google Scholar] [CrossRef]
- Tarleton, R.L. Immune system recognition of Trypanosoma cruzi. Curr. Opin. Immunol. 2007, 19, 430–434. [Google Scholar] [CrossRef]

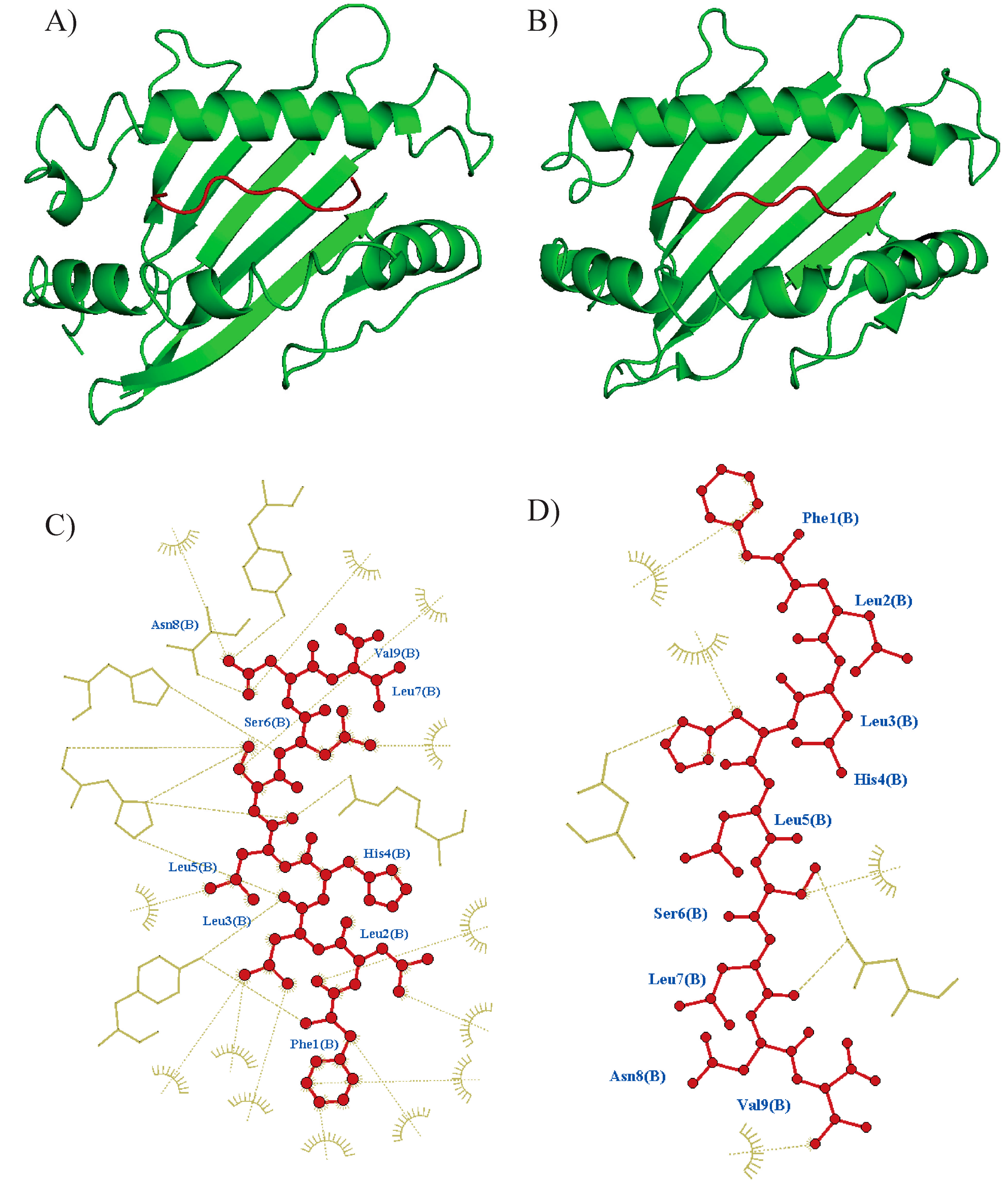


| Conservation | ||||
|---|---|---|---|---|
| ID | Epitope a | Strain | DTU Tc b | Kinetoplastids c |
| Tc07 | FLLHLSLNV | BrazilA4, Dm28c, SylvioX10, YC6, CLB Non-Es, TCC | I, II, III, VI | T. brucei (77), L. mexicana (78), L. major (78), L. braziliensis (88), L. infantum (78), L. donovani (78), L. panamensis (88) |
| Tc11 | ILCDFLLHV | BrazilA4, Dm28c, SylvioX10, G, YC6, CLB Es, CLB Non-Es, TCC | I, II, III, VI | T. brucei (78), T. vivax (78) |
| Tc17 | KLWAFLWSI | BrazilA4, Dm28c, SylvioX10, G, CLB Non-Es, TCC | I, III, VI | --- |
| Tc18 | LLMDCAAYL | CLB Non-Es | III | --- |
| Tc19 | LLMDDFSAV | BrazilA4, Dm28c, SylvioX10, G, YC6, CLB Non-Es, TCC | I, II, III, VI | --- |
| Tc21 | MLLLALAYI | BrazilA4, Dm28c, SylvioX10, YC6, CLB Es, CLB Non-Es, TCC, CLB | I, II, III, VI | T. brucei (78), T. vivax (78), L. mexicana (78), L. major (78), L. braziliensis (78), L. infantum (78), L. donovani (78), L. panamensis (78) |
| Tc29 | VMMPLIFLI | BrazilA4, Dm28c, SylvioX10, YC6, CLB Non-Es, TCC | I, II, III, VI | --- |
| Tc32 | YLIPISLFV | BrazilA4, Dm28c, SylvioX10, YC6, CLB Es, CLB Non-Es, TCC | I, II, III, VI | T. brucei (88), T. vivax (88), L. mexicana (78), L. major (78), L. braziliensis (78), L. infantum (78), L. donovani (78), L. panamensis |
| Tc34 | YLLPLLHTV | BrazilA4, Dm28c, SylvioX10, G, YC6, CLB Es, CLB Non-Es, TCC | I, II, III, VI | T. brucei (88), L. mexicana (78), L. braziliensis (88), L. infantum (78), L. donovani (78), L. panamensis (88) |
| Molecular Docking in CABSDock and FireDock | ||||
|---|---|---|---|---|
| ID | Epitope | RSMD (Å) | Interactions | Binding Affinity (KJ/mol) |
| Tc07 | FLLHLSLNV | 0.42 | 23 | −88.68 |
