Comparative Genomics of Two Newly Sequenced Rodent-Derived and One Previously Reported Tick-Derived Borrelia garinii Strains from South Korea Reveals Plasmid Variation and Virulence Gene Diversity
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Sample Preparation
2.2. Whole-Genome Sequencing, Assembly, and Annotation
2.3. Comparative Genomic Analysis of Korean B. garinii Strains and Other B. garinii Strains
3. Results
3.1. General Genomic Features of the Three Korean B. garinii Strains HN13, HN18, and 935
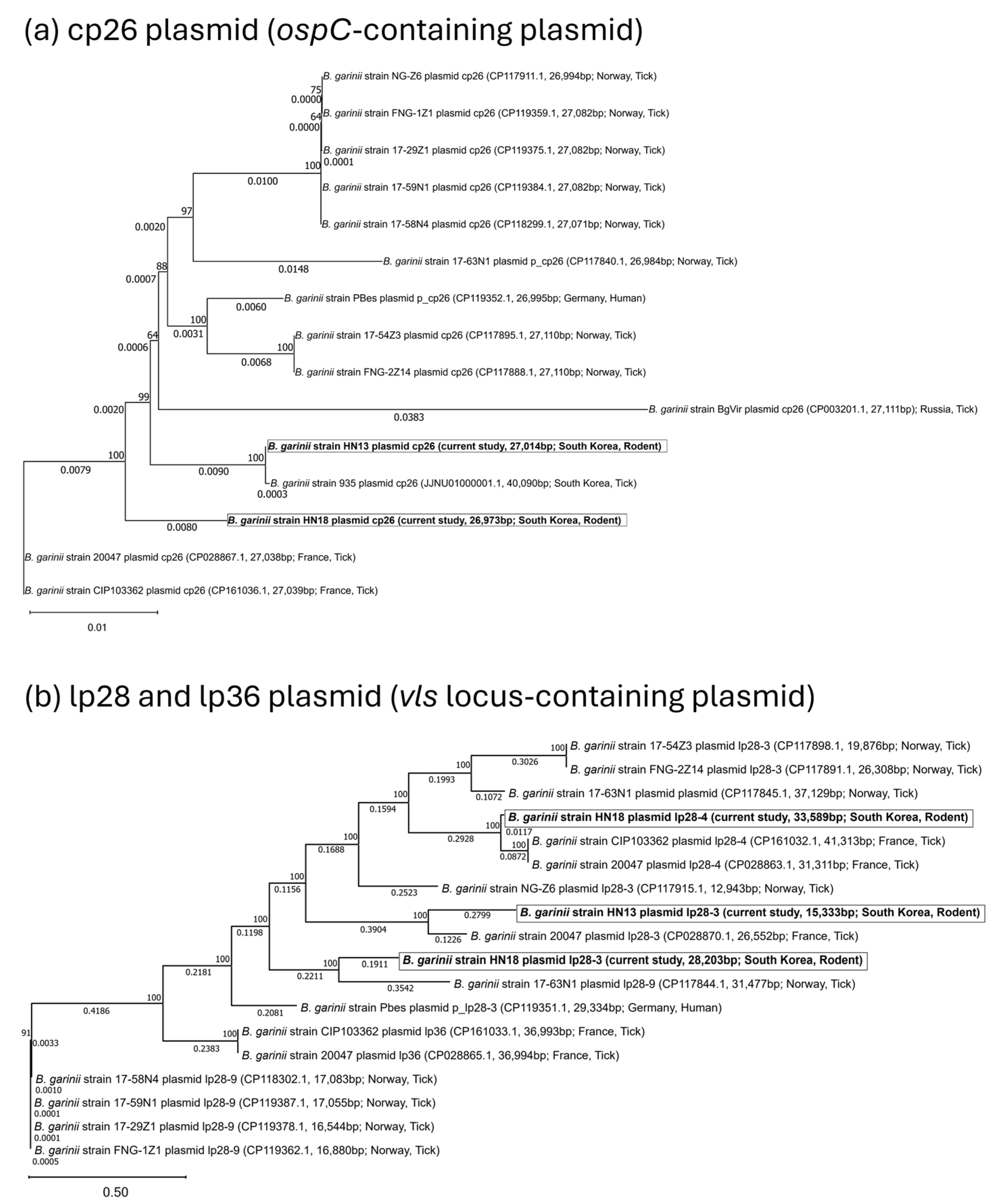
3.2. Phylogeny and Comparative Analysis
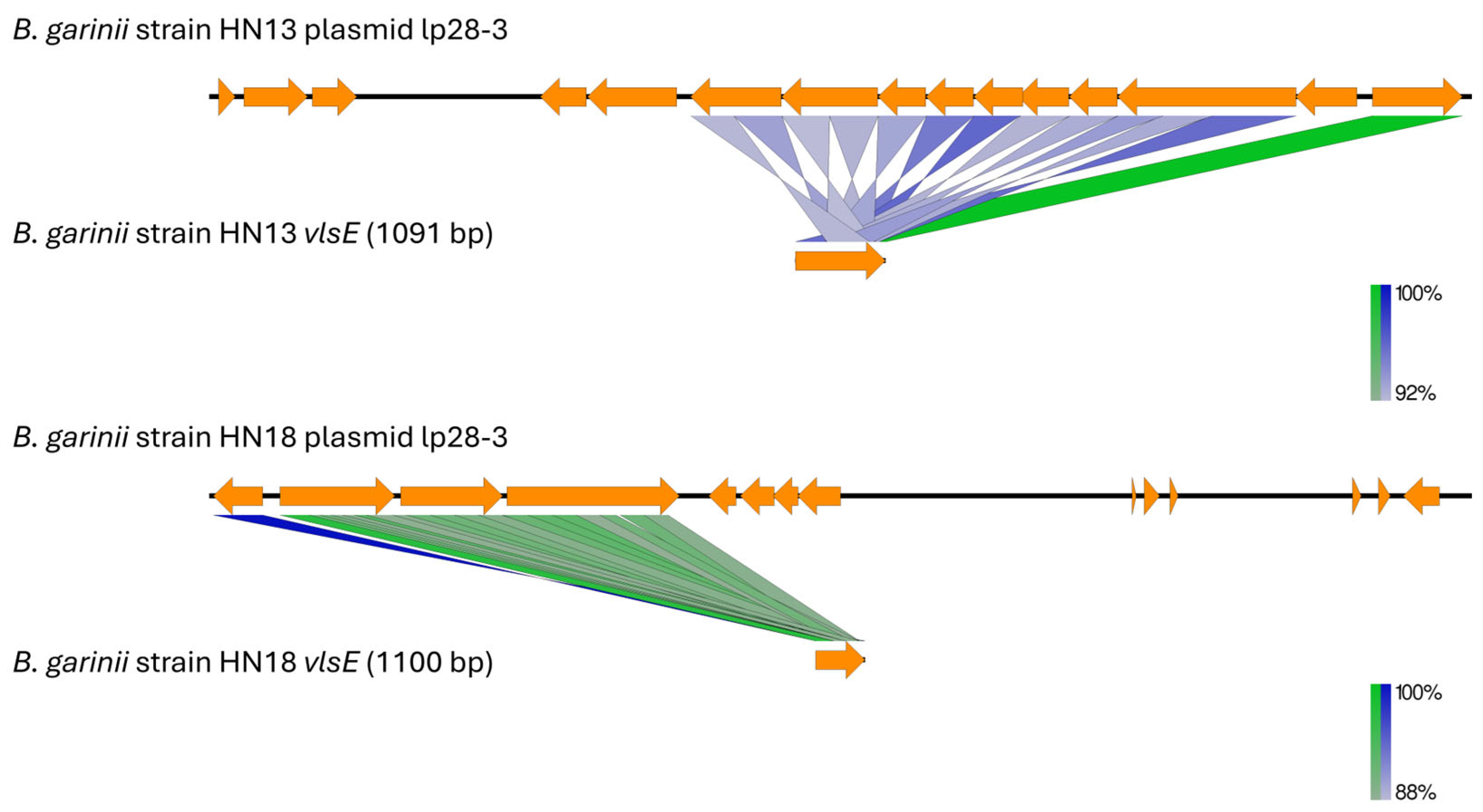
3.3. Plasmid Content, Virulence Gene Distribution, and Comparative Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| WGS | Whole-genome sequencing |
| NCBI | National Center for Biotechnology Information |
| MLST | Multilocus sequence typing |
| PacBio | Pacific Biosciences |
| ST | Sequence type |
| ANI | Average nucleotide identity |
| bp | Base pair |
| CDS | Coding sequence |
| SNP | Single-nucleotide polymorphism |
References
- Parija, S.C. Treponema, Borrelia and Leptospira. In Textbook of Microbiology and Immunology; Parija, S.C., Ed.; Springer Nature Singapore: Singapore, 2023; pp. 617–639. [Google Scholar]
- Strnad, M.; Rudenko, N.; Rego, R.O.M. Pathogenicity and virulence of Borrelia burgdorferi. Virulence 2023, 14, 2265015. [Google Scholar] [CrossRef]
- Petnicki-Ocwieja, T.; Kern, A. Mechanisms of Borrelia burgdorferi internalization and intracellular innate immune signaling. Front. Cell. Infect. Microbiol. 2014, 4, 175. [Google Scholar] [CrossRef] [PubMed]
- Barbour, A.G.; Qiu, W. Borreliella. In Bergey’s Manual of Systematics of Archaea and Bacteria; Barbour, A.G., Qiu, W., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2019. [Google Scholar]
- Bush, L.M.; Vazquez-Pertejo, M.T. Tick borne illness—Lyme disease. Disease-A-Month 2018, 64, 195–212. [Google Scholar] [CrossRef] [PubMed]
- Steere, A.C.; Strle, F.; Wormser, G.P.; Hu, L.T.; Branda, J.A.; Hovius, J.W.R.; Li, X.; Mead, P.S. Lyme borreliosis. Nat. Rev. Dis. Primers 2016, 2, 16090, Erratum in 2017, 3, 17062. [Google Scholar] [CrossRef]
- Baranton, G.; Postic, D.; Saint Girons, I.; Boerlin, P.; Piffaretti, J.-C.; Assous, M.; Grimont, P.A.D. Delineation of Borrelia burgdorferi Sensu Stricto, Borrelia garinii sp. nov., and Group VS461 Associated with Lyme Borreliosis. Int. J. Syst. Evol. Microbiol. 1992, 42, 378–383. [Google Scholar] [CrossRef]
- Lochhead, R.B.; Strle, K.; Arvikar, S.L.; Weis, J.J.; Steere, A.C. Lyme arthritis: Linking infection, inflammation and autoimmunity. Nat. Rev. Rheumatol. 2021, 17, 449–461. [Google Scholar] [CrossRef]
- Fingerle, V.; Schulte-Spechtel, U.C.; Ruzic-Sabljic, E.; Leonhard, S.; Hofmann, H.; Weber, K.; Pfister, K.; Strle, F.; Wilske, B. Epidemiological aspects and molecular characterization of Borrelia burgdorferi s.l. from southern Germany with special respect to the new species Borrelia spielmanii sp. nov. Int. J. Med. Microbiol. 2008, 298, 279–290. [Google Scholar] [CrossRef]
