Insights into Functions of Universal Stress Proteins Encoded by Genomes of Gastric Cancer Pathogen Helicobacter pylori and Related Bacteria
Abstract
1. Introduction
2. Methods
2.1. Retrieval of Helicobacteraceae Genomes Dataset and Genes Encoding Universal Stress Protein Domain Dataset
2.2. Grouping of Helicobacteraceae Species by Patterns of Functional Sites in Universal Stress Protein Sequences
2.3. Similarity of Helicobacteraceae Universal Stress Proteins Sequences
2.4. Genomic Context of Genes for Universal Stress Proteins in Helicobacteraceae Genomes
2.5. Transcriptome and Interactome Evidence for Helicobacter pylori Universal Stress Protein
3. Results
3.1. Universal Stress Protein Proteins Encoded by Helicobacteraceae Genomes
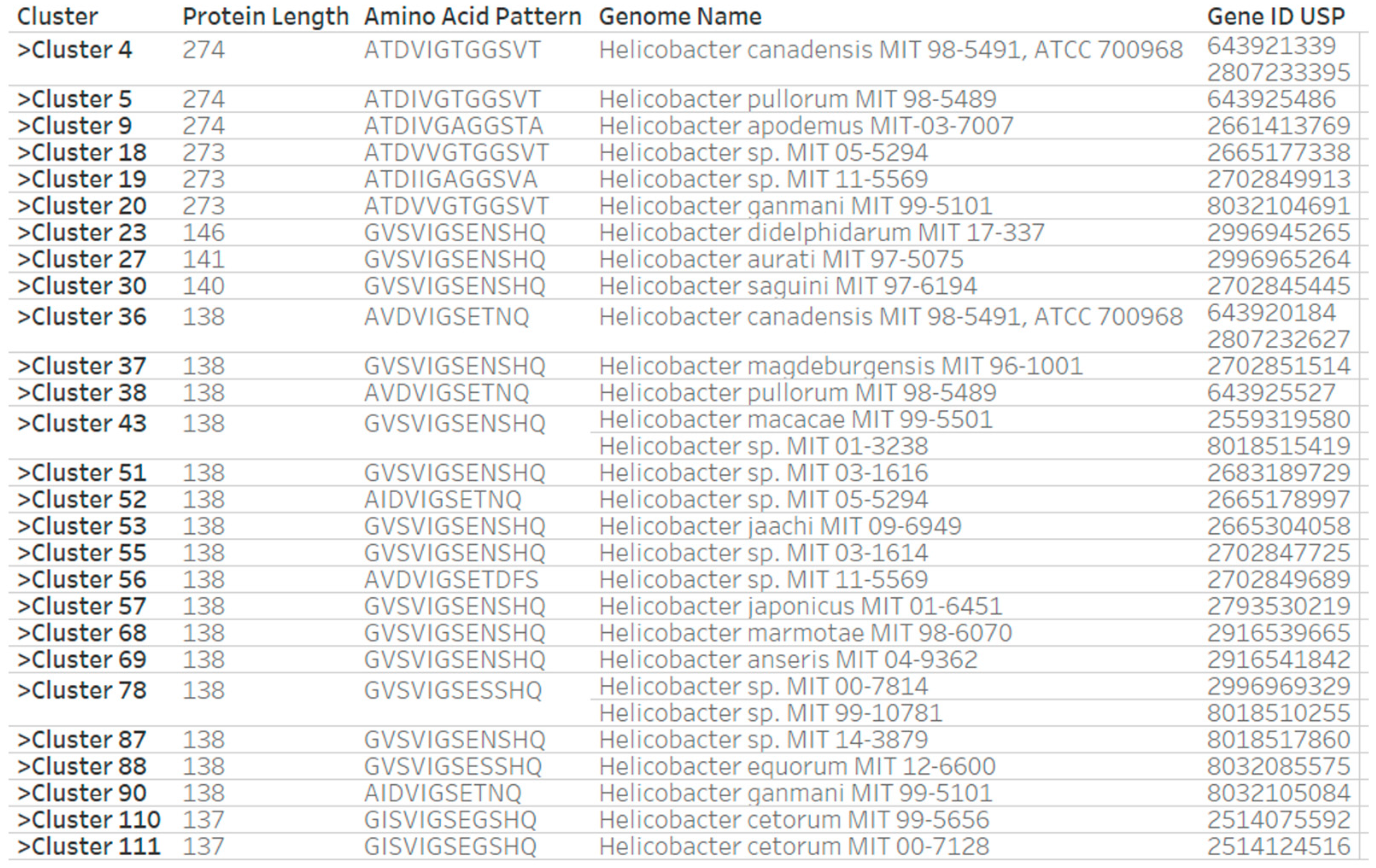
3.2. Grouping of Helicobacteraceae Species by Patterns of Functional Sites in Universal Stress Protein Sequences
3.3. Similarity of Helicobacteraceae Universal Stress Proteins Sequences
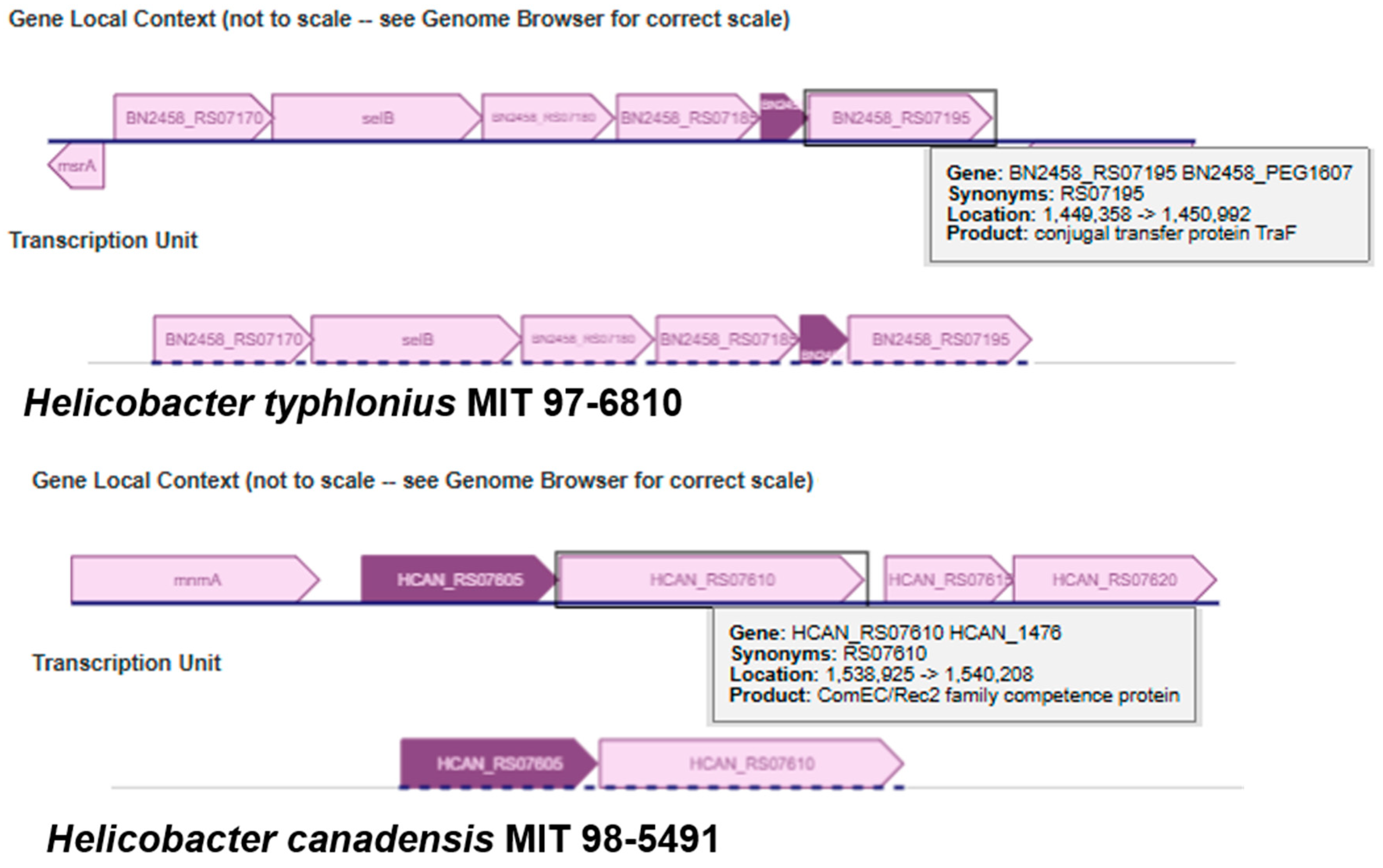
3.4. Predicted Protein Functions for Adjacent Genes of Universal Stress Protein Genes in Helicobacteraceae Genomes
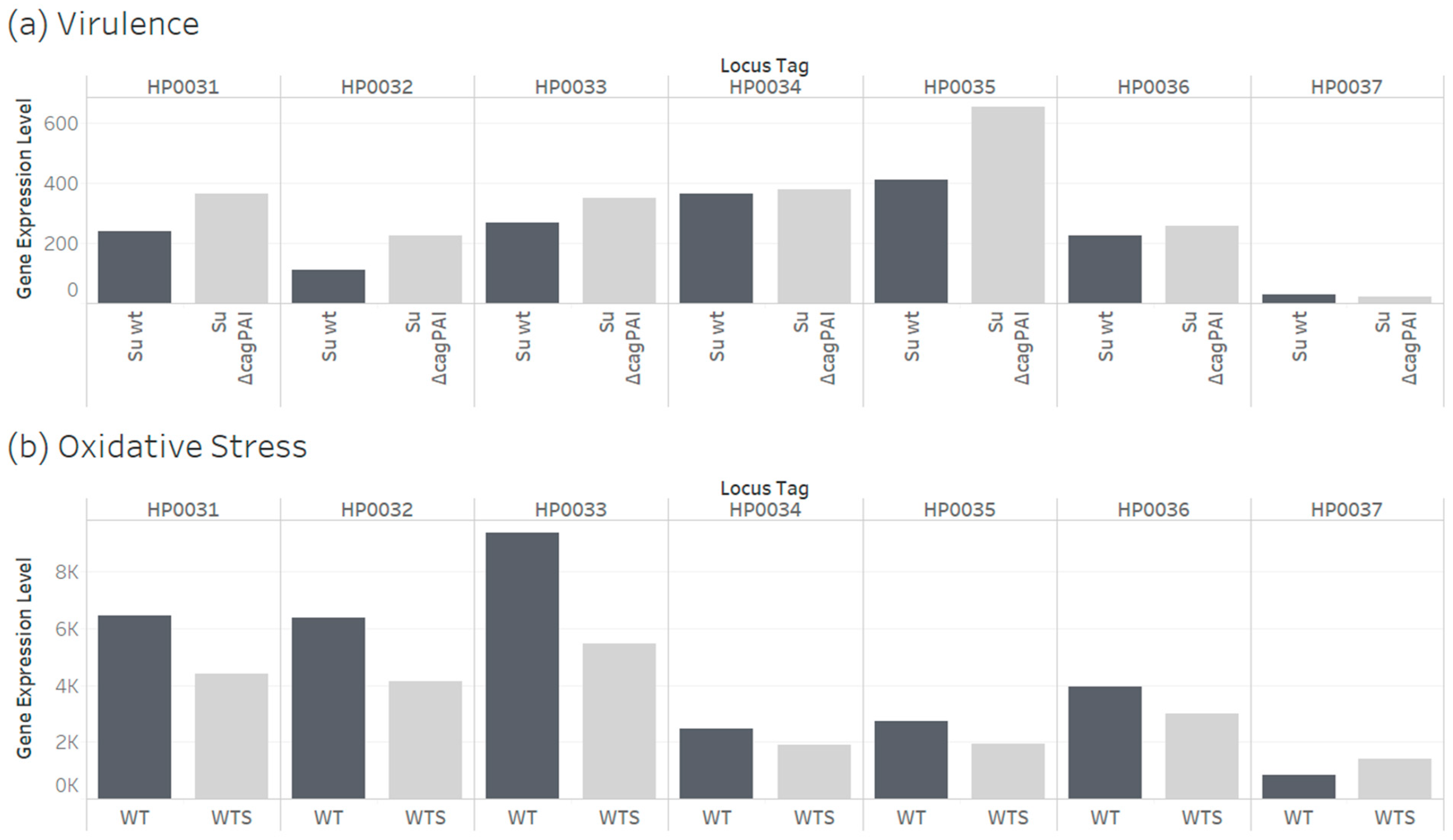
3.5. Transcriptome and Interactome Evidence for Helicobacter pylori Universal Stress Protein
3.6. Interactive Analytics Resources for Investigating Dataset on Helicobacteraceae Genomes and Universal Stress Protein Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AAA+ | ATPases associated with various cellular activities |
| AlpA | Adherence-associated lipoprotein A |
| AhpC | Alkyl hydroperoxide reductase |
| ATP | Adenosine Tri-Phosphate |
| CagA | Cytotoxin-associated gene A |
| DNA | Deoxyribonucleic acid |
| HtrA | High temperature requirement A protease |
| KatA | Catalase |
| NCBI | National Center for Biotechnology Information |
| VacA | Vacuolating cytotoxin A |
| USP | Universal Stress Protein |
References
- Aydin, F.; Saticioglu, I.B.; Ay, H.; Kayman, T.; Karakaya, E.; Abay, S. Description of the two novel species of the genus Helicobacter: Helicobacter anatolicus sp. nov., and Helicobacter kayseriensis sp. nov., isolated from feces of urban wild birds. Syst. Appl. Microbiol. 2022, 45, 126326. [Google Scholar] [CrossRef] [PubMed]
- On, S.L.; Miller, W.G.; Houf, K.; Fox, J.G.; Vandamme, P. Minimal standards for describing new species belonging to the families Campylobacteraceae and Helicobacteraceae: Campylobacter, Arcobacter, Helicobacter and Wolinella spp. Int. J. Syst. Evol. Microbiol. 2017, 67, 5296. [Google Scholar] [CrossRef] [PubMed]
- Varon, C.; Azzi-Martin, L.; Khalid, S.; Seeneevassen, L.; Ménard, A.; Spuul, P. Helicobacters and cancer, not only gastric cancer? Semin. Cancer Biol. 2022, 86, 1138–1154. [Google Scholar] [CrossRef] [PubMed]
- Solnick, J.V.; Schauer, D.B. Emergence of diverse Helicobacter species in the pathogenesis of gastric and enterohepatic diseases. Clin. Microbiol. Rev. 2001, 14, 59–97. [Google Scholar] [CrossRef]
- Mladenova-Hristova, I.; Grekova, O.; Patel, A. Zoonotic potential of Helicobacter spp. J. Microbiol. Immunol. Infect. 2017, 50, 265–269. [Google Scholar] [CrossRef]
- Okoli, A.S.; Wilkins, M.R.; Raftery, M.J.; Mendz, G.L. Response of Helicobacter hepaticus to bovine bile. J. Proteome Res. 2010, 9, 1374–1384. [Google Scholar] [CrossRef]
- Karkhah, A.; Ebrahimpour, S.; Rostamtabar, M.; Koppolu, V.; Darvish, S.; Vasigala, V.K.R.; Validi, M.; Nouri, H.R. Helicobacter pylori evasion strategies of the host innate and adaptive immune responses to survive and develop gastrointestinal diseases. Microbiol. Res. 2019, 218, 49–57. [Google Scholar] [CrossRef]
- Marcus, E.A.; Sachs, G.; Scott, D.R. Colloidal bismuth subcitrate impedes proton entry into Helicobacter pylori and increases the efficacy of growth-dependent antibiotics. Aliment. Pharmacololgy Ther. 2015, 42, 922–933. [Google Scholar] [CrossRef]
- Isokpehi, R.D.; McInnis, D.S.; Destefano, A.M.; Johnson, G.S.; Walker, A.D.; Hall, Y.A.; Mapp, B.W.; Johnson, M.O.; Simmons, S.S. Bioinformatics investigations of universal stress proteins from mercury-methylating Desulfovibrionaceae. Microorganisms 2021, 9, 1780. [Google Scholar] [CrossRef]
- Williams, B.S.; Isokpehi, R.D.; Mbah, A.N.; Hollman, A.L.; Bernard, C.O.; Simmons, S.S.; Ayensu, W.K.; Garner, B.L. Functional annotation analytics of Bacillus genomes reveals stress responsive acetate utilization and sulfate uptake in the biotechnologically relevant Bacillus megaterium. Bioinform. Biol. Insights 2012, 6, 275–286. [Google Scholar] [CrossRef]
