A Begomovirus solanumdelhiense Vector for Virus-Induced Gene Silencing in Melon
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant, Inoculation, and Host Bacterial Materials
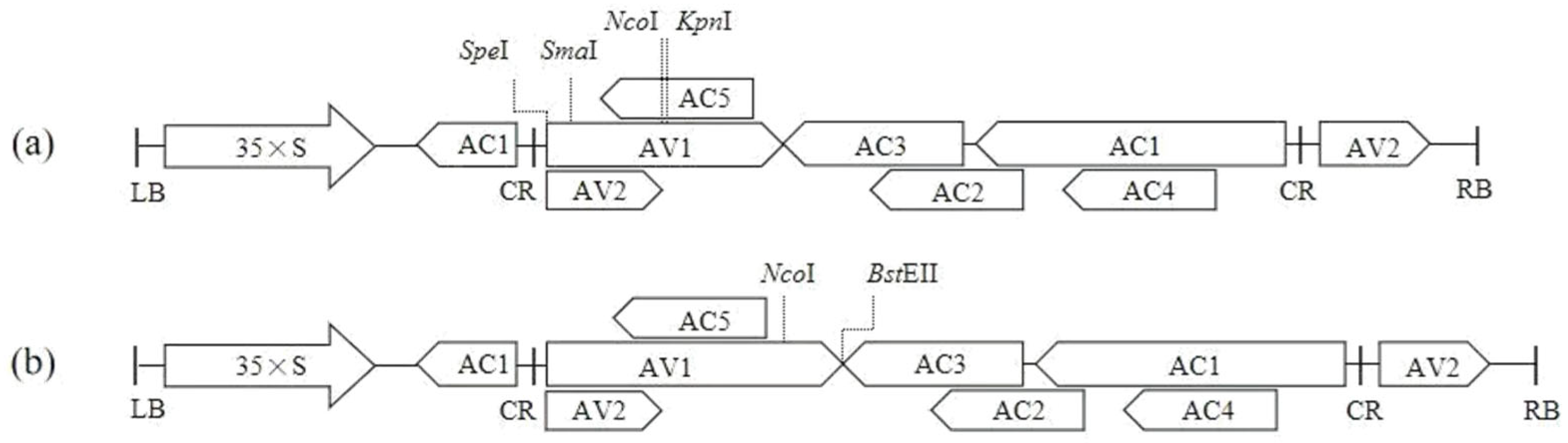
2.2. Design of Target Fragments and Construction of Empty Vectors
2.3. Construction of ToLCNDV-Based VIGS Vectors
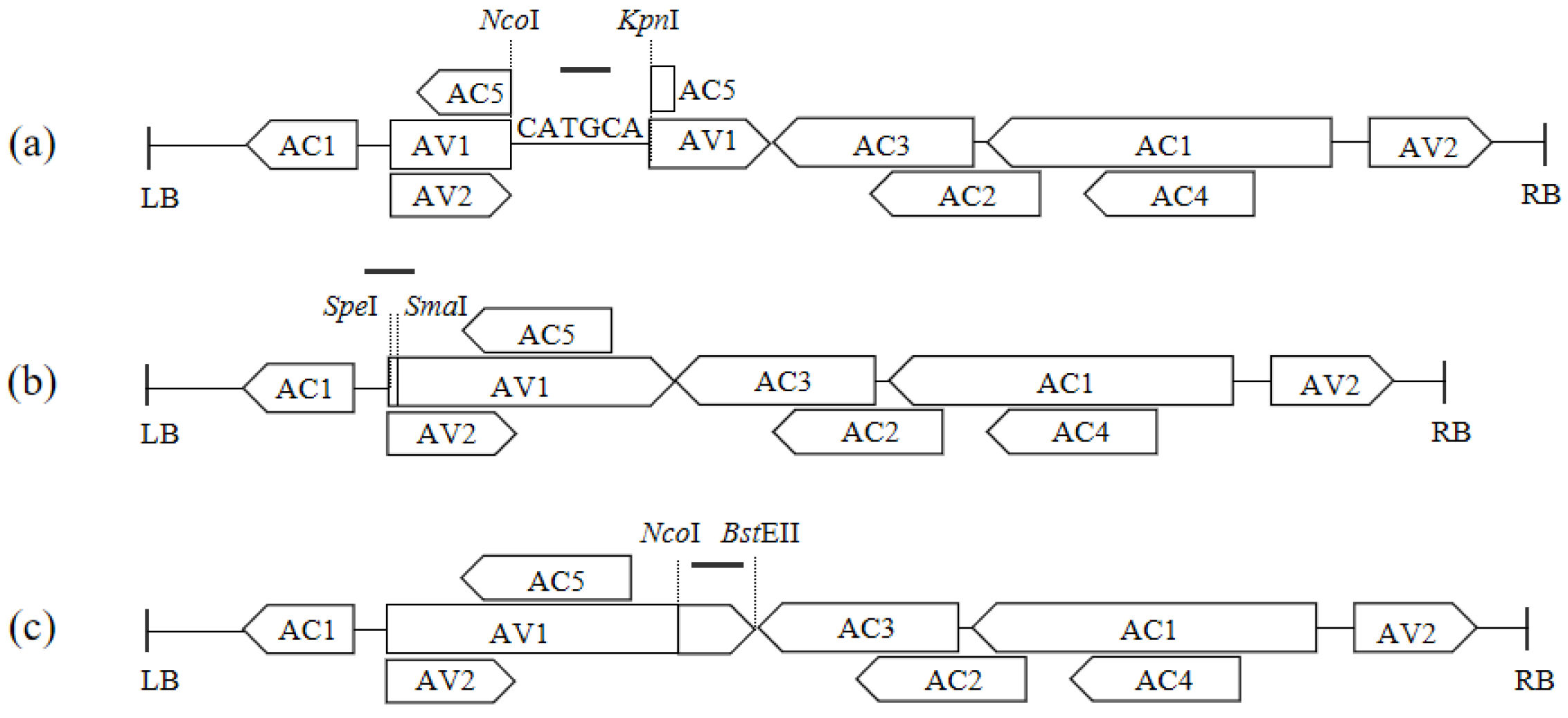
2.3.1. Construction of VIGS Vectors with Different Target Fragment Lengths
2.3.2. Construction of VIGS Vectors with Different Substitution Areas for the CmPDS Fragment
2.3.3. Construction of VIGS Vectors with the Antisense Orientation of CmPDS Fragments
2.4. Agroinfiltration
2.5. Effects of VIGS Vectors on the Phenotypic Traits of Melon Plants
2.6. Effects of VIGS Vectors on PDS Expression in Melon
3. Results
3.1. Design of Target Fragments and Construction of Empty Vectors
3.2. Construction of ToLCNDV-Based VIGS Vectors
3.3. Effects of VIGS Vectors on Melon Leaf Photobleaching
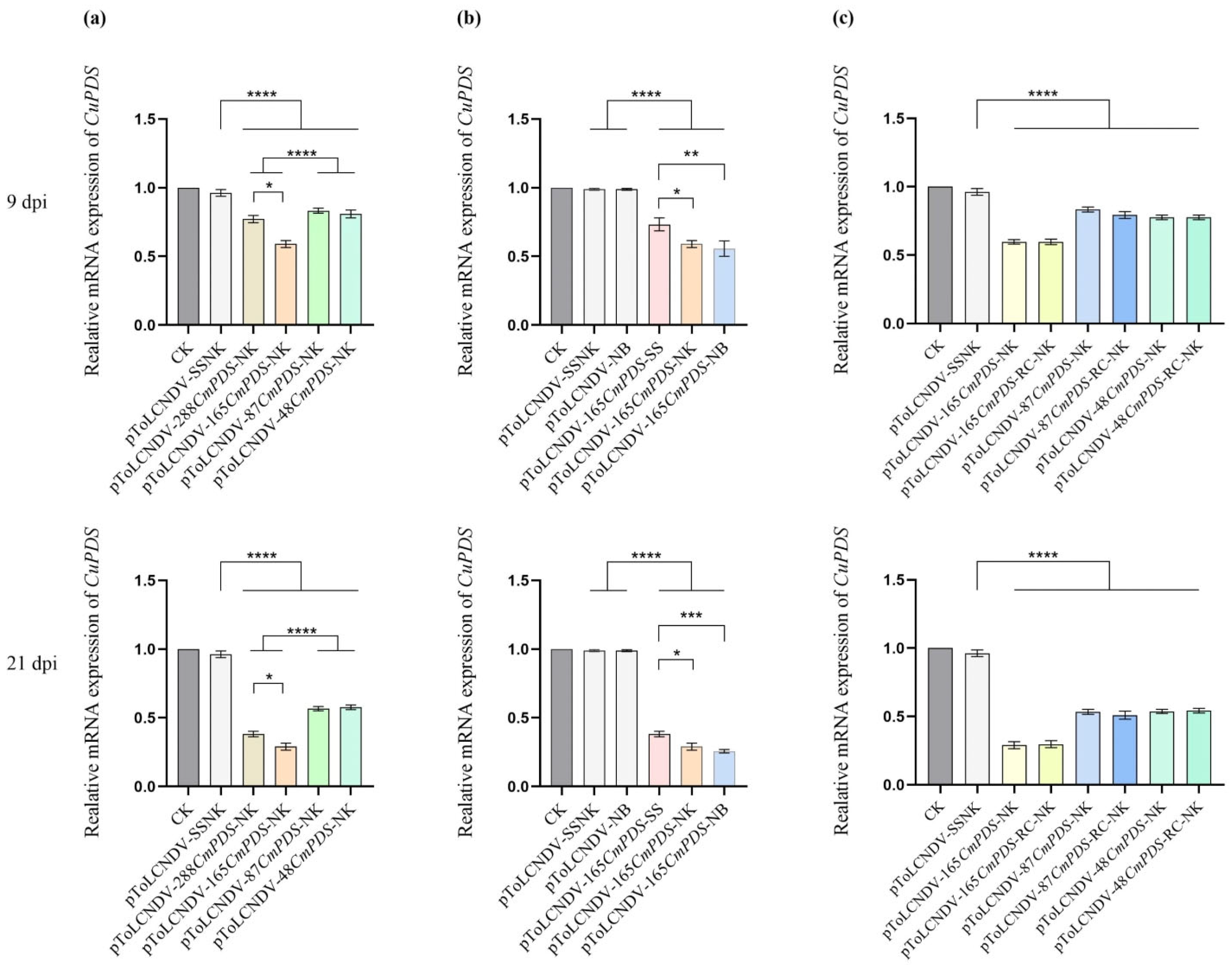
3.4. Influence of VIGS Vectors on PDS Expression in Melon
3.4.1. Silencing Effects of the Length of the CmPDS Fragment
3.4.2. Silencing Effects of the Substitution Area of the CmPDS Fragment
3.4.3. Silencing Effects of CmPDS Fragments in the Antisense Orientation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sun, G.Y.; Ju, Y.Q.; Zhang, C.P.; Li, L.L.; Jiang, X.Q.; Xie, X.M.; Lu, Y.Z.; Wang, K.L.; Li, W. Styrax japonicus functional genomics: An efficient virus induced gene silencing (VIGS) system. Hortic. Plant J. 2024, 10, 252–258. [Google Scholar] [CrossRef]
- Rössner, C.; Lotz, D.; Becker, A. VIGS Goes Viral: How VIGS Transforms Our Understanding of Plant Science. Annu. Rev. Plant Biol. 2022, 73, 703–728. [Google Scholar] [CrossRef]
- Zulfiqar, S.; Farooq, M.A.; Zhao, T.T.; Wang, P.P.; Tabusam, J.; Wang, Y.H.; Xuan, S.X.; Zhao, J.J.; Chen, X.P.; Shen, S.X.; et al. Virus–induced gene silencing(VIGS): A powerful tool for crop improvement and its advancement towards epigenetics. Int. J. Mol. Sci. 2023, 24, 5608. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.C.; Qin, X.; Wei, L.Y.; Jiang, D.D.; Zhang, Q.; Wang, W.Q.; Liang, R.H.; Zhang, R.; Zhang, K.; Liu, G.W.; et al. Engineered geminivirus replicons enable rapid in planta directed evolution. Science 2025, 390, eady2167. [Google Scholar] [CrossRef] [PubMed]
