Metronidazole Activation by a Deeply Entangled Dimeric Malic Enzyme in Entamoeba histolytica
Abstract
1. Introduction
2. Materials and Methods
2.1. Cloning, Over-Expression, and Purification
2.2. Biochemical Characterization
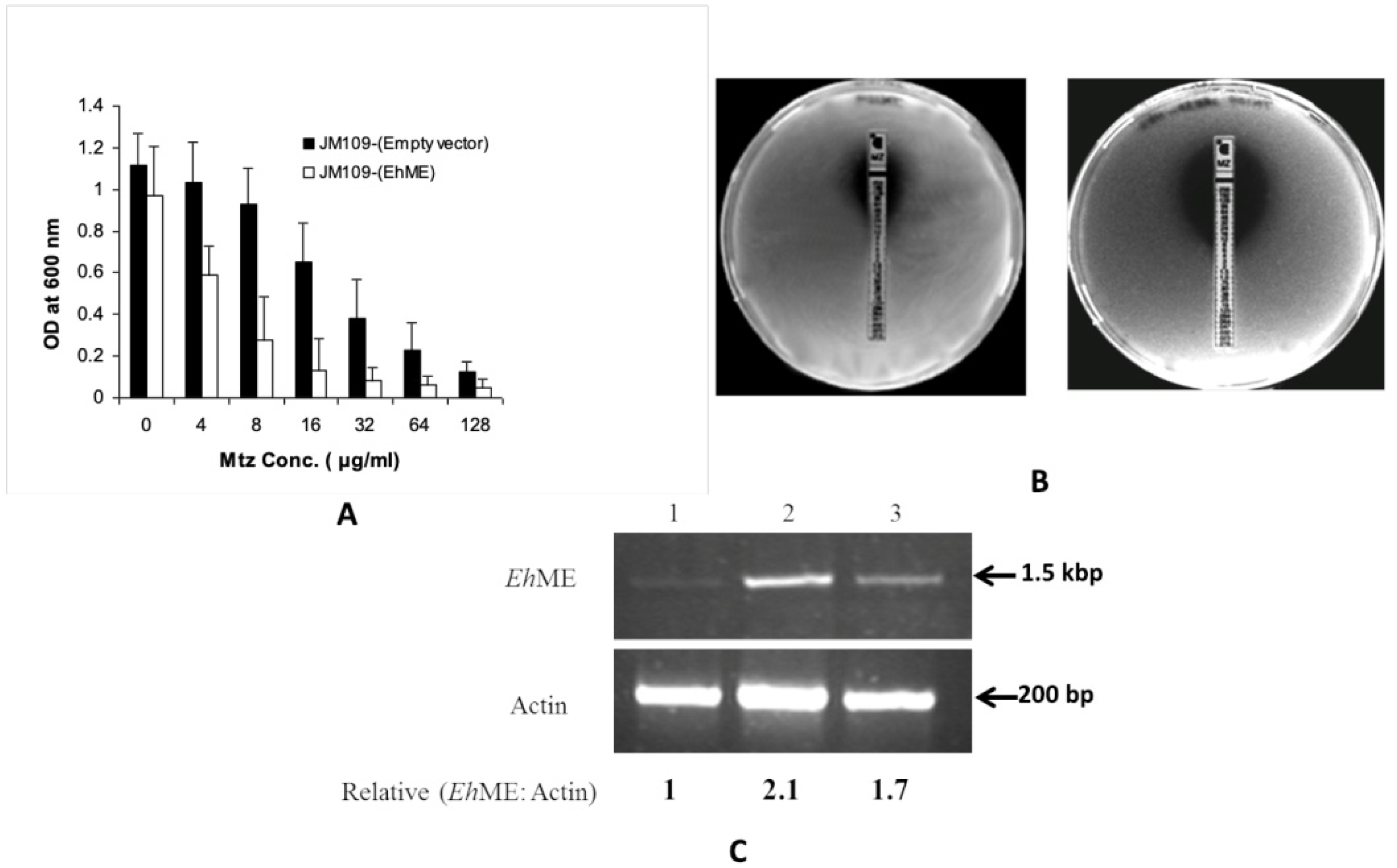
2.3. Metronidazole Susceptibility Assay
2.4. A. Broth Dilution
2.5. B. E-Test Assay
2.6. Cell Culture and Drug Treatment
2.7. Semi-Quantitative RT-PCR
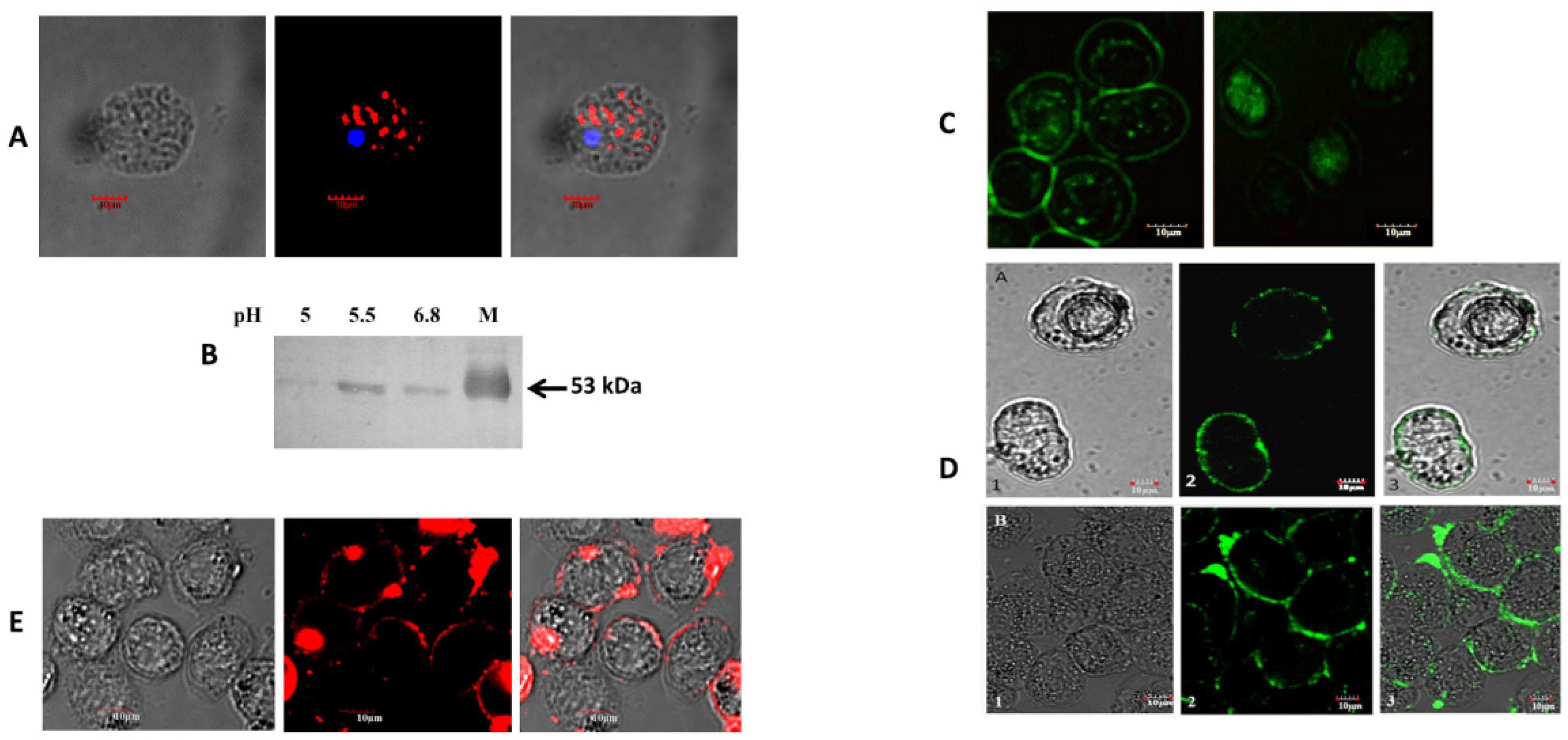
2.8. Western Blot Assay
2.9. Labeling of Recombinant Malic Enzyme
2.10. Preparation of Cell-Free Filtrate
2.11. Detection of Extracellular Released EhME Protein
2.12. Localization of Malic Enzyme in Eh
2.13. Autoligand Assay
2.14. Adherence Assay
2.15. Crystallization
2.16. Data Collection and Processing
2.17. Phasing, Model Building, Refinement, and Validation
2.18. Computational Docking Analysis
3. Results
3.1. Malic Enzyme Is an NADP+ Dependent Enzyme
3.2. Malic Enzyme Is an Alternative Enzyme That Activates Metronidazole
3.3. Malic Enzyme Helps Attach Eh to the Enteric Cell Surface
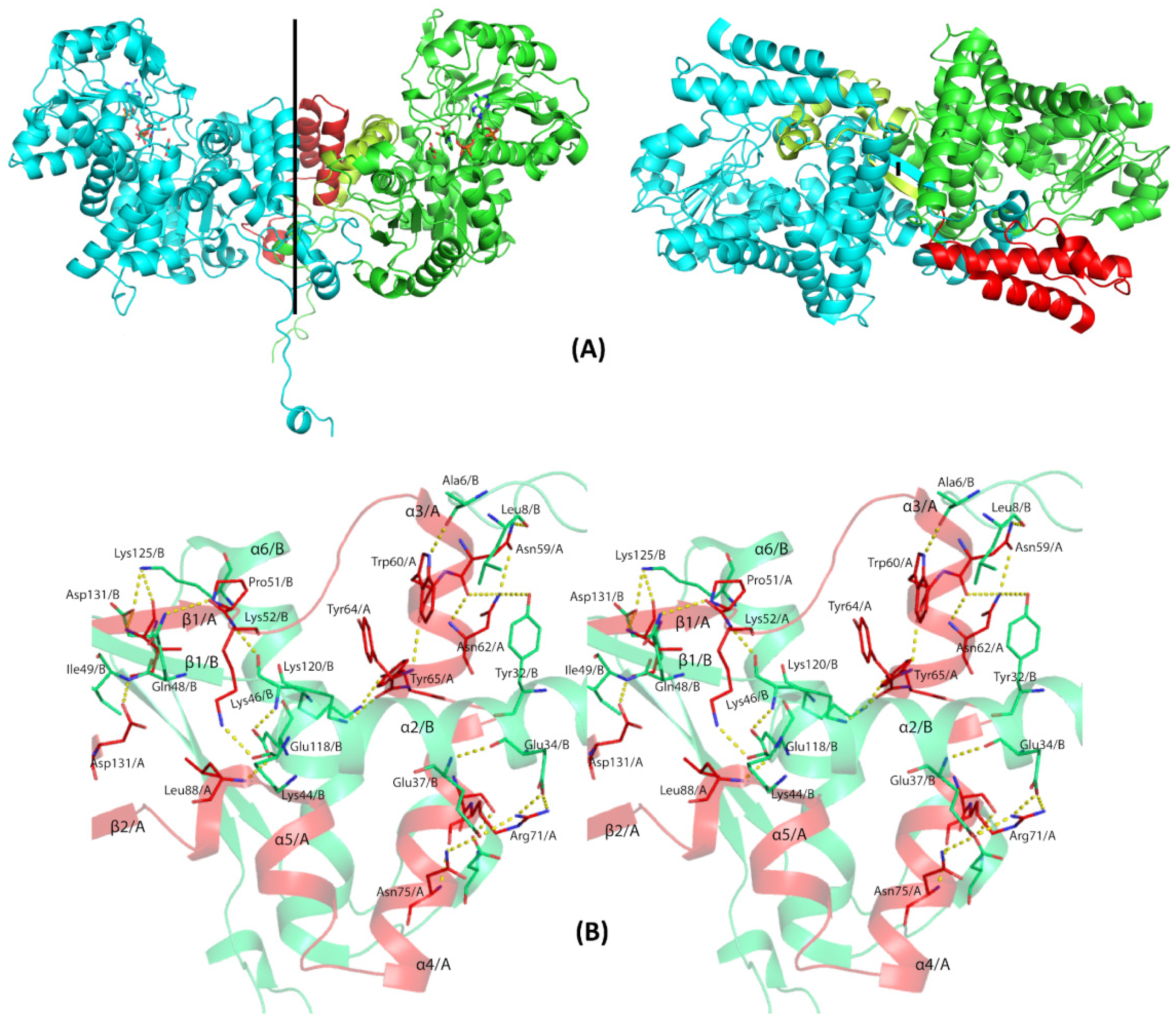
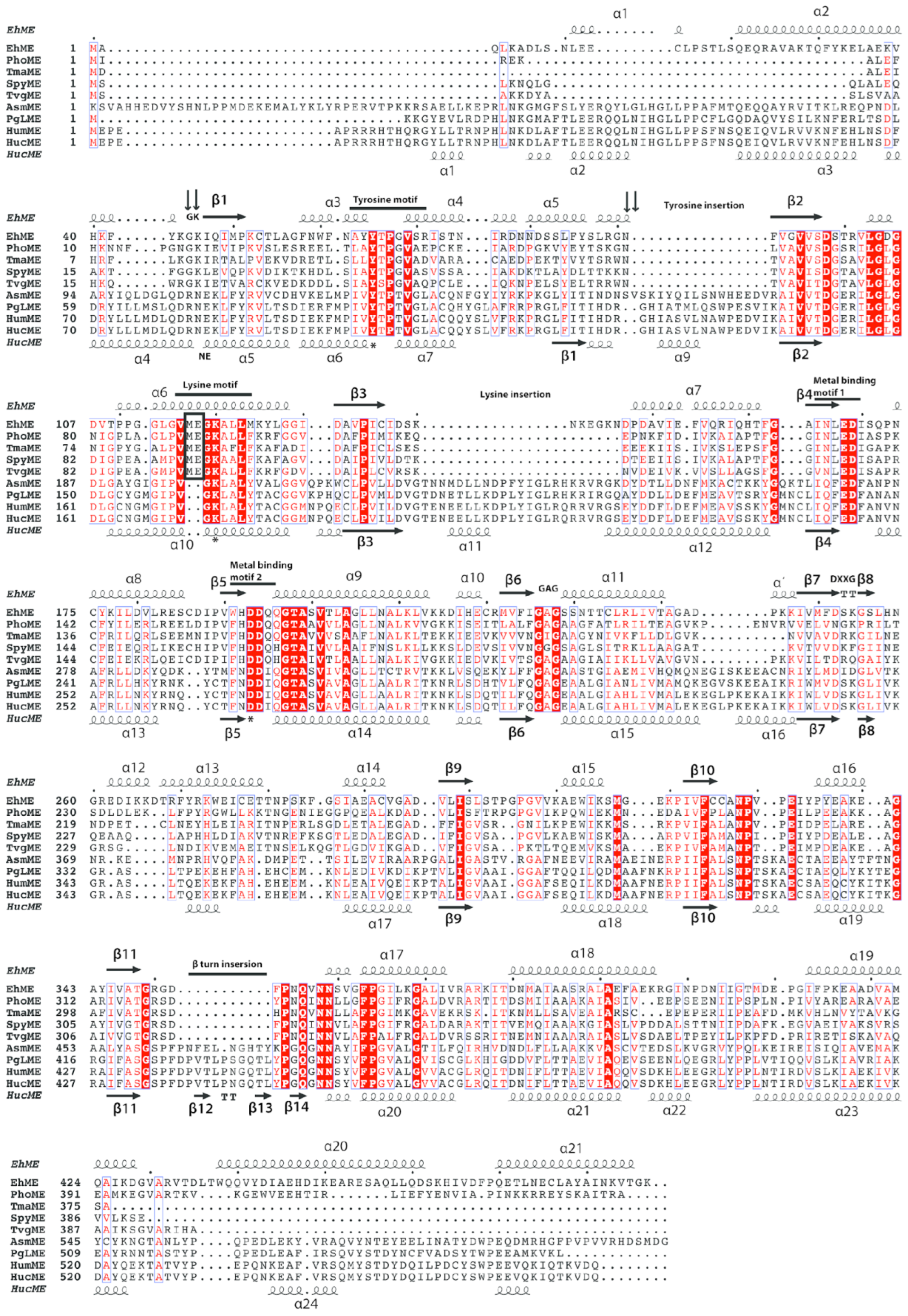
3.4. Overall Structure of EhME
3.5. The Entangled Dimer Interface of EhME Is Distinctive from the Metazoan ME
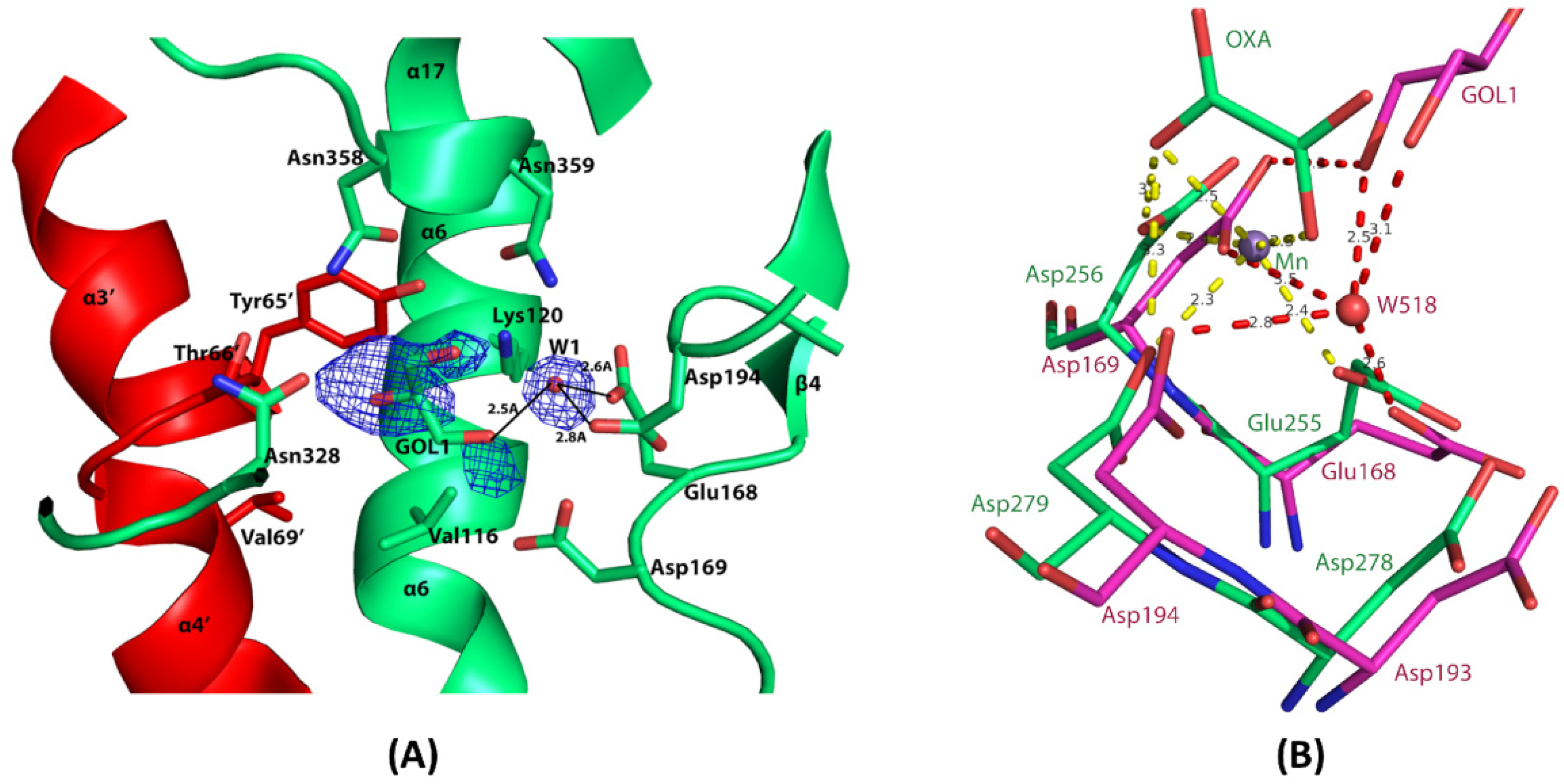
3.6. The Composite Active Site of EhME
3.7. Metal-Binding Residues in EhME
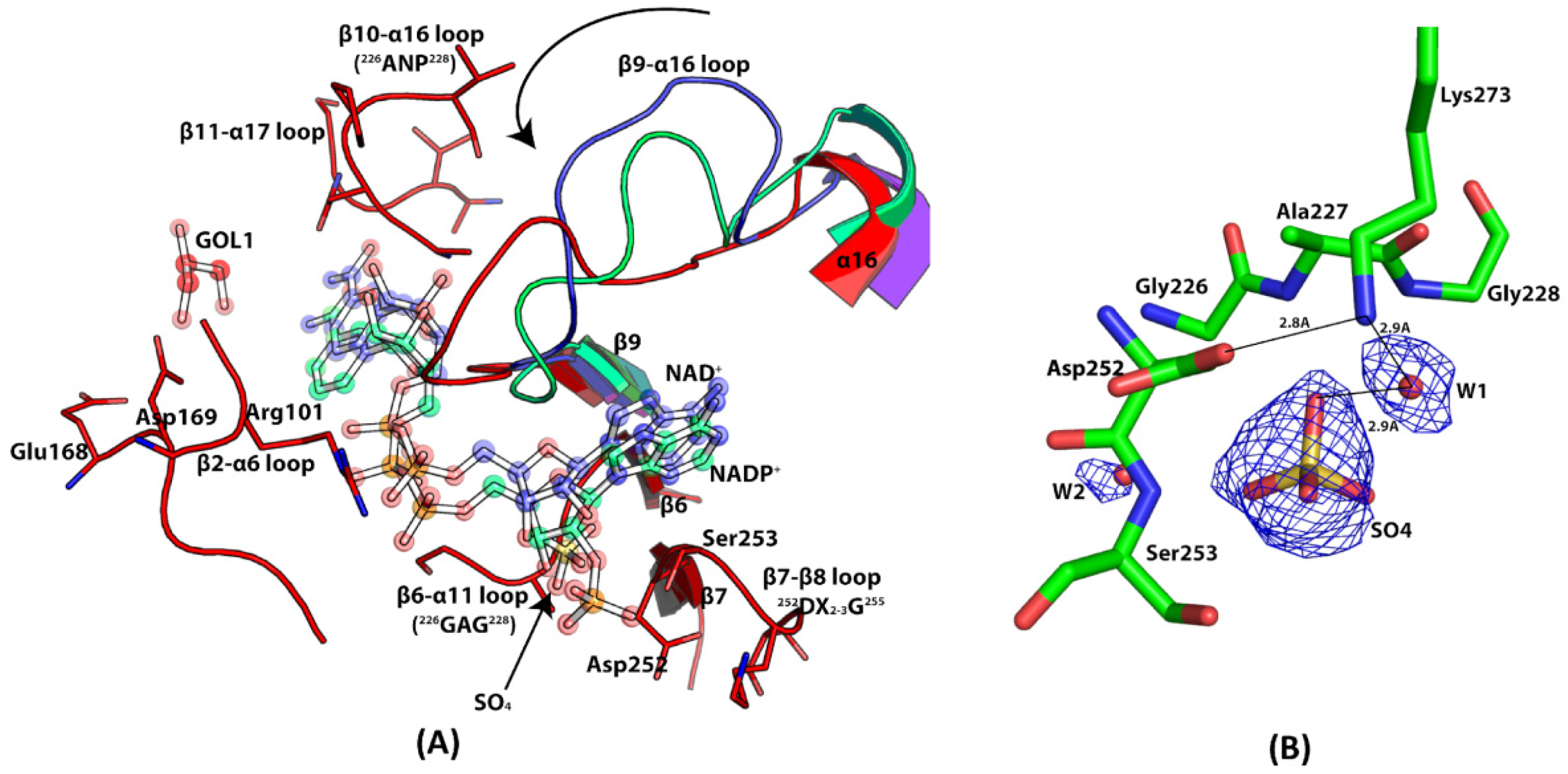
3.8. Coenzyme Specificity of EhME
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Eh | Entamoeba histolytica |
| EhME | Entamoeba histolytica malic enzyme |
| Mtz | Metronidazole |
| FITC | Fluorescein isothiocyanate |
| NADPH | Nicotinamide adenine dinucleotide phosphate (reduced) |
| Ni-NTA | Nickel Nitrilotriacetic acid |
| TRITC | Tetramethylrhodamine isothiocyanate |
| DAPI | 4′,6-diamidino-2-phenylindole |
References
- World Health Organization. Amoebiasis. Wkly. Epidemiol. Rec. 1997, 72, 97–100. [Google Scholar]
- Leitsch, D.; Kolarich, D.; Wilson, I.B.H.; Altmann, F.; Duchêne, M. Nitroimidazole action in Entamoeba histolytica: A central role for thioredoxin reductase. PLoS Biol. 2007, 5, 1820–1834. [Google Scholar] [CrossRef] [PubMed]
- Pal, D.; Banerjee, S.; Cui, J.; Schwartz, A.; Ghosh, S.K.; Samuelson, J. Giardia, Entamoeba, and Trichomonas enzymes activate metronidazole (nitroreductases) and inactivate metronidazole (nitroimidazole reductases). Antimicrob. Agents Chemother. 2009, 53, 458–464. [Google Scholar] [CrossRef]