- Isokpehi, R.D.; Simmons, S.S.; Cohly, H.H.; Ekunwe, S.I.; Begonia, G.B.; Ayensu, W.K. Identification of drought-responsive universal stress proteins in Viridiplantae. Bioinform. Biol. Insights 2011, 5, 41–58. [Google Scholar] [CrossRef]
- Isokpehi, R.D.; Simmons, S.S.; Johnson, M.O.; Payton, M. Genomic evidence for bacterial determinants influencing obesity development. Int. J. Environ. Res. Public Health 2017, 14, 345. [Google Scholar] [CrossRef] [PubMed]
- Isokpehi, R.D.; Mahmud, O.; Mbah, A.N.; Simmons, S.S.; Avelar, L.; Rajnarayanan, R.V.; Udensi, U.K.; Ayensu, W.K.; Cohly, H.H.; Brown, S.D. Developmental regulation of genes encoding universal stress proteins in Schistosoma mansoni. Gene Regul. Syst. Biol. 2011, 5, 61–74. [Google Scholar] [CrossRef] [PubMed]
- Mbah, A.N.; Mahmud, O.; Awofolu, O.R.; Isokpehi, R.D. Inferences on the biochemical and environmental regulation of universal stress proteins from schistosomiasis parasites. Adv. Appl. Bioinform. Chem. 2013, 6, 15–27. [Google Scholar] [PubMed]
- Makolo, A.U. Transcription direction patterns of adjacent genes in Mycobacterium tuberculosis using GENAVIS. Comput. Biol. Bioinform. 2019, 7, 1–4. [Google Scholar] [CrossRef]
- Sousa, M.C.; McKay, D.B. Structure of the universal stress protein of Haemophilus influenzae. Structure 2001, 9, 1135–1141. [Google Scholar] [CrossRef]
- Tkaczuk, K.L.; I, A.S.; Chruszcz, M.; Evdokimova, E.; Savchenko, A.; Minor, W. Structural and functional insight into the universal stress protein family. Evol. Appl. 2013, 6, 434–449. [Google Scholar] [CrossRef]
- Zarembinski, T.I.; Hung, L.W.; Mueller-Dieckmann, H.J.; Kim, K.K.; Yokota, H.; Kim, R.; Kim, S.H. Structure-based assignment of the biochemical function of a hypothetical protein: A test case of structural genomics. Proc. Natl. Acad. Sci. USA 1998, 95, 15189–15193. [Google Scholar] [CrossRef]
- Schweikhard, E.S.; Kuhlmann, S.I.; Kunte, H.J.; Grammann, K.; Ziegler, C.M. Structure and function of the universal stress protein TeaD and its role in regulating the ectoine transporter TeaABC of Halomonas elongata DSM 2581(T). Biochemistry 2010, 49, 2194–2204. [Google Scholar] [CrossRef]
- Nachin, L.; Nannmark, U.; Nystrom, T. Differential roles of the universal stress proteins of Escherichia coli in oxidative stress resistance, adhesion, and motility. J. Bacteriol. 2005, 187, 6265–6272. [Google Scholar] [CrossRef]
- Seifart Gomes, C.; Izar, B.; Pazan, F.; Mohamed, W.; Mraheil, M.A.; Mukherjee, K.; Billion, A.; Aharonowitz, Y.; Chakraborty, T.; Hain, T. Universal stress proteins are important for oxidative and acid stress resistance and growth of Listeria monocytogenes EGD-e in vitro and in vivo. PLoS ONE 2011, 6, e24965. [Google Scholar] [CrossRef]
- Nystrom, T.; Gustavsson, N. Maintenance energy requirement: What is required for stasis survival of Escherichia coli? Biochim. Biophys. Acta 1998, 1365, 225–231. [Google Scholar] [CrossRef] [PubMed]
- Nystrom, T.; Neidhardt, F.C. Expression and role of the universal stress protein, UspA, of Escherichia coli during growth arrest. Mol. Microbiol. 1994, 11, 537–544. [Google Scholar] [CrossRef] [PubMed]
- Gustavsson, N.; Diez, A.; Nystrom, T. The universal stress protein paralogues of Escherichia coli are co-ordinately regulated and co-operate in the defence against DNA damage. Mol. Microbiol. 2002, 43, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Heermann, R.; Lippert, M.L.; Jung, K. Domain swapping reveals that the N-terminal domain of the sensor kinase KdpD in Escherichia coli is important for signaling. BMC Microbiol. 2009, 9, 133. [Google Scholar] [CrossRef]
- Drumm, J.E.; Mi, K.; Bilder, P.; Sun, M.; Lim, J.; Bielefeldt-Ohmann, H.; Basaraba, R.; So, M.; Zhu, G.; Tufariello, J.M. Mycobacterium tuberculosis universal stress protein Rv2623 regulates bacillary growth by ATP-Binding: Requirement for establishing chronic persistent infection. PLoS Pathog. 2009, 5, e1000460. [Google Scholar] [CrossRef]
- Jia, Q.; Hu, X.; Shi, D.; Zhang, Y.; Sun, M.; Wang, J.; Mi, K.; Zhu, G. Universal stress protein Rv2624c alters abundance of arginine and enhances intracellular survival by ATP binding in mycobacteria. Sci. Rep. 2016, 6, 35462. [Google Scholar] [CrossRef]
- Sharma, C.M.; Hoffmann, S.; Darfeuille, F.; Reignier, J.; Findeiß, S.; Sittka, A.; Chabas, S.; Reiche, K.; Hackermüller, J.; Reinhardt, R. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 2010, 464, 250–255. [Google Scholar] [CrossRef]