- Bu, R.; Wang, R.; Wei, Q.; Hu, H.Y.; Sun, H.L.; Song, P.W.; Yu, Y.A.; Liu, Q.L.; Zheng, Z.C.; Li, T.; et al. Silencing of glycerol–3–phosphate acyltransferase 6 (GPAT6) gene using a newly established virus induced gene silencing (VIGS) system in cucumber alleviates autotoxicity mimicked by cinnamic acid (CA). Plant Soil 2019, 438, 329–346. [Google Scholar] [CrossRef]
- Liao, J.J.; Wang, C.H.; Xing, Q.J.; Li, Y.P.; Liu, X.F.; Qi, H.Y. Overexpression and VIGS system for functional gene validation in oriental melon (Cucumis melo var. makuwa Makino). Plant Cell. Tissue Organ Cult. (PCTOC) 2019, 137, 275–284. [Google Scholar] [CrossRef]
- Zhao, F.M.; Lim, S.; Igori, D.; Yoo, R.H.; Kwon, S.; Moon, J.S. Development of tobacco ringspot virus–based vectors for foreign gene expression and virus–induced gene silencing in a variety of plants. Virology 2016, 492, 166–178. [Google Scholar] [CrossRef]
- Liu, M.; Liang, Z.; Aranda, M.A.; Hong, N.; Liu, L.M.; Kang, B.S. A cucumber green mottle mosaic virus vector for virus–induced gene silencing in cucurbit plants. Plant Methods 2020, 16, 9. [Google Scholar] [CrossRef]
- Burch–Smith, T.M.; Anderson, J.C.; Martin, G.B.; Dinesh–Kumar, S.P. Applications and advantages of virus–induced gene silencing for gene function studies in plants. Plant J. 2004, 39, 734–746. [Google Scholar] [CrossRef]
- Zaidi, S.S.; Mansoor, S. Viral vectors for plant genome engineering. Front. Plant Sci. 2017, 8, 539. [Google Scholar] [CrossRef]
- Li, F.F.; Xu, X.B.; Huang, C.J.; Gu, Z.H.; Cao, L.G.; Hu, T.; Ding, M.; Li, Z.H.; Zhou, X.P. The AC5 protein encoded by Mungbean yellow mosaic India virus is a pathogenicity determinant that suppresses RNA silencing-based antiviral defenses. New Phytol. 2015, 208, 555–569. [Google Scholar] [CrossRef]
- Domingo, E.; Holland, J.J. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 1997, 51, 151–178. [Google Scholar] [CrossRef]
- Fofana, I.B.F.; Sangare, A.; Collier, R.; Taylor, C.; Fauquet, C.M. A geminivirus–induced gene silencing system for gene function validation in cassava. Plant Mol. Biol. 2004, 56, 613–624. [Google Scholar] [CrossRef] [PubMed]