- Rasoloson, D.; Tomkova, E.; Hrdy, I.; Tachezy, J.; Kulda, J. Mechanisms of in vitro development of resistance to metronidazole in Trichomonas vaginalis. Microbiology 2002, 148, 2467–2477. [Google Scholar] [CrossRef]
- Hrdy, I.; Cammack, R.; Stopka, P.; Kulda, J.; Tachezy, J. Alternative pathway of metronidazole activation in Trichomonas vaginalis hydrogenosomes. Antimicrob. Agents Chemother. 2005, 49, 5033–5036. [Google Scholar] [CrossRef]
- Ludlum, D.B.; Colinas, R.J.; Kirk, M.C.; Mehta, J.R. Reaction of reduced metronidazole with guanosine to form an unstable adduct. Carcinogenesis 1988, 9, 593–596. [Google Scholar] [CrossRef]
- Samuelson, J. Why Metronidazole Is Active against both bacteria and parasites. Antimicrob. Agents Chemother. 1999, 43, 1533–1541. [Google Scholar] [CrossRef]
- Bartoluccis, S.; Rellab, R.; Guagliardis, A.; Raiab, C.A.; Gambacortaq, A.; De Rosaq, M. Malic enzyme from Archaebacterium Sulfolobus solfuturicus. Purification, structure, and kinetic properties. J. Biol. Chem. 1987, 262, 7725–7731. [Google Scholar] [CrossRef]
- Alvarez, C.E.; Trajtenberg, F.; Larrieux, N.; Saigo, M.; Golic, A.; Andreo, C.S.; Hogenhout, S.A.; Mussi, M.A.; Drincovich, M.F.; Buschiazzo, A. The crystal structure of the malic enzyme from Candidatus Phytoplasma reveals the minimal structural determinants for a malic enzyme. Acta Crystallogr. D Struct. Biol. 2018, 74, 332–340. [Google Scholar] [CrossRef]
- Tronconi, M.A.; Andreo, C.S.; Drincovich, M.F. Chimeric Structure of Plant Malic Enzyme Family: Different Evolutionary Scenarios for NAD- and NADP-Dependent Isoforms. Front. Plant Sci. 2018, 9, 565. [Google Scholar] [CrossRef]
- Loeber, G.; Maurer-Fogy, I.; Schwendenwein, R. Purification, cDNA cloning and heterologous expression of the human mitochondrial NADP+ -dependent malic enzyme. Biochem. J. 1994, 692, 687–692. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Bhargava, G.; Wu, H.; Loeber, G.; Tong, L. Crystal structure of human mitochondrial NAD(P)+ -dependent malic enzyme: A new class of oxidative decarboxylases. Structure 1999, 7, 877–889. [Google Scholar] [CrossRef] [PubMed]