- Duffau, P.; Korbi, S.; Guillotin, V.; Talagrand-Reboul, E.; Ménard, A.; Peuchant, O. An unexpected case of Borrelia garinii liver infection. Ann. Clin. Microbiol. Antimicrob. 2022, 21, 15. [Google Scholar] [CrossRef]
- Hansford, K.M.; Wheeler, B.W.; Tschirren, B.; Medlock, J.M. Questing Ixodes ricinus ticks and Borrelia spp. in urban green space across Europe: A review. Zoonoses Public Health 2022, 69, 153–166. [Google Scholar] [CrossRef]
- Cinco, M.; Murgia, R.; Costantini, C. Prevalence of IgG reactivity in Lyme borreliosis patients versus Borrelia garinii and Borrelia afzelii in a restricted area of Northern Italy. FEMS Immunol. Med. Microbiol. 1995, 12, 217–222. [Google Scholar] [CrossRef]
- Ruivo, M.; Kovács, N.Z.; Schötta, A.-M.; Stelzer, T.; Hermann, L.; Mündler, V.; Bergthaler, A.; Reiter, M.; Wijnveld, M. Optimising Transformation Efficiency in Borrelia: Unravelling the Role of the Restriction-Modification System of Borrelia afzelii and Borrelia garinii. Int. J. Mol. Sci. 2024, 25, 11343. [Google Scholar] [CrossRef] [PubMed]
- Brenner, E.V.; Kurilshikov, A.M.; Stronin, O.V.; Fomenko, N.V. Whole-Genome Sequencing of Borrelia garinii BgVir, isolated from Taiga ticks (Ixodes persulcatus). J. Bacteriol. 2012, 194, 5713. [Google Scholar] [CrossRef] [PubMed]
- Wilske, B.; Preac-Mursic, V.; Gobel, U.B.; Graf, B.; Jauris, S.; Soutschek, E.; Schwab, E.; Zumstein, G. An OspA serotyping system for Borrelia burgdorferi based on reactivity with monoclonal antibodies and OspA sequence analysis. J. Clin. Microbiol. 1993, 31, 340–350. [Google Scholar] [CrossRef] [PubMed]
- Nohlmans, L.M.K.E.; De Boer, R.; Van Den Bogaard, A.E.J.M.; Van Boven, C.P.A. Genotypic and phenotypic analysis of Borrelia burgdorferi isolates from The Netherlands. J. Clin. Microbiol. 1995, 33, 119–125. [Google Scholar] [CrossRef]
- MÄKinen, J.; Vuorinen, I.; Oksi, J.; Peltomaa, M.; He, Q.; MarjamÄKi, M.; Viljanen, M.K. Prevalence of granulocytic Ehrlichia and Borrelia burgdorferi sensu lato in Ixodes ricinus ticks collected from Southwestern Finland and from Vormsi Island in Estonia. J. Pathol. Microbiol. Immunol. 2003, 111, 355–362. [Google Scholar] [CrossRef]
- Rudenko, N.; Golovchenko, M.; Horak, A.; Grubhoffer, L.; Mongodin, E.F.; Fraser, C.M.; Qiu, W.; Luft, B.J.; Morgan, R.G.; Casjens, S.R.; et al. Genomic Confirmation of Borrelia garinii, United States. Emerg. Infect. Dis. 2023, 29, 64–69. [Google Scholar] [CrossRef]
- Smith, J.R.P.; Muzaffar, S.B.; Lavers, J.; Lacombe, E.H.; Cahill, B.K.; Lubelczyk, C.B.; Kinsler, A.; Mathers, A.J.; Rand, P.W. Borrelia garinii in seabird ticks (Ixodes uriae), Atlantic Coast, North America. Emerg. Infect. Dis. 2006, 12, 1909–1912. [Google Scholar] [CrossRef]
- Jiang, B.; Yao, H.; Tong, Y.; Yang, X.; Huang, Y.; Jiang, J.; Cao, W. Genome Sequence of Borrelia garinii Strain NMJW1, Isolated from China. J. Bacteriol. 2012, 194, 6660–6661. [Google Scholar] [CrossRef]
- Uesaka, K.; Maezawa, M.; Inokuma, H. Serological survey of Borrelia infection of dogs in Sapporo, Japan, where Borrelia garinii infection was previously detected. J. Vet. Med. Sci. 2016, 78, 463–465. [Google Scholar] [CrossRef]