- Krenz, B.; Wege, C.; Jeske, H. Cell–free construction of disarmed abutilon mosaic virus–based gene silencing vectors. J. Virol. Methods 2010, 169, 129–137. [Google Scholar] [CrossRef]
- Tao, X.; Zhou, X. A modified viral satellite DNA that suppresses gene expression in plants. Plant J. 2004, 38, 850–860. [Google Scholar] [CrossRef] [PubMed]
- Tuttle, J.R.; Idris, A.M.; Brown, J.K.; Haigler, C.H.; Robertson, D. Geminivirus–mediated gene silencing from cotton leaf crumple virus is enhanced by low temperature in cotton. Plant Physiol. 2008, 148, 41–50. [Google Scholar] [CrossRef]
- Peele, C.; Jordan, C.V.; Muangsan, N.; Turnage, M.; Robertson, D. Silencing of a meristematic gene using geminivirus–derived vectors. Plant J. 2010, 27, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Jeyabharathy, C.; Shakila, H.; Usha, R. Development of a VIGS vector based on the beta–satellite DNA associated with bhendi yellow vein mosaic virus. Virus Res. 2015, 195, 73–78. [Google Scholar] [CrossRef]
- Tang, Y.; Wang, F.; Zhao, J.; Xie, K.; Hong, Y.; Liu, Y. Virus–based microRNA expression for gene functional analysis in plants. Plant Physiol. 2010, 153, 632–641. [Google Scholar] [CrossRef]
- Romero, J.; Aguado, E.; Martinez, C.; Garcia, A.; Cebrian, C.; Paris, H.S.; Jamilena, M. A novel dominant resistance gene for ToLCNDV in Cucurbita spp. Acta Hortic. 2020, 1294, 233–238. [Google Scholar] [CrossRef]
- Vo, T.T.B.; Troiano, E.; Lal, A.; Hoang, P.T.; Kil, E.J.; Lee, S.K.; Parrella, G. ToLCNDV–ES infection in tomato is enhanced by TYLCV: Evidence from field survey and agroinoculation. Front. Microbiol. 2022, 13, 954460. [Google Scholar] [CrossRef] [PubMed]
- Renukadevi, P.; Devi, G.R.; Jothika, C.; Karthikeyan, G.; Malathi, V.G.; Balakrishnan, N.; Rajagopal, B.; Nakkeeran, S.; Abd–Allah, E.F. Genomic distinctiveness and recombination in tomato leaf curl New Delhi virus (ToLCNDV–BG) isolates infecting bitter gourd. 3 Biotech 2024, 14, 184. [Google Scholar] [CrossRef] [PubMed]
- Martinez, C.; Manzano, S.; Megias, Z.; Garcia, A.; Garrido, R.; Paris, H.S.; Jamilena, M. Screening of Cucurbita germplasm for ToLCNDV resistance under natural greenhouse conditions. Acta Hortic. 2017, 1151, 57–62. [Google Scholar] [CrossRef]