- Winning, B.M.; Bourguignons, J.; Leave, C.J. Plant mitochondrial NAD+-dependent malic enzyme. cDNA cloning, deduced primary structure of the 59- and 62-kDa subunits, import, gene complexity and expression analysis. J. Biol. Chem. 1994, 269, 4780–4786. [Google Scholar] [CrossRef] [PubMed]
- Loebers, G.; Anthony, A.; Maurer-fogy, I.; Krystek, E.; Dworkin, M.B. Human NAD(+)-dependent mitochondrial malic enzyme. cDNA cloning, primary structure, and expression in Escherichia coli. J. Biol. Chem. 1991, 266, 3016–3021. [Google Scholar] [CrossRef]
- Hassel, B. Pyruvate carboxylation in neurons. J. Neurosci. Res. 2001, 66, 755–762. [Google Scholar] [CrossRef]
- Moreadith, R.W.; Lehninger, A.L. The Pathways of glutamate and glutamine oxidation by tumor cell mitochondria. J. Biol. Chem. 1984, 259, 6215–6221. [Google Scholar] [CrossRef]
- Field, J.; Rosenthal, B.; Samuelson, J. Early lateral transfer of genes encoding malic enzyme, acetyl-CoA synthetase and alcohol dehydrogenases from anaerobic prokaryotes to Entamoeba histolytica. Mol. Microbiol. 2000, 38, 446–455. [Google Scholar] [CrossRef]
- Addis, M.F.; Rappelli, P.; Cappuccinelli, P.; Fiori, P.L. Extracellular release by Trichomonas vaginalis of a NADP+ dependent malic enzyme involved in pathogenicity. Microb. Pathog. 1997, 23, 55–61. [Google Scholar] [CrossRef]
- Garcia, A.F.; Alderete, J.F. Characterization of the Trichomonas vaginalis surface associated AP65 and binding domain interacting with trichomonads and host cells. BMC Microbiol. 2007, 7, 116. [Google Scholar] [CrossRef]
- Pflugrath, J.W. The finer things in X-ray diffraction data collection. Acta Crystallogr. Sect. D Struct. Biol. 1999, 55, 1718–1725. [Google Scholar] [CrossRef]
- Schneider, T.R.; Sheldrick, G.M. Substructure solution with SHELXD. Acta Crystallogr. Sect. D Struct. Biol. 2002, 58, 1772–1779. [Google Scholar] [CrossRef]
- Vonrhein, C.; Blanc, E.; Roversi, P.; Bricogne, G. Automated structure solution with autoSHARP. Methods Mol. Biol. 2006, 364, 215–230. [Google Scholar] [CrossRef]
- Abrahams, J.P.; Leslie, A.G. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. Sect. D Struct. Biol. 1996, 52, 30–42. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics research papers. Acta Crystallogr. Sect. D Struct. Biol. 2004, 60, 2126–2132. [Google Scholar] [CrossRef] [PubMed]