- Fomenko, N.V.; Stronin, O.V.; Khasnatinov, M.N.; Danchinova, G.A.; Bataa, J.; Gol’tsova, N.A. Heterogeneity of the ospA gene structure from isolates of Borrelia garinii and Borrelia afzelii from Western Siberia and Mongolia. Mol. Genet. Microbiol. Virol. 2009, 24, 183–188. [Google Scholar] [CrossRef]
- Chao, L.-L.; Lu, C.-F.; Shih, C.-M. Molecular detection and genetic identification of Borrelia garinii and Borrelia afzelii from patients presenting with a rare skin manifestation of prurigo pigmentosa in Taiwan. Int. J. Infect. Dis. 2013, 17, e1141–e1147. [Google Scholar] [CrossRef]
- Oh, W.; Kim, J.; Choi, Y.-J.; Kang, T.; Park, H.-J.; Lee, K.; Jang, W.-J. Human Co-infection with Rickettsia spp., Borrelia garinii, and Orientia tsutsugamushi in South Korea. Syst. Appl. Acarol. 2024, 29, 1201–1206. [Google Scholar] [CrossRef]
- Noh, Y.; Kim, S.Y.; Lee, Y.S.; Kim, D.-W.; Kwon, T.; Hwang, K.-J. Whole-Genome Sequence of Borrelia garinii Strain 935T Isolated from Ixodes persulcatus in South Korea. Genome Announc. 2014, 2, e01298-14. [Google Scholar] [CrossRef] [PubMed]
- Park, K.-H.; Choi, Y.-J.; Kim, J.; Park, H.-J.; Song, D.; Jang, W.-J. Reclassification of Borrelia spp. Isolated in South Korea Using Multilocus Sequence Typing. Jpn. J. Infect. Dis. 2018, 71, 350–353. [Google Scholar] [CrossRef] [PubMed]
- Park, K.H.; Chang, W.H.; Schwan, T.G. Identification and characterization of lyme disease spirochetes, Borrelia burgdorferi sensu lato, isolated in Korea. J. Clin. Microbiol. 1993, 31, 1831–1837. [Google Scholar] [CrossRef]
- Kim, K.G.; Hwang, D.J.; Park, J.W.; Ryu, M.G.; Kim, Y.; Yang, S.-J.; Lee, J.-E.; Lee, G.S.; Lee, J.H.; Park, J.S.; et al. Distribution and pathogen prevalence of field-collected ticks from south-western Korea: A study from 2019 to 2022. Sci. Rep. 2024, 14, 12336. [Google Scholar] [CrossRef]
- Park, J.W.; Lee, S.H.; Lee, G.S.; Seo, J.J.; Chung, J.K. Epidemiological Characteristics of Field Tick-Borne Pathogens in Gwang-ju Metropolitan Area, South Korea, from 2014 to 2018. Osong Public Health Res. Perspect. 2020, 11, 177–184. [Google Scholar] [CrossRef]
- Kim, S.Y.; Kim, T.-K.; Kim, T.Y.; Lee, H.I. Geographical Distribution of Borrelia burgdorferi sensu lato in Ticks Collected from Wild Rodents in the Republic of Korea. Pathogens 2020, 9, 866. [Google Scholar] [CrossRef]
- Lee, H.; Lee, S.-H.; Shin, S.; Kwak, D. Molecular Identification of Borrelia spp. from Ticks in Pastures Nearby Livestock Farms in Korea. Insects 2021, 12, 1011. [Google Scholar] [CrossRef]
- Limbach, F.X.; Jaulhac, B.; Puechal, X.; Monteil, H.; Kuntz, J.L.; Piemont, Y.; Sibilia, J. Treatment resistant Lyme arthritis caused by Borrelia garinii. Ann. Rheum. Dis. 2001, 60, 284–286. [Google Scholar] [CrossRef]
- Rožič, M.; Lah, L.L.; Ružić-Sabljić, E.; Kastrin, A.; Arnež, M. Lyme Neuroborreliosis in Children: Etiology and Comparison of Clinical Findings of Lyme Neuroborreliosis Caused by Borrelia garinii and Borrelia afzelii. Pediatr. Infect. Dis. J. 2019, 38, e279–e284. [Google Scholar] [CrossRef]