- Naveena, E.; Rajasree, V.; Behara, T.K.; Karthikeyan, G.; Kavitha, M.; Rameshkumar, D. Molecular confirmation of ToLCNDV resistance in cucumber through agroinoculation and field screening. J. Anim. Plant Sci. Japs 2024, 34, 1582–1593. [Google Scholar] [CrossRef]
- Arif, M. Cross–species substitution matrix comparison of Tomato leaf curl New Delhi virus (ToLCNDV) with medicinal plant isolates. J. Plant Dis. Prot. 2024, 131, 1925–1934. [Google Scholar] [CrossRef]
- Iqbal, Z.; Khurshid, M. Immunocapture PCR Detection of ToLCNDV from Plant Extract by using Heterologous Virus Coat Protein Antisera. Pak. J. Zool. 2017, 49, 1025–1031. [Google Scholar] [CrossRef]
- Belén, R.; Pedro, G.; Dirk, J.; Ruiz, L. Insights into the Key Genes in Cucumis melo and Cucurbita moschata ToLCNDV Resistance. Horticulturae 2023, 9, 231. [Google Scholar] [CrossRef]
- Prasad, A.; Prasad, M. Interaction of ToLCNDV TrAP with SlATG8f marks it susceptible to degradation by autophagy. Cell. Mol. Life Sci. 2022, 79, 241. [Google Scholar] [CrossRef]
- Pérez–Moro, C.; Sáez, C.; Sifres, A.; Lopez, C.; Dhillon, N.P.S.; Picó, B.; Pérez–de–Castro, A. Genetic Dissection of ToLCNDV Resistance in Resistant Sources of Cucumis melo. Int. J. Mol. Sci. 2024, 25, 8880. [Google Scholar] [CrossRef]
- Ruiz, M.T.; Voinnet, O.; Baulcomb, D.C. Initiation and maintenance of virus–induced gene silencing. Plant Cell 1998, 10, 937–946. [Google Scholar] [CrossRef]
- Lacomme, C.; Hrubikova, K.; Ingo, H. Enhancement of virus–induced gene silencing through viral–based production of inverted–repeats. Plant J. Cell Mol. Biol. 2003, 34, 543–553. [Google Scholar] [CrossRef]
- Bennypaul, H.S.; Mutti, J.S.; Rustgi, S.; Kumar, N.; Okubara, P.A.; Gill, K.S. Virusinduced gene silencing (VIGS) of genes expressed in root, leaf, and meiotic tissues of wheat. Funct. Integr. Genom. 2012, 12, 143–156. [Google Scholar] [CrossRef] [PubMed]
- Pahlavan, A.; Solouki, M.; Fakheri, B.; Nasab, B.F. Using Morphological and Phytochemical Traits and ITS (1, 4) and rbcl DNA Barcodes in the Assessment of Different Malva sylvestris L. Genotypes. J. Med. Plants By-Prod. 2021, 10, 19–55. [Google Scholar] [CrossRef]
- Haseeb, M.; Amir, A.; Ikram, A. In Silico Analysis of SARS-CoV-2 Spike Proteins of Different Field Variants. Vaccines 2023, 11, 736. [Google Scholar] [CrossRef]