- Murshudov, G.N.; Vagin, A.A.; Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. Sect. D Struct. Biol. 1997, 53, 240–255. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. Autodock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2010, 30, 2785–2791. [Google Scholar] [CrossRef]
- Grosdidier, A.; Zoete, V.; Michielin, O. Fast docking using the CHARMM force field with EADock DSS. J. Comput. Chem. 2011, 32, 2149–2159. [Google Scholar] [CrossRef]
- Alvarez, C.E.; Bovdilova, A.; Höppner, A.; Wolff, C.C.; Saigo, M.; Trajtenberg, F.; Zhang, T.; Buschiazzo, A.; Nagel-Steger, L.F.; Drincovich, M.F.; et al. Molecular adaptations of NADP-malic enzyme for its function in C4 photosynthesis in grasses. Nat. Plants 2019, 5, 755–765. [Google Scholar] [CrossRef]
- Burley, K.H.; Cuthbert, B.J.; Basu, P.; Newcombe, J.; Irimpan, E.M.; Quechol, R.; Foik, I.P.; Mobley, D.L.; Beste, D.J.V.; Goulding, C.W. Structural and Molecular Dynamics of Mycobacterium tuberculosis Malic Enzyme, a Potential Anti-TB Drug Target. ACS Infect. Dis. 2021, 7, 174–188. [Google Scholar] [CrossRef]
- Andrä, J.; Herbst, R.; Leippe, M. Amoebapores, archaic effector peptides of protozoan origin, are discharged into phagosomes and kill bacteria by permeabilizing their membranes. Dev. Comp. Immunol. 2003, 27, 291–304. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.; Chou, W.; Chan, G.G. Effect of metal binding on the structural stability of pigeon liver malic enzyme. J. Biol. Chem. 2002, 277, 4663–4671. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Tong, L. Structural studies of a human malic enzyme. Protein Pept. Lett. 2000, 7, 287–296. [Google Scholar]
- Tao, X.; Yang, Z.; Tong, L. Crystal structures of substrate complexes of malic enzyme and insights into the catalytic mechanism. Structure 2003, 11, 1141–1150. [Google Scholar] [CrossRef]
- Chang, G.; Tong, L. Structure and function of malic enzymes, a new class of oxidative decarboxylases. Biochemistry 2003, 42, 12721–12733. [Google Scholar] [CrossRef]
- Hsieh, J.; Hung, H. Engineering of the cofactor specificities and isoform-specific inhibition of malic enzyme. J. Biol. Chem. 2009, 284, 4536–4544. [Google Scholar] [CrossRef]
- Aktas, D.F.; Cook, P.F. Proper positioning of the nicotinamide ring is crucial for the Ascaris suum malic enzyme reaction. Biochemistry 2008, 47, 2539–2546. [Google Scholar] [CrossRef]