| Tc11 | ILCDFLLHV | 0.49 | 28 | −133.28 |
| Tc17 | KLWAFLWSI | 3.00 a | 31 | −82.02 |
| Tc18 | LLMDCAAYL | 1.46 | 23 | −64.49 a |
| TC19 | LLMDDFSAV | 2.81 | 30 | −126.11 |
| Tc21 | MLLLALAYI | 1.77 | 28 | −95.53 |
| Tc29 | VMMPLIFLI | 1.43 | 33 | −110.7 |
| Tc32 | YLIPISLFV | 2.64 | 22 a | −112.07 |
| Tc34 | YLLPLLHTV | 2.85 | 24 | −99.21 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Teh-Poot, C.F.; Alfaro-Chacón, A.; Pech-Pisté, L.M.; Rosado-Vallado, M.E.; Asojo, O.A.; Villanueva-Lizama, L.E.; Dumonteil, E.; Cruz-Chan, J.V. Immunogenicity of Trypanosoma cruzi Multi-Epitope Recombinant Protein as an Antigen Candidate for Chagas Disease Vaccine in Humans. Pathogens 2025, 14, 342. https://doi.org/10.3390/pathogens14040342
Teh-Poot CF, Alfaro-Chacón A, Pech-Pisté LM, Rosado-Vallado ME, Asojo OA, Villanueva-Lizama LE, Dumonteil E, Cruz-Chan JV. Immunogenicity of Trypanosoma cruzi Multi-Epitope Recombinant Protein as an Antigen Candidate for Chagas Disease Vaccine in Humans. Pathogens. 2025; 14(4):342. https://doi.org/10.3390/pathogens14040342
Chicago/Turabian StyleTeh-Poot, Christian F., Andrea Alfaro-Chacón, Landy M. Pech-Pisté, Miguel E. Rosado-Vallado, Oluwatoyin Ajibola Asojo, Liliana E. Villanueva-Lizama, Eric Dumonteil, and Julio Vladimir Cruz-Chan. 2025. "Immunogenicity of Trypanosoma cruzi Multi-Epitope Recombinant Protein as an Antigen Candidate for Chagas Disease Vaccine in Humans" Pathogens 14, no. 4: 342. https://doi.org/10.3390/pathogens14040342
APA StyleTeh-Poot, C. F., Alfaro-Chacón, A., Pech-Pisté, L. M., Rosado-Vallado, M. E., Asojo, O. A., Villanueva-Lizama, L. E., Dumonteil, E., & Cruz-Chan, J. V. (2025). Immunogenicity of Trypanosoma cruzi Multi-Epitope Recombinant Protein as an Antigen Candidate for Chagas Disease Vaccine in Humans. Pathogens, 14(4), 342. https://doi.org/10.3390/pathogens14040342