- Tran, N.; Milewski, M.D. Arthrofibrosis Associated with the Surgical Treatment of Chronic Lyme Arthritis and a Concomitant Medial Meniscal Tear: A Case Report. JBJS Case Connect. 2017, 7, e6. [Google Scholar] [CrossRef]
- Stewart, P.E.; Wang, X.; Bueschel, D.M.; Clifton, D.R.; Grimm, D.; Tilly, K.; Carroll, J.A.; Weis, J.J.; Rosa, P.A. Delineating the Requirement for the Borrelia burgdorferi Virulence Factor OspC in the Mammalian Host. Infect. Immun. 2006, 74, 3547–3553. [Google Scholar] [CrossRef]
- Lone, A.G.; Bankhead, T. The Borrelia burgdorferi VlsE Lipoprotein Prevents Antibody Binding to an Arthritis-Related Surface Antigen. Cell Rep. 2020, 30, 3663–3670.e5. [Google Scholar] [CrossRef] [PubMed]
- Embers, M.E.; Liang, F.T.; Howell, J.K.; Jacobs, M.B.; Purcell, J.E.; Norris, S.J.; Johnson, B.J.B.; Philipp, M.T. Antigenicity and recombination of VlsE, the antigenic variation protein of Borrelia burgdorferi, in rabbits, a host putatively resistant to long-term infection with this spirochete. FEMS Immunol. Med. Microbiol. 2007, 50, 421–429. [Google Scholar] [CrossRef] [PubMed]
- Wilske, B.; Busch, U.; Eiffert, H.; Fingerle, V.; Pfister, H.W.; RÖSsler, D.; Preac-Mursic, V. Diversity of OspA and OspC among cerebrospinal fluid isolates of Borrelia burgdorferi sensu lato from patients with neuroborreliosis in Germany. Med. Microbiol. Immunol. 1996, 184, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Izac, J.R.; Camire, A.C.; Earnhart, C.G.; Embers, M.E.; Funk, R.A.; Breitschwerdt, E.B.; Marconi, R.T. Analysis of the antigenic determinants of the OspC protein of the Lyme disease spirochetes: Evidence that the C10 motif is not immunodominant or required to elicit bactericidal antibody responses. Vaccine 2019, 37, 2401–2407. [Google Scholar] [CrossRef]
- Rogovskyy, A.S.; Casselli, T.; Tourand, Y.; Jones, C.R.; Owen, J.P.; Mason, K.L.; Scoles, G.A.; Bankhead, T. Evaluation of the Importance of VlsE Antigenic Variation for the Enzootic Cycle of Borrelia burgdorferi. PLoS ONE 2015, 10, e0124268. [Google Scholar] [CrossRef]
- Heikkilä, T.; Seppälä, I.; Saxen, H.; Panelius, J.; Yrjänäinen, H.; Lahdenne, P. Species-Specific Serodiagnosis of Lyme Arthritis and Neuroborreliosis Due to Borrelia burgdorferi Sensu Stricto, B. afzelii, and B.garinii by Using Decorin Binding ProteinA. J. Clin. Microbiol. 2002, 40, 453–460. [Google Scholar] [CrossRef]
- Brangulis, K.; Akopjana, I.; Petrovskis, I.; Kazaks, A.; Tars, K. Structural analysis of the outer surface proteins from Borrelia burgdorferi paralogous gene family 54 that are thought to be the key players in the pathogenesis of Lyme disease. J. Struct. Biol. 2020, 210, 107490. [Google Scholar] [CrossRef]
- Glöckner, G.; Lehmann, R.; Romualdi, A.; Pradella, S.; Schulte-Spechtel, U.; Schilhabel, M.; Wilske, B.; Sühnel, J.; Platzer, M. Comparative analysis of the Borrelia garinii genome. Nucleic Acids Res. 2004, 32, 6038–6046. [Google Scholar] [CrossRef]