- Rao, G.S.J.; Coleman, D.E.; Karsten, W.E.; Cook, P.F.; Harris, B.G. Crystallographic studies on Ascaris suum NAD+-malic enzyme bound to reduced cofactor andn identification of an effector Site. J. Biol. Chem. 2003, 278, 38051–38058. [Google Scholar] [CrossRef]
- Bhattacharyya, S.; Dutta, A.; Dutta, D.; Ghosh, A.K.; Das, A.K. Structural elucidation of the NADP(H) phosphatase activity of staphylococcal dual-specific IMPase/NADP(H) phosphatase. Acta Crystallogr. D Struct. Biol. 2016, 72, 281–290. [Google Scholar] [CrossRef]
- Lin, H.; Chu, L.J.; Huang, P.; Cheng, W.; Zheng, Y.; Huang, C.; Hong, S.; Chen, L.; Lin, H.; Wang, J.; et al. Proteomic signatures of metronidazole-resistant Trichomonas vaginalis reveal novel proteins associated with drug resistance. Parasit. Vectors 2020, 13, 274. [Google Scholar] [CrossRef]
- Štáfková, J.; Rada, P.; Meloni, D.; Žárský, V.; Smutná, T.; Zimmann, N.; Harant, K.; Pompach, P.; Hrdý, I.; Tachezy, J. Dynamic secretome of Trichomonas vaginalis: Case study of β-amylases. Mol. Cell Proteom. 2018, 17, 304–320. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Lanks, C.W.; Tong, L. Molecular mechanism for the regulation of human mitochondrial NAD(P)+ -dependent malic enzyme by ATP and fumarate. Structure 2002, 10, 951–960. [Google Scholar] [CrossRef] [PubMed]
- Coleman, D.E.; Rao, G.S.J.; Goldsmith, E.J.; Cook, P.F.; Harris, B.G. Crystal structure of the malic enzyme from Ascaris suum complexed with with nicotinamide adenine dinucleotide at 2.3 Å resolution. Biochemistry 2002, 41, 6928–6938. [Google Scholar] [CrossRef]
- Doležal, P.; Vaňáčová, S.; Tachezy, J.; Hrdy, I. Malic enzymes of Trichomonas vaginalis: Two enzyme families, two distinct origins. Gene 2004, 329, 81–92. [Google Scholar] [CrossRef]
- Pancholi, V.; Chhatwal, G.S. Housekeeping enzymes as virulence factors for pathogens. Int. J. Med. Microbiol. 2003, 293, 391–401. [Google Scholar] [CrossRef]
- Kucknoor, A.S.; Mundodi, V.; Alderete, J.F. The proteins secreted by Trichomonas vaginalis and vaginal epithelial cell response to secreted and episomally expressed AP65. Cell Microbiol. 2007, 9, 2586–2597. [Google Scholar] [CrossRef]
- Ralston, K.S. Chew on this: Amoebic trogocytosis and host cell killing by Entamoeba histolytica. Trends Parasitol. 2015, 31, 442–452. [Google Scholar] [CrossRef]