- Singh, S.; Guttula, P.K.; Guruprasad, L. Structure based annotation of Helicobacter pylori strain 26695 proteome. PLoS ONE 2014, 9, e115020. [Google Scholar] [CrossRef]
- Luo, D.; Wu, Z.; Bai, Q.; Zhang, Y.; Huang, M.; Huang, Y.; Li, X. Universal stress proteins: From gene to function. Int. J. Mol. Sci. 2023, 24, 4725. [Google Scholar] [CrossRef]
- Samanta, S.; Biswas, P.; Banerjee, A.; Bose, A.; Siddiqui, N.; Nambi, S.; Saini, D.K.; Visweswariah, S.S. A universal stress protein in Mycobacterium smegmatis sequesters the cAMP-regulated lysine acyltransferase and is essential for biofilm formation. J. Biol. Chem. 2020, 295, 1500–1516. [Google Scholar] [CrossRef] [PubMed]
- Chen, I.-M.A.; Chu, K.; Palaniappan, K.; Ratner, A.; Huang, J.; Huntemann, M.; Hajek, P.; Ritter, S.J.; Webb, C.; Wu, D. The IMG/M data management and analysis system v. 7: Content updates and new features. Nucleic Acids Res. 2023, 51, D723–D732. [Google Scholar] [CrossRef] [PubMed]
- Kehrer, J.; Hauser, H. Visualization and visual analysis of multifaceted scientific data: A survey. IEEE Trans. Vis. Comput. Graph. 2012, 19, 495–513. [Google Scholar] [CrossRef] [PubMed]
- Beard, L.; Aghassibake, N. Tableau (version 2020.3). J. Med. Libr. Assoc. 2021, 109, 159. [Google Scholar] [CrossRef]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A. The conserved domain database in 2023. Nucleic Acids Res. 2023, 51, D384–D388. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. CD-HIT: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- The Galaxy Community. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2022 update. Nucleic Acids Res. 2022, 50, W345–W351. [Google Scholar] [CrossRef]
- Thorell, K.; Muñoz-Ramírez, Z.Y.; Wang, D.; Sandoval-Motta, S.; Boscolo Agostini, R.; Ghirotto, S.; Torres, R.C.; Falush, D.; Camargo, M.C. The Helicobacter pylori Genome Project: Insights into H. pylori population structure from analysis of a worldwide collection of complete genomes. Nat. Commun. 2023, 14, 8184. [Google Scholar] [CrossRef]
- Rozewicki, J.; Li, S.; Amada, K.M.; Standley, D.M.; Katoh, K. MAFFT-DASH: Integrated protein sequence and structural alignment. Nucleic Acids Res. 2019, 47, W5–W10. [Google Scholar] [CrossRef]
- Sayers, E.W.; Beck, J.; Bolton, E.E.; Bourexis, D.; Brister, J.R.; Canese, K.; Comeau, D.C.; Funk, K.; Kim, S.; Klimke, W. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2021, 49, D10. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Serra, F.; Bork, P. ETE 3: Reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol. 2016, 33, 1635–1638. [Google Scholar] [CrossRef]
- Kountz, D.J.; Balskus, E.P. Leveraging microbial genomes and genomic context for chemical discovery. Acc. Chem. Res. 2021, 54, 2788–2797. [Google Scholar] [CrossRef]
- Mavromatis, K.; Chu, K.; Ivanova, N.; Hooper, S.D.; Markowitz, V.M.; Kyrpides, N.C. Gene context analysis in the Integrated Microbial Genomes (IMG) data management system. PLoS ONE 2009, 4, e7979. [Google Scholar] [CrossRef] [PubMed]
- Seitzer, P.; Yao, A.I.; Cisneros, A.; Facciotti, M.T. The exploration of novel regulatory relationships drives Haloarchaeal operon-like structural dynamics over short evolutionary distances. Microorganisms 2020, 8, 1900. [Google Scholar] [CrossRef]
- Lim, H.N.; Lee, Y.; Hussein, R. Fundamental relationship between operon organization and gene expression. Proc. Natl. Acad. Sci. USA 2011, 108, 10626–10631. [Google Scholar] [CrossRef]
- Li, X.-Q.; Du, D. Gene direction in living organisms. Sci. Rep. 2012, 2, 982. [Google Scholar] [CrossRef]
- Karp, P.D.; Ivanova, N.; Krummenacker, M.; Kyrpides, N.; Latendresse, M.; Midford, P.; Ong, W.K.; Paley, S.; Seshadri, R. A comparison of microbial genome web portals. Front. Microbiol. 2019, 10, 208. [Google Scholar] [CrossRef] [PubMed]
- Galperin, M.Y.; Makarova, K.S.; Wolf, Y.I.; Koonin, E.V. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 2015, 43, D261–D269. [Google Scholar] [CrossRef]
- Karp, P.D.; Billington, R.; Caspi, R.; Fulcher, C.A.; Latendresse, M.; Kothari, A.; Keseler, I.M.; Krummenacker, M.; Midford, P.E.; Ong, Q. The BioCyc collection of microbial genomes and metabolic pathways. Brief. Bioinform. 2019, 20, 1085–1093. [Google Scholar] [CrossRef]
- Mannion, A.; Shen, Z.; Fox, J.G. Comparative genomics analysis to differentiate metabolic and virulence gene potential in gastric versus enterohepatic Helicobacter species. BMC Genom. 2018, 19, 830. [Google Scholar] [CrossRef]