- Casjens, S.R.; Mongodin, E.F.; Qiu, W.-G.; Dunn, J.J.; Luft, B.J.; Fraser-Liggett, C.M.; Schutzer, S.E. Whole-Genome Sequences of Two Borrelia afzelii and Two Borrelia garinii Lyme Disease Agent Isolates. J. Bacteriol. 2011, 193, 6995–6996. [Google Scholar] [CrossRef]
- Luciani, M.; Krasteva, I.; Schirone, M.; D’Onofrio, F.; Iannetti, L.; Torresi, M.; Di Pancrazio, C.; Perletta, F.; Valentinuzzi, S.; Tittarelli, M.; et al. Adaptive strategies of Listeria monocytogenes: An in-depth analysis of the virulent strain involved in an outbreak in Italy through quantitative proteomics. Int. J. Food Microbiol. 2025, 427, 110951. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Kim, B.J.; Kim, J.H.; Park, K.H.; Yeo, S.J.; Kim, S.J.; Kook, Y.H. Characterization of Borrelia burgdorferi strains isolated from Korea by 16S rDNA sequence analysis and PCR-RFLP analysis of rrf (5S)-rrl (23S) intergenic spacer amplicons. Int. J. Syst. Evol. Microbiol. 2000, 50 Pt 2, 857–863. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Concepcion, G.T.; Feng, X.; Zhang, H.; Li, H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods 2021, 18, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhang, Y.; Wang, A.Y.; Gao, M.; Chong, Z. Accurate long-read de novo assembly evaluation with Inspector. Genome Biol. 2021, 22, 312. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Huerta-Cepas, J.; Szklarczyk, D.; Forslund, K.; Cook, H.; Heller, D.; Walter, M.C.; Rattei, T.; Mende, D.R.; Sunagawa, S.; Kuhn, M.; et al. EGGNOG 4.5: A hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016, 44, D286–D293. [Google Scholar] [CrossRef] [PubMed]
- Robertson, J.; Nash, J.H.E. MOB-suite: Software tools for clustering, reconstruction and typing of plasmids from draft assemblies. Microb. Genom. 2018, 4, e000206. [Google Scholar] [CrossRef] [PubMed]
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Čech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.A.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Croucher, N.J.; Page, A.J.; Connor, T.R.; Delaney, A.J.; Keane, J.A.; Bentley, S.D.; Parkhill, J.; Harris, S.R. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2014, 43, e15. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2014, 32, 268–274. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Suleski, M.; Sanderford, M.; Sharma, S.; Tamura, K. MEGA12: Molecular Evolutionary Genetic Analysis Version 12 for Adaptive and Green Computing. Mol. Biol. Evol. 2024, 41, msae263. [Google Scholar] [CrossRef]
- Margos, G.; Gatewood, A.G.; Aanensen, D.M.; Hanincová, K.; Terekhova, D.; Vollmer, S.A.; Cornet, M.; Piesman, J.; Donaghy, M.; Bormane, A.; et al. MLST of housekeeping genes captures geographic population structure and suggests a European origin of Borrelia burgdorferi. Proc. Natl. Acad. Sci. USA 2008, 105, 8730–8735. [Google Scholar] [CrossRef]
- National Library of Medicine: National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/genbank/ (accessed on 16 October 2025).
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef]