- Zhao, Y.; Cieplak, M. Stability of structurally entangled protein dimers. Proteins 2018, 86, 945–955. [Google Scholar] [CrossRef]
- Taylor, W.R. A deeply knotted protein structure and how it might fold. Nature 2000, 406, 916–919. [Google Scholar] [CrossRef]
- Budiman, C.; Angkawidjaja, C.; Motoike, H.; Koga, Y.; Takano, K.; Kanaya, S. Crystal structure of N-domain of FKBP22 from Shewanella sp. SIB1: Dimer dissociation by disruption of Val-Leu knot. Protein Sci. 2011, 20, 1755–1764. [Google Scholar] [CrossRef]
- Banerjee, R.; Nath, S.; Ranjan, A.; Khamrui, S.; Pani, B.; Sen, R.; Sen, U. The first structure of polarity suppression protein, Psu from enterobacteria phage P4, reveals a novel fold and a knotted dimer. J. Biol. Chem. 2012, 287, 44667–44675. [Google Scholar] [CrossRef] [PubMed]
- Virnau, P.; Mirny, L.A.; Kardar, M. Intricate knots in proteins: Function and evolution. PLoS Comput. Biol. 2006, 2, e122. [Google Scholar] [CrossRef] [PubMed]
- King, N.P.; Yeates, E.O.; Yeates, T.O. Identification of rare slipknots in proteins and their implications for stability and folding. J. Mol. Biol. 2007, 373, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Sayre, T.C.; Lee, T.M.; King, N.P.; Yeates, T.O. Protein stabilization in a highly knotted protein polymer. Protein Eng. Des. Sel. 2011, 24, 627–630. [Google Scholar] [CrossRef]
- Bornschlögl, T.; Anstrom, D.M.; Mey, E.; Dzubiella, J.; Rief, M.; Forest, K.T. Tightening the knot in phytochrome by single-molecule atomic force microscopy. Biophys. J. 2009, 96, 1508–1514. [Google Scholar] [CrossRef]
 ), 0.5 mM (
), 0.5 mM ( ), and 0.8 mM (
), and 0.8 mM ( ) are kept fixed to determine the reaction velocities with respect to the different L-malate concentrations, (ii) uncompetitive inhibition of the EhME by ATP with respect to malate, (iii) non-competitive inhibition of the EhME by ATP with respect to NADP+. Malic enzyme activity was measured at different concentrations of NADP+ in various concentrations of ATP (indicated by different color). The X-axis indicates the concentration of the substrate, and the Y-axis indicates the velocity of the reaction.