- O’Toole, P.W.; Snelling, W.J.; Canchaya, C.; Forde, B.M.; Hardie, K.R.; Josenhans, C.; Graham, R.L.; McMullan, G.; Parkhill, J.; Belda, E. Comparative genomics and proteomics of Helicobacter mustelae, an ulcerogenic and carcinogenic gastric pathogen. BMC Genom. 2010, 11, 164. [Google Scholar] [CrossRef]
- Gilbert, M.J.; Duim, B.; Timmerman, A.J.; Zomer, A.L.; Wagenaar, J.A. Whole genome-based phylogeny of reptile-associated Helicobacter indicates independent niche adaptation followed by diversification in a poikilothermic host. Sci. Rep. 2017, 7, 8387. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Clark, K.; Gevorgyan, R.; Gorelenkov, V.; Gribov, E.; Karsch-Mizrachi, I.; Kimelman, M.; Pruitt, K.D.; Resenchuk, S.; Tatusova, T. BioProject and BioSample databases at NCBI: Facilitating capture and organization of metadata. Nucleic Acids Res. 2012, 40, D57–D63. [Google Scholar] [CrossRef] [PubMed]
- Jeon, W.J.; Dong, H.-J.; Shin, J.H.; Kim, I.Y.; Ho, H.; Oh, S.H.; Yoon, Y.M.; Choi, Y.-K.; Suh, J.G.; Nam, K.-H. Helicobacter apodemus sp. nov., a new Helicobacter species identified from the gastrointestinal tract of striped field mice in Korea. J. Vet. Sci. 2015, 16, 475–481. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Cantillo, M.; Vidal-Veuthey, B.; Mella, A.; de la Haba, R.R.; Collado, L. Helicobacter ibis sp. nov., isolated from faecal droppings of black-faced ibis (Theristicus melanopis). Int. J. Syst. Evol. Microbiol. 2023, 73, 005983. [Google Scholar] [CrossRef]
- Kersulyte, D.; Bertoli, M.T.; Tamma, S.; Keelan, M.; Munday, R.; Geary, J.; Veldhuyzen van Zanten, S.; Goodman, K.J.; Berg, D.E. Complete genome sequences of two Helicobacter pylori strains from a Canadian Arctic Aboriginal community. Genome Announc. 2015, 3, e00209-15. [Google Scholar] [CrossRef]
- Mukherjee, S.; Stamatis, D.; Li, C.T.; Ovchinnikova, G.; Bertsch, J.; Sundaramurthi, J.C.; Kandimalla, M.; Nicolopoulos, P.A.; Favognano, A.; Chen, I.-M.A. Twenty-five years of Genomes OnLine Database (GOLD): Data updates and new features in v. 9. Nucleic Acids Res. 2023, 51, D957–D963. [Google Scholar] [CrossRef]
- Didelot, X.; Nell, S.; Yang, I.; Woltemate, S.; Van der Merwe, S.; Suerbaum, S. Genomic evolution and transmission of Helicobacter pylori in two South African families. Proc. Natl. Acad. Sci. USA 2013, 110, 13880–13885. [Google Scholar] [CrossRef]
- Shen, Z.; Mannion, A.; Lin, M.; Esmail, M.; Bakthavatchalu, V.; Yang, S.; Ho, C.; Feng, Y.; Smith, B.; Elliott, J. Helicobacter monodelphidis sp. nov. and Helicobacter didelphidarum sp. nov., isolated from grey short-tailed opossums (Monodelphis domestica) with endemic cloacal prolapses. Int. J. Syst. Evol. Microbiol. 2020, 70, 6032–6043. [Google Scholar] [CrossRef]
- Schwarz, S.; Morelli, G.; Kusecek, B.; Manica, A.; Balloux, F.; Owen, R.J.; Graham, D.Y.; Van der Merwe, S.; Achtman, M.; Suerbaum, S. Horizontal versus familial transmission of Helicobacter pylori. PLoS Pathog. 2008, 4, e1000180. [Google Scholar] [CrossRef]
- Hu, L.; Zeng, X.; Ai, Q.; Liu, C.; Zhang, X.; Chen, Y.; Liu, L.; Li, G.-Q. Long-read-and short-read-based whole-genome sequencing reveals the antibiotic resistance pattern of Helicobacter pylori. Microbiol. Spectr. 2023, 11, e04522. [Google Scholar] [CrossRef] [PubMed]
- Gunaletchumy, S.P.; Teh, X.; Khosravi, Y.; Ramli, N.S.K.; Chua, E.G.; Kavitha, T.; Mason, J.N.; Lee, H.T.; Alias, H.; Zaidan, N.Z. Draft genome sequences of Helicobacter pylori isolates from Malaysia, cultured from patients with functional dyspepsia and gastric cancer. J. Bacteriol. 2012, 194, 5695–5696. [Google Scholar] [CrossRef]
- Rehvathy, V.; Tan, M.H.; Gunaletchumy, S.P.; Teh, X.; Wang, S.; Baybayan, P.; Singh, S.; Ashby, M.; Kaakoush, N.O.; Mitchell, H.M.; et al. Multiple genome sequences of Helicobacter pylori strains of diverse disease and antibiotic resistance backgrounds from Malaysia. Genome Announc. 2013, 1, e00687-13. [Google Scholar] [CrossRef]
- Blanchard, T.G.; Czinn, S.J.; Correa, P.; Nakazawa, T.; Keelan, M.; Morningstar, L.; Santana-Cruz, I.; Maroo, A.; McCracken, C.; Shefchek, K. Genome sequences of 65 Helicobacter pylori strains isolated from asymptomatic individuals and patients with gastric cancer, peptic ulcer disease, or gastritis. Pathog. Dis. 2013, 68, 39–43. [Google Scholar] [CrossRef]
- Gutiérrez-Escobar, A.J.; Velapatiño, B.; Borda, V.; Rabkin, C.S. Identification of new Helicobacter pylori subpopulations in native Americans and mestizos from Peru. Front. Microbiol. 2020, 11, 601839. [Google Scholar] [CrossRef] [PubMed]