- Posit Team. RStudio: Integrated Development Environment for R, 2025.05.0.496; Posit Team: Boston, MA, USA, 2025. [Google Scholar]
- Wickham, H. ggplot2: Elegant graphics for data analysis. In Use R! Wickham, H., Ed.; Springer Nature Link: Houston, TX, USA, 2016. [Google Scholar]
- Wickham, H. Reshaping Data with the reshape Package. J. Stat. Softw. 2007, 21, i12. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef] [PubMed]
- Rudenko, N.; Golovchenko, M.; Horak, A.; Grubhoffer, L.; Mongodin, E.F.; Fraser, C.M.; Qiu, W.; Luft, B.J.; Morgan, R.G.; Casjens, S.R.; et al. Detection of Borrelia garinii in the USA. 2022. preprint. [Google Scholar] [CrossRef]
- Bankhead, T.; Chaconas, G. The role of VlsE antigenic variation in the Lyme disease spirochete: Persistence through a mechanism that differs from other pathogens. Mol. Microbiol. 2007, 65, 1547–1558. [Google Scholar] [CrossRef]
- Margos, G.; Hofmann, M.; Casjens, S.; Dupraz, M.; Heinzinger, S.; Hartberger, C.; Hepner, S.; Schmeusser, M.; Sing, A.; Fingerle, V.; et al. Genome diversity of Borrelia garinii in marine transmission cycles does not match host associations but reflects the strains evolutionary history. Infect. Genet. Evol. 2023, 115, 105502. [Google Scholar] [CrossRef]
- Sun, Y.; Zhang, Y.; Fang, R.; Wang, J.; Yang, X.; Wang, L.; Guo, Y.; Miao, L.; Li, S. Geographical Distribution and Prevalence of Borrelia Genospecies in Eurasian Ticks. Sci. Data 2025, 12, 1530. [Google Scholar] [CrossRef]
- Comstedt, P.; Bergström, S.; Olsen, B.; Garpmo, U.; Marjavaara, L.; Mejlon, H.; Barbour, A.G.; Bunikis, J. Migratory passerine birds as reservoirs of Lyme borreliosis in Europe. Emerg. Infect. Dis. 2006, 12, 1087–1102. [Google Scholar] [CrossRef]
- Olsen, B.; Jaenson, T.G.T.; Noppa, L.; Bunikis, J.; Bergstrom, S. Lyme borreliosis cycle in seabirds and Ixodes uriae ticks. Nature 1993, 362, 340–342. [Google Scholar] [CrossRef] [PubMed]
- Margos, G.; Fingerle, V.; Reynolds, S. Borrelia bavariensis: Vector Switch, Niche Invasion, and Geographical Spread of a Tick-Borne Bacterial Parasite. Front. Ecol. Evol. 2019, 7, e00401. [Google Scholar] [CrossRef]
- Comstedt, P.; Jakobsson, T.; Bergström, S. Global ecology and epidemiology of Borrelia garinii spirochetes. Infect. Ecol. Epidemiol. 2011, 1, 9545. [Google Scholar] [CrossRef]
- Kurtenbach, K.; Hanincová, K.; Tsao, J.I.; Margos, G.; Fish, D.; Ogden, N.H. Fundamental processes in the evolutionary ecology of Lyme borreliosis. Nat. Rev. Microbiol. 2006, 4, 660–669. [Google Scholar] [CrossRef]
- Pedersen, B.N.; Jenkins, A.; Kjelland, V. Tick-borne pathogens in Ixodes ricinus ticks collected from migratory birds in southern Norway. PLoS ONE 2020, 15, e0230579. [Google Scholar] [CrossRef]
- Miyamoto, K.; Nakao, M.; Fujita, H.; Sato, F. The ixodid ticks on migratory birds in Japan and the isolation of Lyme disease spirochetes from bird-feeding ticks. Med. Entomol. Zool. 1993, 44, 315–326. [Google Scholar] [CrossRef]
- Nakao, M.; Miyamoto, K.; Fukunaga, M. Lyme Disease Spirochetes in Japan: Enzootic Transmission Cycles in Birds, Rodents, And Ixodes Persulcatus Ticks. J. Infect. Dis. 1994, 170, 878–882. [Google Scholar] [CrossRef]