) are kept fixed to determine the reaction velocities with respect to the different L-malate concentrations, (ii) uncompetitive inhibition of the EhME by ATP with respect to malate, (iii) non-competitive inhibition of the EhME by ATP with respect to NADP+. Malic enzyme activity was measured at different concentrations of NADP+ in various concentrations of ATP (indicated by different color). The X-axis indicates the concentration of the substrate, and the Y-axis indicates the velocity of the reaction.
 ), 0.5 mM (
), 0.5 mM ( ), and 0.8 mM (
), and 0.8 mM ( ) are kept fixed to determine the reaction velocities with respect to the different L-malate concentrations, (ii) uncompetitive inhibition of the EhME by ATP with respect to malate, (iii) non-competitive inhibition of the EhME by ATP with respect to NADP+. Malic enzyme activity was measured at different concentrations of NADP+ in various concentrations of ATP (indicated by different color). The X-axis indicates the concentration of the substrate, and the Y-axis indicates the velocity of the reaction.
) are kept fixed to determine the reaction velocities with respect to the different L-malate concentrations, (ii) uncompetitive inhibition of the EhME by ATP with respect to malate, (iii) non-competitive inhibition of the EhME by ATP with respect to NADP+. Malic enzyme activity was measured at different concentrations of NADP+ in various concentrations of ATP (indicated by different color). The X-axis indicates the concentration of the substrate, and the Y-axis indicates the velocity of the reaction.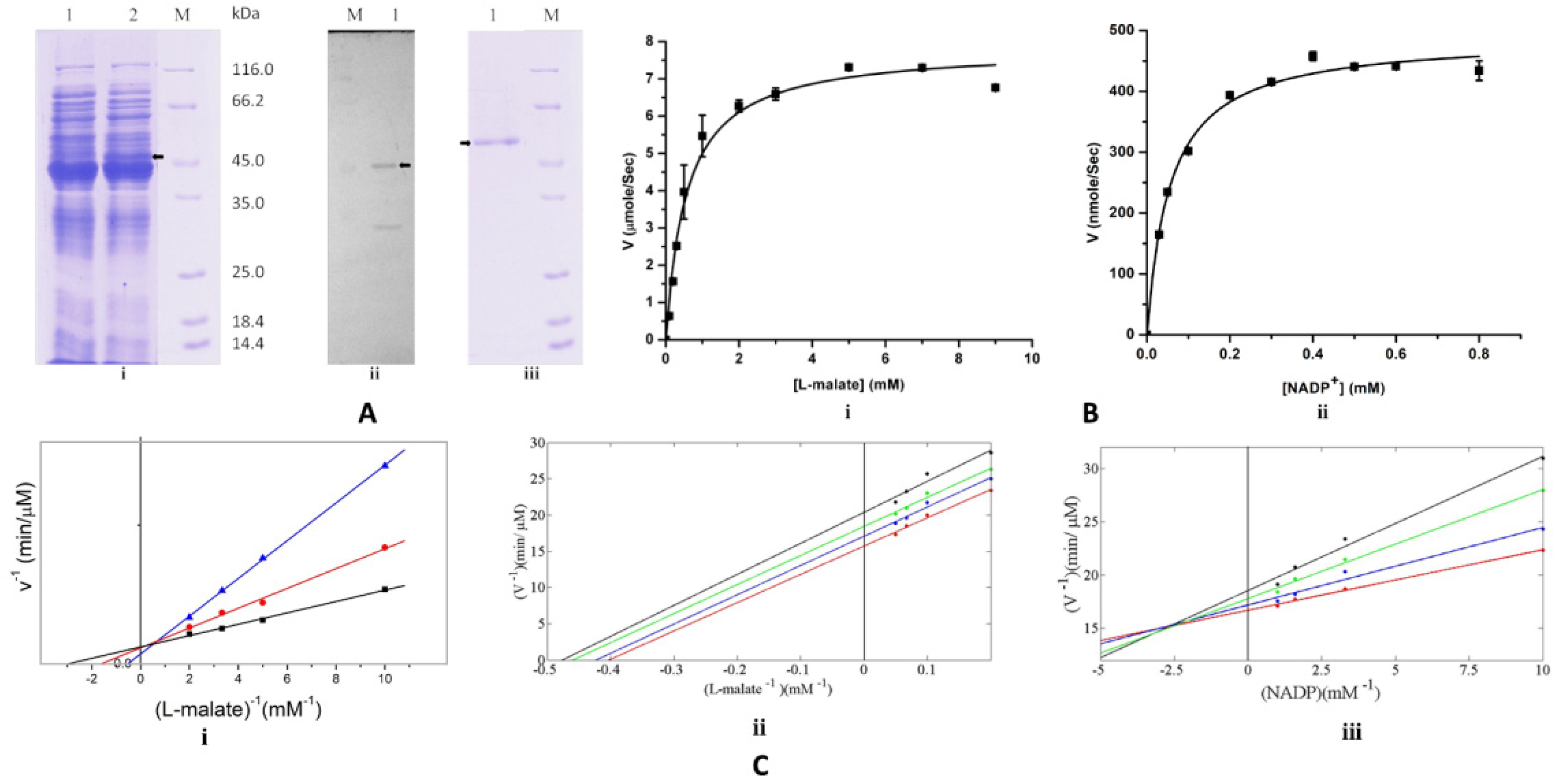

| Parameters | Native Data | Platinum Data |
|---|---|---|
| Data collection and cell parameters | ||
| Wavelength (Å) | 1.5418 | 1.5418 |
| Resolution Range (Å) 1 | 19.85–2.25 (2.33–2.25) | 44.45–2.90 (3.00–2.90) |
| Space Group (Å) | P43212 | P43212 |
| Unit Cell Parameters (Å, °) | a = b = 117.73, c = 157.62 | a = b = 117.93, c = 153.63 |
| Total number of reflections 1 | 1,394,718 (103,047) | 535,258 |
| Unique number of reflections 1 | 53,238 (5225) | 25,354 |
| Completeness (%) 1 | 100 (100) | 100 (100) |
| Rmerge (%) 1 | 6.3 (24.2) | 6.9 (19.0) |
| I/σ(I) | 32.3 (12.1) | 18.2 (8.4) |
| Redundancy | 26.20 (19.49) | 21.11 (20.34) |
| Number of monomers in asymmetric unit (Z) | 2 | 2 |
| Phasing | ||
| Number of Pt sites | 4 | |
| Rcullis (%) | 88.2 | |
| Rano (%) | 91 | |