- Hauke, M.; Metz, F.; Rapp, J.; Faass, L.; Bats, S.H.; Radziej, S.; Link, H.; Eisenreich, W.; Josenhans, C. Helicobacter pylori modulates heptose metabolite biosynthesis and heptose-dependent innate immune host cell activation by multiple mechanisms. Microbiol. Spectr. 2023, 11, e03132-22. [Google Scholar] [CrossRef] [PubMed]
- Noszka, M.; Strzałka, A.; Muraszko, J.; Kolenda, R.; Meng, C.; Ludwig, C.; Stingl, K.; Zawilak-Pawlik, A. Profiling of the Helicobacter pylori redox switch HP1021 regulon using a multi-omics approach. Nat. Commun. 2023, 14, 6715. [Google Scholar] [CrossRef]
- Olbermann, P.; Josenhans, C.; Moodley, Y.; Uhr, M.; Stamer, C.; Vauterin, M.; Suerbaum, S.; Achtman, M.; Linz, B. A global overview of the genetic and functional diversity in the Helicobacter pylori cag pathogenicity island. PLoS Genet. 2010, 6, e1001069. [Google Scholar] [CrossRef]
- Rain, J.-C.; Selig, L.; De Reuse, H.; Battaglia, V.; Reverdy, C.; Simon, S.; Lenzen, G.; Petel, F.; Wojcik, J.; Schächter, V. The protein–protein interaction map of Helicobacter pylori. Nature 2001, 409, 211–215. [Google Scholar] [CrossRef]
- Malfertheiner, P.; Selgrad, M.; Wex, T.; Romi, B.; Borgogni, E.; Spensieri, F.; Zedda, L.; Ruggiero, P.; Pancotto, L.; Censini, S. Efficacy, immunogenicity, and safety of a parenteral vaccine against Helicobacter pylori in healthy volunteers challenged with a Cag-positive strain: A randomised, placebo-controlled phase 1/2 study. Lancet Gastroenterol. Hepatol. 2018, 3, 698–707. [Google Scholar] [CrossRef]
- Linz, B.; Windsor, H.M.; McGraw, J.J.; Hansen, L.M.; Gajewski, J.P.; Tomsho, L.P.; Hake, C.M.; Solnick, J.V.; Schuster, S.C.; Marshall, B.J. A mutation burst during the acute phase of Helicobacter pylori infection in humans and rhesus macaques. Nat. Commun. 2014, 5, 4165. [Google Scholar] [CrossRef]
- Kumar, S.; Karmakar, B.C.; Nagarajan, D.; Mukhopadhyay, A.K.; Morgan, R.D.; Rao, D.N. N4-cytosine DNA methylation regulates transcription and pathogenesis in Helicobacter pylori. Nucleic Acids Res. 2018, 46, 3429–3445. [Google Scholar] [CrossRef]
- Clancy, C.D.; Forde, B.M.; Moore, S.A.; O’Toole, P.W. Draft genome sequences of Helicobacter pylori strains 17874 and P79. J. Bacteriol. 2012, 194, 2402. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yang, T.; Liao, T.; Debowski, A.W.; Nilsson, H.-O.; Fulurija, A.; Haslam, S.M.; Mulloy, B.; Dell, A.; Stubbs, K.A. The redefinition of Helicobacter pylori lipopolysaccharide O-antigen and core-oligosaccharide domains. PLoS Pathog. 2017, 13, e1006280. [Google Scholar] [CrossRef] [PubMed]
- Covacci, A.; Censini, S.; Bugnoli, M.; Petracca, R.; Burroni, D.; Macchia, G.; Massone, A.; Papini, E.; Xiang, Z.; Figura, N. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc. Natl. Acad. Sci. USA 1993, 90, 5791–5795. [Google Scholar] [CrossRef]
- Xiang, Z.; Censini, S.; Bayeli, P.F.; Telford, J.L.; Figura, N.; Rappuoli, R.; Covacci, A. Analysis of expression of CagA and VacA virulence factors in 43 strains of Helicobacter pylori reveals that clinical isolates can be divided into two major types and that CagA is not necessary for expression of the vacuolating cytotoxin. Infect. Immun. 1995, 63, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Ashaolu, J.O.; Tsai, Y.-J.; Liu, C.-C.; Ji, D.-D. Prevalence, diversity and public health implications of Helicobacter species in pet and stray dogs. One Health 2022, 15, 100430. [Google Scholar] [CrossRef]
- Foret, S.; Seneca, F.; de Jong, D.; Bieller, A.; Hemmrich, G.; Augustin, R.; Hayward, D.C.; Ball, E.E.; Bosch, T.C.; Agata, K.; et al. Phylogenomics reveals an anomalous distribution of USP genes in metazoans. Mol. Biol. Evol. 2011, 28, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Masamba, P.; Kappo, A.P. Parasite survival and disease persistence in cystic fibrosis, schistosomiasis and pathogenic bacterial diseases: A role for universal stress proteins? Int. J. Mol. Sci. 2021, 22, 10878. [Google Scholar] [CrossRef]
- Matarredona, L.; Zafrilla, B.; Rubio-Portillo, E.; Bonete, M.-J.; Esclapez, J. Deepening the knowledge of universal stress proteins in Haloferax mediterranei. Appl. Microbiol. Biotechnol. 2024, 108, 124. [Google Scholar] [CrossRef]
- Corbinais, C.; Mathieu, A.; Damke, P.P.; Kortulewski, T.; Busso, D.; Prado-Acosta, M.; Radicella, J.P.; Marsin, S. ComB proteins expression levels determine Helicobacter pylori competence capacity. Sci. Rep. 2017, 7, 41495. [Google Scholar] [CrossRef] [PubMed]