- Naing, H.A.; Song, Y.H.; Lee, M.J.; Lim, K.B.; Kim, C.K. Development of an efficient virus–induced gene silencing method in petunia using the pepper phytoene desaturase (PDS) gene. Plant Cell. Tissue Organ Cult. (PCTOC) 2019, 138, 507–515. [Google Scholar] [CrossRef]
- Avesani, L.; Marconi, G.; Morandini, F.; Albertini, E.; Bruschetta, M.; Bortesi, L.; Pezzotti, M.; Porceddu, A. Stability of Potato virus X expression vectors is related to insert size: Implications for replication models and risk assessment. Transgenic Res. 2007, 16, 587–597. [Google Scholar] [CrossRef]
- Gleba, Y.; Klimyuk, V.; Marillonnet, S. Viral vectors for the expression of proteins in plants. Curr. Opin. Biotechnol. 2007, 18, 134–141. [Google Scholar] [CrossRef]
- Peyret, H.; Lomonossoff, G.P. When plant virology met Agrobacterium: The rise of the deconstructed clones. Plant Biotechnol. J. 2015, 13, 1121–1135. [Google Scholar] [CrossRef]
- García, A.; Aragonés, V.; Gioiosa, S.; Herraiz, F.; Ortiz-García, P.; Prohens, J.; Daros, J.; Pasin, F. Comparative genomics-driven design of virus-delivered short RNA inserts triggering robust gene silencing. Plant Biotechnol. J. 2025, 23, 4930–4932. [Google Scholar] [CrossRef] [PubMed]
- Bruun–Rasmussen, M.; Madsen, C.T.; Jessing, S.; Albrechtsen, M. Stability of barley stripe mosaic virus–induced gene silencing in barley. Mol. Plant Microbe Interact. 2007, 20, 1323–1331. [Google Scholar] [CrossRef] [PubMed]
- Cakir, C.; Tör, M. Factors influencing barley stripe mosaic virus–mediated gene silencing in wheat. Physiol. Mol. Plant Pathol. 2010, 74, 246–253. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, Y.; Xu, H.; Amilijiang, W.; Wang, H. Developing and applying a virus–induced gene silencing system for functional genomics in walnut (Juglans regia L.) mediated by tobacco rattle virus. Gene 2025, 936, e936149087. [Google Scholar] [CrossRef]
- Deng, K.; Lu, Z.; Yang, H.; Chen, S.; Li, C.; Cao, D.; Wang, H.; Hao, Q.; Chen, H.; Shan, Z. Efficient Virus–Induced Gene Silencing (VIGS) Method for Discovery of Resistance Genes in Soybean. Plants 2025, 14, 1547–1563. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.Z.; Zhang, Y.; Wang, M.S.; Cheng, A.C.; Yang, Q.; Wu, Y.; Jia, R.Y.; Liu, M.F.; Zhu, D.K.; Chen, S.; et al. Structures and Functions of the 3′ Untranslated Regions of Positive–Sense Single–Stranded RNA Viruses Infecting Humans and Animals. Front. Cell. Infect. Microbiol. 2020, 10, 453. [Google Scholar] [CrossRef]