| Phasing power iso (acentric/centric) | 0.746/0.708 | |
| Phasing power ano | 0.638 | |
| FOM iso (acentric/centric) | 0.176/0.207 | |
| Refinement Statistics | ||
| Wilson B factor (Å2) | 32.6 | |
| Rwork/Rfree | 17.9/24.4 | |
| R.M.S.D. Bond lengths (Å) | 0.02 | |
| R.M.S.D. Bond angle (°) | 1.83 | |
| Average B factor (Å2) | 29.0 | |
| Number of Protein atoms | 7466 | |
| Number of Ligand atoms | 137 | |
| Number of Solvent atoms | 582 | |
| Ramachandran Plot | ||
| Favored (%) | 95.9 | |
| Additionally allowed (%) | 3.7 | |
| Generously allowed (%) | 0.0 | |
| Disallowed (%) | 0.4 |
| Chain B | Chain A | Distance (Å) |
|---|---|---|
| B/Ala6/O | A/Trp60/NE1 | 2.9 |
| B/Leu8/O | A/Asn59/ND2 | 3.0 |
| B/Tyr32/OH | A/Asn62/ND2 | 3.6 |
| B/Glu34/OE1 | A/Arg71/NH2 | 3.2 |
| B/Glu37/OE1 | A/Arg71/NH1 | 2.8 |
| B/Glu37/OE1 | A/Asn75/ND2 | 2.8 |
| B/Lys44/O | A/Lys52/NZ | 2.8 |
| B/Lys46/O | A/Lys52/N | 2.9 |
| B/Lys46/NZ | A/Tyr64/O | 2.8 |
| B/Gln48/NE2 | A/Pro51/O | 3.4 |
| B/Ile49/N | A/Asp131/OD1 | 2.9 |
| B/Asp105/OD1 | A/Ser70/OG | 2.9 |
| B/Glu118/OE1 | A/Leu88/N | 3.1 |
| B/Lys120/NZ | A/Tyr65/OH | 2.8 |
| B/Lys125/NZ | A/Ile49/O | 3.3 |
| B/Asp131/OD2 | A/Ile49/O | 2.9 |
| B/Thr161/OG1 | A/Cys136/N | 2.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chakrabarty, A.; Dutta, D.; Baidya, M.; Dutta, A.; Das, A.K.; Ghosh, S.K. Metronidazole Activation by a Deeply Entangled Dimeric Malic Enzyme in Entamoeba histolytica. Pathogens 2025, 14, 277. https://doi.org/10.3390/pathogens14030277
Chakrabarty A, Dutta D, Baidya M, Dutta A, Das AK, Ghosh SK. Metronidazole Activation by a Deeply Entangled Dimeric Malic Enzyme in Entamoeba histolytica. Pathogens. 2025; 14(3):277. https://doi.org/10.3390/pathogens14030277
Chicago/Turabian StyleChakrabarty, Arindam, Debajyoti Dutta, Mithu Baidya, Anirudha Dutta, Amit Kumar Das, and Sudip K. Ghosh. 2025. "Metronidazole Activation by a Deeply Entangled Dimeric Malic Enzyme in Entamoeba histolytica" Pathogens 14, no. 3: 277. https://doi.org/10.3390/pathogens14030277
APA StyleChakrabarty, A., Dutta, D., Baidya, M., Dutta, A., Das, A. K., & Ghosh, S. K. (2025). Metronidazole Activation by a Deeply Entangled Dimeric Malic Enzyme in Entamoeba histolytica. Pathogens, 14(3), 277. https://doi.org/10.3390/pathogens14030277