- Sen, A.; Zhou, Y.; Imlay, J.A. During oxidative stress the Clp proteins of Escherichia coli ensure that iron pools remain sufficient to reactivate oxidized metalloenzymes. J. Bacteriol. 2020, 202, e00235-20. [Google Scholar] [CrossRef]
- Dougan, D.A.; Reid, B.G.; Horwich, A.L.; Bukau, B. ClpS, a substrate modulator of the ClpAP machine. Mol. Cell 2002, 9, 673–683. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-H.; Chiou, S.-H. Proteomic analysis of upregulated proteins in Helicobacter pylori under oxidative stress induced by hydrogen peroxide. Kaohsiung J. Med. Sci. 2011, 27, 544–553. [Google Scholar] [CrossRef]
- Tegtmeyer, N.; Wessler, S.; Backert, S. Role of the cag-pathogenicity island encoded type IV secretion system in Helicobacter pylori pathogenesis. FEBS J. 2011, 278, 1190–1202. [Google Scholar] [CrossRef]
- Diez, A.; Gustavsson, N.; Nyström, T. The universal stress protein A of Escherichia coli is required for resistance to DNA damaging agents and is regulated by a RecA/FtsK-dependent regulatory pathway. Mol. Microbiol. 2000, 36, 1494–1503. [Google Scholar] [CrossRef] [PubMed]
- Gu, H. Role of flagella in the pathogenesis of Helicobacter pylori. Curr. Microbiol. 2017, 74, 863–869. [Google Scholar] [CrossRef]
- Shaik, M.M.; Cendron, L.; Percudani, R.; Zanotti, G. The structure of Helicobacter pylori HP0310 reveals an atypical peptidoglycan deacetylase. PLoS ONE 2011, 6, e19207. [Google Scholar] [CrossRef]
- Zarzecka, U.; Matkowska, D.; Backert, S.; Skorko-Glonek, J. Importance of two PDZ domains for the proteolytic and chaperone activities of Helicobacter pylori serine protease HtrA. Cell. Microbiol. 2021, 23, e13299. [Google Scholar] [CrossRef]
- Kumar, S.; Patel, G.K.; Ghoshal, U.C. Helicobacter pylori-induced inflammation: Possible factors modulating the risk of gastric cancer. Pathogens 2021, 10, 1099. [Google Scholar] [CrossRef]
- Masamba, P.; Adenowo, A.F.; Oyinloye, B.E.; Kappo, A.P. Universal stress proteins as new targets for environmental and therapeutic interventions of Schistosomiasis. Int. J. Environ. Res. Public Health 2016, 13, 972. [Google Scholar] [CrossRef]
- Wong, E.H.J.; Ng, C.G.; Chua, E.G.; Tay, A.C.Y.; Peters, F.; Marshall, B.J.; Ho, B.; Goh, K.L.; Vadivelu, J.; Loke, M.F. Comparative genomics revealed multiple Helicobacter pylori genes associated with biofilm formation in vitro. PLoS ONE 2016, 11, e0166835. [Google Scholar] [CrossRef]
- Njenga, R.; Boele, J.; Öztürk, Y.; Koch, H.-G. Coping with stress: How bacteria fine-tune protein synthesis and protein transport. J. Biol. Chem. 2023, 299, 105163. [Google Scholar] [CrossRef] [PubMed]
- Krüger, N.-J.; Knüver, M.-T.; Zawilak-Pawlik, A.; Appel, B.; Stingl, K. Genetic diversity as consequence of a microaerobic and neutrophilic lifestyle. PLoS Pathog. 2016, 12, e1005626. [Google Scholar] [CrossRef]
- Dorer, M.S.; Sessler, T.H.; Salama, N.R. Recombination and DNA repair in Helicobacter pylori. Annu. Rev. Microbiol. 2011, 65, 329–348. [Google Scholar] [CrossRef] [PubMed]
- O’Rourke, E.J.; Chevalier, C.; Pinto, A.V.; Thiberge, J.M.; Ielpi, L.; Labigne, A.; Radicella, J.P. Pathogen DNA as target for host-generated oxidative stress: Role for repair of bacterial DNA damage in Helicobacter pylori colonization. Proc. Natl. Acad. Sci. USA 2003, 100, 2789–2794. [Google Scholar] [CrossRef] [PubMed]
- Roman-Hernandez, G.; Hou, J.Y.; Grant, R.A.; Sauer, R.T.; Baker, T.A. The ClpS adaptor mediates staged delivery of N-end rule substrates to the AAA+ ClpAP protease. Mol. Cell 2011, 43, 217–228. [Google Scholar] [CrossRef]
- Kim, S.; Fei, X.; Sauer, R.T.; Baker, T.A. AAA+ protease-adaptor structures reveal altered conformations and ring specialization. Nat. Struct. Mol. Biol. 2022, 29, 1068–1079. [Google Scholar] [CrossRef]
- Loughlin, M.F.; Arandhara, V.; Okolie, C.; Aldsworth, T.G.; Jenks, P.J. Helicobacter pylori mutants defective in the clpP ATP-dependant protease and the chaperone clpA display reduced macrophage and murine survival. Microb. Pathog. 2009, 46, 53–57. [Google Scholar] [CrossRef]
- Vegge, C.S.; Brøndsted, L.; Ligowska-Marzęta, M.; Ingmer, H. Natural transformation of Campylobacter jejuni occurs beyond limits of growth. PLoS ONE 2012, 7, e45467. [Google Scholar] [CrossRef] [PubMed]
- Vorn, J. Biophysical Characterization of Unusual ClpS Proteolytic Adapters. Bachelor’s Thesis, University of Delaware, Newark, DE, USA, 2020. [Google Scholar]