- Peng, T.Y.; Yang, F.Y.; Yang, F.; Cao, W.J.; Zheng, H.X.; Zhu, Z.X. Structural diversity and biological role of the 5′ untranslated regions of picornavirus. RNA Biol. 2023, 20, 548–562. [Google Scholar] [CrossRef]
- El–Mohtar, C.; Dawson, W.O. Exploring the limits of vector construction based on citrus tristeza virus. Virology 2014, 448, 274–283. [Google Scholar] [CrossRef] [PubMed]
- Qiao, W.; Falk, B. Efficient protein expression and virus–induced gene silencing in plants using a crinivirus–derived vector. Viruses 2018, 10, 216. [Google Scholar] [CrossRef]
- Zhang, C.; Bradshaw, J.D.; Whitham, S.; Hill, J.H. The development of an efficient multipurpose bean pod mottle virus viral vector set for foreign gene expression and RNA silencing. Plant Physiol. 2010, 153, 52–65. [Google Scholar] [CrossRef]
- Pacak, A.; Strozycki, P.M.; Barciszewska–Pacak, M.; Alejska, M.; Lacomme, C.; Jarmolowski, A.; Szweykowska-Kulinska, Z.; Figlerowicz, M. The brome mosaic virus–based recombination vector triggers a limited gene silencing response depending on the orientation of the inserted sequence. Arch. Virol. 2010, 155, 169–179. [Google Scholar] [CrossRef]
- Killiny, N.; Nehela, Y.; Hijaz, F.; Ben–Mahmoud, S.K.; Hajeri, S.; Gowda, S. Citrus tristeza virus–based induced gene silencing of phytoene desaturase is more efficient when antisense orientation is used. Plant Biotechnol. Rep. 2019, 13, 179–192. [Google Scholar] [CrossRef]
- Kant, R.; Dasgupta, I. Phenotyping of VIGS–mediated gene silencing in rice using a vector derived from a DNA virus. Plant Cell Rep. 2017, 36, 1159–1170. [Google Scholar] [CrossRef] [PubMed]
- Hiriart, J.B.; Lehto, K.; Tyystjärvi, E.; Junttila, T.; Aro, E.M. Suppression of a key gene involved in chlorophyll biosynthesis by means of virus–inducing gene silencing. Plant Mol. Biol. 2002, 50, 213–224. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Xing, M.; Liu, X.; Fang, Z.; Yang, L.; Zhang, Y.; Wang, Y.; Zhuang, M.; Lv, H. An efficient virus–induced gene silencing (VIGS) system for functional genomics in Brassicas using a cabbage leaf curl virus (CaLCuV)–based vector. Planta 2020, 252, 1–12. [Google Scholar] [CrossRef]