- Norris, S.J.; Brangulis, K. Meta-analysis of the Vmp-like sequences of Lyme disease Borrelia: Evidence for the evolution of an elaborate antigenic variation system. Front. Microbiol. 2024, 15, 1469411. [Google Scholar] [CrossRef]
- Tilly, K.; Bestor, A.; Rosa, P.A. Lipoprotein succession in Borrelia burgdorferi: Similar but distinct roles for OspC and VlsE at different stages of mammalian infection. Mol. Microbiol. 2013, 89, 216–227. [Google Scholar] [CrossRef]


| B. garinii Strain HN13 | B. garinii Strain HN18 | B. garinii Strain 935 * | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Contigs | Size (bp) | GC (%) | Depth (x) | Size (bp) | GC (%) | Depth (x) | Size (bp) | GC (%) | Depth (x) |
| Chromosome | 906,429 | 28.4 | 381 | 906,083 | 28.4 | 1251 | 918,136 | 28.4 | - |
| lp54 | 52,486 | 26.9 | 2141 | 59,237 | 26.6 | 4813 | - | - | - |
| lp36 | 25,219 | 23.6 | 1498 | 24,716 | 23.6 | 4286 | 40,674 | 23.9 | - |
| cp32-6 | 29,083 | 29.1 | 1195 | 29,466 | 28.8 | 669 | 36,850 | 28.5 | - |
| lp32-10 | 26,630 | 25.5 | 710 | 32,156 | 26.7 | 3690 | - | - | - |
| lp28-3 | 15,333 | 33.8 | 391 | 28,203 | 31.4 | 1088 | 66,309 | 24.8 | - |
| lp28-4 | 40,508 | 25.4 | 1033 | 33,589 | 25.1 | 2574 | - | - | - |
| lp28-7 | - | - | - | 28,225 | 32.2 | 4472 | - | - | - |
| cp26 | 27,014 | 25.9 | 1552 | 26,973 | 25.9 | 846 | 40,090 | 25.4 | - |
| lp17 | 17,940 | 22.9 | 1582 | 22,602 | 24.2 | 6414 | 29,571 | 23.2 | - |
| lp54_lp32-10 | - | - | - | - | - | - | 67,109 | 26.6 | - |
| Strains | Linear-Form Plasmids | Circular-Form Plasmids | Other * | Total | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| lp54 | lp36 | lp32 | lp28 | lp25 | lp17 | cp32 | cp26 | cp9 | |||
| HN13 | lp54 | lp36 | lp32-10 | lp28-3 lp28-4 | - | lp17 | cp32-6 | cp26 | - | - | 8 |
| HN18 | lp54 | lp36 | lp32-10 | lp28-3 lp28-4 lp28-7 | - | lp17 | cp32-6 | cp26 | - | - | 9 |
| 935 | - | lp36 | - | lp28-3 | - | lp17 | cp32-6 | cp26 | - | lp54_lp32-10 | 6 |
| BgVir | lp54 | - | - | - | - | - | - | cp26 | - | - | 2 |
| 20047 | lp54 | lp36 | lp32-10 | lp28-3 lp28-4 lp28-7 | - | lp17 | cp32-3 cp32-6 | cp26 | - | - | 10 |
| CIP103362 | lp54 | lp36 | lp32-10 | lp28-4 lp28-7 | - | lp17 | cp32-3 cp32-6 | cp26 | - | - | 9 |
| PBes | p_lp54 | p_lp36 | p_lp32-5 p_lp32-9 p_lp32-10 | p_lp28-3 p_lp28-7 | p_lp25 | p_lp17 | - | p_cp26 | p_cp9 | - | 11 |
| NMJW1 | - | - | - | - | - | - | - | - | - | - | 0 |
| SZ | - | - | - | - | - | - | - | - | - | - | 0 |
| 17-63N1 | lp54 | - | - | lp28-9 | lp25 | - | p_cp32-3 p_cp32-6 | p_cp26 | - | lp17_32-10 Plasmid | 8 |
| NG-Z6 | lp54 | - | - | lp28-2 lp28-3 | lp25 | lp17 | - | cp26 | cp9 | Plasmid | 8 |
| FNG-1Z1 | lp54 | lp36 | lp32-10 | lp28-9 | lp25 | lp17 | - | cp26 | - | - | 7 |
| 17-29Z1 | lp54 | lp36 | lp32-10 | lp28-9 | lp25 | lp17 | - | cp26 | - | - | 7 |
| 17-54Z3 | lp54 | - | lp32-10 | lp28-3 | lp25 | lp17 | - | cp26 | - | Plasmid | 7 |
| 17-59N1 | lp54 | lp36 | lp32-10 | lp28-9 | lp25 | lp17 | - | cp26 | - | - | 7 |