- Bullock, K.K.; Shaffer, C.L.; Brooks, A.W.; Secka, O.; Forsyth, M.H.; McClain, M.S.; Cover, T.L. Genetic signatures for Helicobacter pylori strains of West African origin. PLoS ONE 2017, 12, e0188804. [Google Scholar] [CrossRef]
- Njenga, P.; Njau, A.; Moloo, Z.; Revathi, G.; Tshibangu, E.; Yamaoka, Y. Pattern and trends of Helicobacter pylori genotypes in gastric cancer: A Kenyan 8-year study. Front. Med. 2023, 10, 1119513. [Google Scholar] [CrossRef]
- Dammann, O. Data, information, evidence, and knowledge: A proposal for health informatics and data science. Online J. Public Health Inform. 2018, 10, e224. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.I.; Ajayi, A.; Jolaiya, T.; Onyekwere, C.; Setshedi, M.; Schulz, C.; Otegbayo, J.A.; Ndip, R.; Dieye, Y.; Alboraie, M. Helicobacter pylori infection in Africa: Update of the current situation and challenges. Dig. Dis. 2022, 40, 535–544. [Google Scholar] [CrossRef]
- Smith, S.I.; Schulz, C.; Ugiagbe, R.; Ndip, R.; Dieye, Y.; Leja, M.; Onyekwere, C.; Ndububa, D.; Ajayi, A.; Jolaiya, T.F. Helicobacter pylori diagnosis and treatment in Africa: The First Lagos Consensus Statement of the African Helicobacter and Microbiota Study Group. Dig. Dis. 2024, 42, 240–256. [Google Scholar] [CrossRef]
- Goffredi, S.K.; Panossian, B.; Brzechffa, C.; Field, N.; King, C.; Moggioli, G.; Rouse, G.W.; Martín-Durán, J.M.; Henry, L.M. A dynamic epibiont community associated with the bone-eating polychaete genus Osedax. MBio 2023, 14, e03140-22. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, Z.; Chen, C.-T.A.; Tang, K.; Su, J.; Jiao, N. Sulfur metabolizing microbes dominate microbial communities in andesite-hosted shallow-sea hydrothermal systems. PLoS ONE 2012, 7, e44593. [Google Scholar] [CrossRef] [PubMed]
- Olson, R.D.; Assaf, R.; Brettin, T.; Conrad, N.; Cucinell, C.; Davis, J.J.; Dempsey, D.M.; Dickerman, A.; Dietrich, E.M.; Kenyon, R.W. Introducing the bacterial and viral bioinformatics resource center (BV-BRC): A resource combining PATRIC, IRD and ViPR. Nucleic Acids Res. 2023, 51, D678–D689. [Google Scholar] [CrossRef]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L. InterPro in 2022. Nucleic Acids Res. 2023, 51, D418–D427. [Google Scholar] [CrossRef]




| Helicobacter Group by Anatomic Niche | Host Organism 1 | Helicobacter Species |
|---|---|---|
| Enterohepatic | Bird | H. anatolicus, H. anseris, H. pullorum, H. valdiviensis |
| Dog | H. canis | |
| Hamster | H. aurati, H. mesocricetorum | |
| Horse | H. equorum | |
| Human | H. bilis, H. burdigaliensis, H. canadensis, H. cinaedi, H. fennelliae, H. labetoulli, H. rappini, H. trogontum, H. winghamensis | |
| Marmot | H. marmotae, H. himalayensis | |
| Marmoset | H. jaachi | |
| Monkey | H. macacae, H. saguini | |
| Mouse | H. apodemus, H. ganmani, H. hepaticus, H. japonicas, H. magdeburgensis, H. rodentium, H. typhlonius | |
| Opossum | H. didelphidarum | |
| Pig | H. colisuis | |
| Rat | H. muridarum | |
| Squirrel | H. turcicus | |
| Enterohepatic and Gastric | Ferret | H. mustelae |
| Gastric | Cat | H. ailurogastricus, H. baculiformis, H. felis, H. heilmannii |
| Cheetah, Lion, Tiger | H. acinonychis | |
| Dog | H. bizzozeronii, H. cynogastricus, H. salomonis | |
| Dolphin | H. cetorum | |
| Fox | H. mehlei, H. vulpis | |
| Human | H. pylori | |
| Pig | H. suis |
| Strain Group 1 | Country of Isolation | Count of Strains | Count of Universal Stress Protein Sequences | Count of Sequence Clusters | Reference for Strain Group |
|---|---|---|---|---|---|
| GAM | Gambia | 45 | 45 | 4 | |
| SA | South Africa | 97 | 97 | 11 | [59,61] |
| Hpfe | China | 95 | 95 | 21 | [62] |
| UM | Malaysia | 61 | 68 | 17 | [63,64] |
| CPY | Japan | 9 | 9 | 5 | [65] |
| PUNO | Peru | 9 | 9 | 4 | [66] |
| Hp | USA | 74 | 74 | 11 | [65] |
| Universal Stress Protein Sequence Length Category | Biological Process Category of Predicted Function of Adjacent Gene to Universal Stress Protein Gene | Helicobacter Species |
|---|---|---|
| <200 aa | Energy production and conversion | H. aurati, H. muridarum |
| Amino acid transport and metabolism | H. bilis, H. canadensis, H. colisuis, H. pullorum, H. macacae | |
| Coenzyme transport and metabolism | H. anatolicus, H. cinaedi, H. equorum, H. hepaticus, H. himalayensis, H. jaachi, H. japonicus, H. labetoulli, H. magdeburgensis, H. marmotae, H. mesocricetorum, H. typhlonius, H. valdiviensis | |