- Mo, Z.H.; Chen, Y.; Zhai, M.; Zhu, K.K.; Xuan, J.P.; Hu, L.J. Development and application of a virus–induced gene silencing system for functional genomics in pecan (Carya illinoinensis). Sci. Hortic. 2023, 310, e111759. [Google Scholar] [CrossRef]


| VIGS Systems (with pCambia-ToLCNDV-1.6B) | Systemically Silenced Plants/Total Infiltrated Plants | Incidence of Silenced Plants in Average (%) | Photobleaching Index in Average |
|---|---|---|---|
| pToLCNDV-288CmPDS-NK | 14/16, 14/16, 15/16 | 89.58 abc | 43.50 c |
| pToLCNDV-165CmPDS-NK | 15/16, 16/16, 16/16 | 97.92 a | 61.75 b |
| pToLCNDV-87CmPDS-NK | 13/16, 14/16, 13/16 | 83.33 bc | 29.34 d |
| pToLCNDV-48CmPDS-NK | 14/16, 13/16, 12/16 | 81.25 c | 25.67 d |
| pToLCNDV-165CmPDS-NB | 16/16, 16/16, 16/16 | 100 a | 76.50 a |
| pToLCNDV-165CmPDS-SS | 13/16, 12/16, 13/16 | 79.17 c | 48.13 c |
| pToLCNDV-165CmPDS-RC-NK | 15/16, 15/16, 16/16 | 95.83 ab | 60.58 b |
| pToLCNDV-87CmPDS-RC-NK | 14/16, 12/16, 13/16 | 81.25 c | 27.91 d |
| pToLCNDV-48CmPDS-RC-NK | 13/16, 13/16, 14/16 | 83.33 bc | 26.29 d |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, Y.; Liao, Q.; Gao, P.; Zhang, L.; Wan, B.; Xu, L.; Gao, S.; Song, Z.; Dai, F.; Zeng, R. A Begomovirus solanumdelhiense Vector for Virus-Induced Gene Silencing in Melon. Pathogens 2025, 14, 1269. https://doi.org/10.3390/pathogens14121269
Han Y, Liao Q, Gao P, Zhang L, Wan B, Xu L, Gao S, Song Z, Dai F, Zeng R. A Begomovirus solanumdelhiense Vector for Virus-Induced Gene Silencing in Melon. Pathogens. 2025; 14(12):1269. https://doi.org/10.3390/pathogens14121269
Chicago/Turabian StyleHan, Yufei, Qiansheng Liao, Ping Gao, Liqing Zhang, Bingqian Wan, Lihui Xu, Shigang Gao, Zhiwei Song, Fuming Dai, and Rong Zeng. 2025. "A Begomovirus solanumdelhiense Vector for Virus-Induced Gene Silencing in Melon" Pathogens 14, no. 12: 1269. https://doi.org/10.3390/pathogens14121269
APA StyleHan, Y., Liao, Q., Gao, P., Zhang, L., Wan, B., Xu, L., Gao, S., Song, Z., Dai, F., & Zeng, R. (2025). A Begomovirus solanumdelhiense Vector for Virus-Induced Gene Silencing in Melon. Pathogens, 14(12), 1269. https://doi.org/10.3390/pathogens14121269