| 17-58N4 | lp54 | lp36 | lp32-10 | lp28-9 | lp25 | lp17 | - | cp26 | - | - | 7 |
| FNG-2Z14 | lp54 | - | lp32-10 | lp28-3 | lp25 | lp17 | - | cp26 | - | - | 6 |
| ospA | ospB | ospC | dbpA | dbpB | vlsE/vls Silent Cassettes | PFam54/60 (Bbcrasp-1 Domain) ** | |
|---|---|---|---|---|---|---|---|
| HN13 | lp54 | lp54 | cp26 | - | lp54 | lp28-3/lp28-3 | lp54, lp36, lp32-10, lp28-4 |
| HN18 | lp54 | - | cp26 | lp54 | lp54 | lp28-3/lp28-3, lp28-4 | lp54; lp36, lp32-10; lp28-4 |
| 935 | lp54_lp32-10 * | lp54_lp32-10 * | cp26 | - | lp54_lp32-10 * | -/- | lp54_lp32-10 *; lp36, lp28-3 |
| BgVir | lp54 | lp54 | cp26 | lp54 | lp54 | -/- | lp54 |
| 20047 | lp54 | - | cp26 | lp54 | lp54 | lp36/lp36, lp28-3, lp28-4 | lp54, lp36, lp32-10, lp28-4, |
| CIP103362 | lp54 | - | cp26 | lp54 | lp54 | lp36/lp36, lp28-4 | lp54, lp36, lp32-10, lp28-4, |
| PBes | p_lp54 | - | p_cp26 | - | p_lp54 | p_lp28-3 | p_lp54, p_lp32-10, p_lp28-3, p_lp28-5, p_lp25 |
| NMJW1 | No plasmid | ||||||
| SZ | No plasmid | ||||||
| 17-63N1 | lp54 | - | p_cp26 | lp54 | lp54 | lp28-9, plasmid | lp54, p_cp32-3, lp28-9, lp25, plasmid |
| NG-Z6 | lp54 | - | cp26 | - | lp54 | lp28-3 | lp54, lp25, plasmid |
| FNG-1Z1 | lp54 | - | cp26 | lp54 | lp54 | lp28-9 | lp54, lp36, lp32-10, lp25 |
| 17-29Z1 | lp54 | - | cp26 | lp54 | lp54 | lp28-9 | lp54, lp36, lp32-10, lp25 |
| 17-54Z3 | lp54 | - | cp26 | lp54 | lp54 | lp28-3 | lp54, lp32-10, lp28-3, lp25 |
| 17-59N1 | lp54 | - | cp26 | lp54 | lp54 | lp28-9 | lp54, lp36, lp32-10, lp25 |
| 17-58N4 | lp54 | - | cp26 | lp54 | lp54 | lp28-9 | lp54, lp36, lp32-10, lp25 |
| FNG-2Z14 | lp54 | - | cp26 | lp54 | lp54 | lp28-3 | lp54, lp32-10, lp28-3, lp25 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, H.; Choi, Y.-J.; Park, J.-Y.; Lee, K.; Jang, W.-J. Comparative Genomics of Two Newly Sequenced Rodent-Derived and One Previously Reported Tick-Derived Borrelia garinii Strains from South Korea Reveals Plasmid Variation and Virulence Gene Diversity. Pathogens 2025, 14, 1182. https://doi.org/10.3390/pathogens14111182
Kang H, Choi Y-J, Park J-Y, Lee K, Jang W-J. Comparative Genomics of Two Newly Sequenced Rodent-Derived and One Previously Reported Tick-Derived Borrelia garinii Strains from South Korea Reveals Plasmid Variation and Virulence Gene Diversity. Pathogens. 2025; 14(11):1182. https://doi.org/10.3390/pathogens14111182
Chicago/Turabian StyleKang, Hyungsuk, Yeon-Joo Choi, Ji-Young Park, Kwangjun Lee, and Won-Jong Jang. 2025. "Comparative Genomics of Two Newly Sequenced Rodent-Derived and One Previously Reported Tick-Derived Borrelia garinii Strains from South Korea Reveals Plasmid Variation and Virulence Gene Diversity" Pathogens 14, no. 11: 1182. https://doi.org/10.3390/pathogens14111182
APA StyleKang, H., Choi, Y.-J., Park, J.-Y., Lee, K., & Jang, W.-J. (2025). Comparative Genomics of Two Newly Sequenced Rodent-Derived and One Previously Reported Tick-Derived Borrelia garinii Strains from South Korea Reveals Plasmid Variation and Virulence Gene Diversity. Pathogens, 14(11), 1182. https://doi.org/10.3390/pathogens14111182