| Replication, recombination, and repair | H. anatolicus, H. mustelae | |
| Posttranslational modification, protein turnover, and chaperones | H. acinonychis, H. ailurogastricus, H. baculiformis, H. bizzozeronii, H. cetorum, H. cynogastricus, H. felis, H. heilmannii, H. mehlei, H. mustelae, H. pylori, H. salomonis, H. suis, H. vulpis | |
| DNA Uptake | H. canis, H. cinaedi, H. fennelliae, H. hepaticus, H. jaachi, H. japonicus, H. labetoulli, macacae, H. magdeburgensis, H. marmotae, H. typhlonius | |
| Mediation of protein–protein interactions | H. ganmani | |
| >200 aa | DNA Uptake | H. canadensis, H. colisuis, H. ganmani, H. pullorum, H. rodentium, H. turcicus, H. valdiviensis, H. winghamensis |
| Membrane transport | H. ganmani, H. rodentium |
| Locus Tag * | Gene Symbol | Protein Name | Function Category |
|---|---|---|---|
| HP0006 | panC | Pantoate—beta-alanine ligase | Coenzyme transport and metabolism |
| HP0066 | DNA translocase FtsK | Cell cycle control, cell division, and chromosome partitioning | |
| HP0281 | tgt | tRNA guanosine(34) transglycosylase Tgt | Nucleotide transport and metabolism |
| HP1041 | flhA | Flagellar biosynthesis protein FlhA | Cell motility |
| HP1513 | L-seryl-tRNA(Sec) selenium transferase | Translation, ribosomal structure, and biogenesis | |
| HP1567 | Ribosome biogenesis GTP-binding protein YihA/YsxC | Cell cycle control, cell division, and chromosome partitioning |
| Gene Adjacency Distances * | Sequence Cluster | H. pylori Population or County and Strains in Sequence Cluster | Notes and References |
|---|---|---|---|
| 8:171 | Cluster 22 | hpEurope: BCM-300, HE101/09, HE132/09, HE136/09, HE141/09, HE142/09, HE143/09, HE147/09, HE170/09, HE171/09, HE178/09 | HE labeled strains are re-isolates from human volunteers experimentally challenged with strain BCM-300 [71]. |
| 10:111 | Cluster 22 | hpEurope: J166, J166output_1moA, J166output_1moB, J166output_1moC, J166output_1wkA, J166output_1wkB, J166output_1wkC, J166output_2moA, J166output_2moB, J166output_2moC, J166output_6moA, J166output_6moB, J166output_6moC | J166 output isolates are from time points after experimental infection of a rhesus macaque with J166 [72]. |
| 31:109 | Cluster 22 | hpEurope: 26695, 26695 dRdM2addM2, 26695-1, 26695-1CH, 26695-1CL, 26695-1MET, 26695-dR, 26695-dRdM1dM2, 26695-dRdM2, dRdM1, G27, HP2RS, Rif1, Rif2 | The strains are derived from or closely related to strain 26695 [73]. HP2RS indicates H. pylori 26695-related sequence [74]. G27 and 26695 have the same fundamental structure of the lipopolysaccharide of the outer membrane protein, a key factor for colonization and persistence in gastric niche [75]. |
| 31:109 | Cluster 102 | Australia: JCM 12093, CCUG 17874 hpEAsia: Hpfe0001 | JCM 12093, CCUG 17874, NCTC 11637, and ATCC 43504 are equivalent strains [58]. CCUG 17874 is a CagA and VacA producing strain [76,77]. There is genomic sequence evidence that Hpfe0001 is closely related to CCUG 17874 and ATCC 43504 [78]. |
| 44:126 | Cluster 103 | hpAfrica2: SA47A, SA47C | H. pylori isolates from atrium (SA47A) and corpus (SA47C) stomach regions of same individual [59,61]. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Isokpehi, R.D.; Simmons, S.S.; Makolo, A.U.; Hollman, A.L.; Adesida, S.A.; Ojo, O.O.; Abioye, A.O. Insights into Functions of Universal Stress Proteins Encoded by Genomes of Gastric Cancer Pathogen Helicobacter pylori and Related Bacteria. Pathogens 2025, 14, 275. https://doi.org/10.3390/pathogens14030275
Isokpehi RD, Simmons SS, Makolo AU, Hollman AL, Adesida SA, Ojo OO, Abioye AO. Insights into Functions of Universal Stress Proteins Encoded by Genomes of Gastric Cancer Pathogen Helicobacter pylori and Related Bacteria. Pathogens. 2025; 14(3):275. https://doi.org/10.3390/pathogens14030275
Chicago/Turabian StyleIsokpehi, Raphael D., Shaneka S. Simmons, Angela U. Makolo, Antoinesha L. Hollman, Solayide A. Adesida, Olabisi O. Ojo, and Amos O. Abioye. 2025. "Insights into Functions of Universal Stress Proteins Encoded by Genomes of Gastric Cancer Pathogen Helicobacter pylori and Related Bacteria" Pathogens 14, no. 3: 275. https://doi.org/10.3390/pathogens14030275
APA StyleIsokpehi, R. D., Simmons, S. S., Makolo, A. U., Hollman, A. L., Adesida, S. A., Ojo, O. O., & Abioye, A. O. (2025). Insights into Functions of Universal Stress Proteins Encoded by Genomes of Gastric Cancer Pathogen Helicobacter pylori and Related Bacteria. Pathogens, 14(3), 275. https://doi.org/10.3390/pathogens14030275





